- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
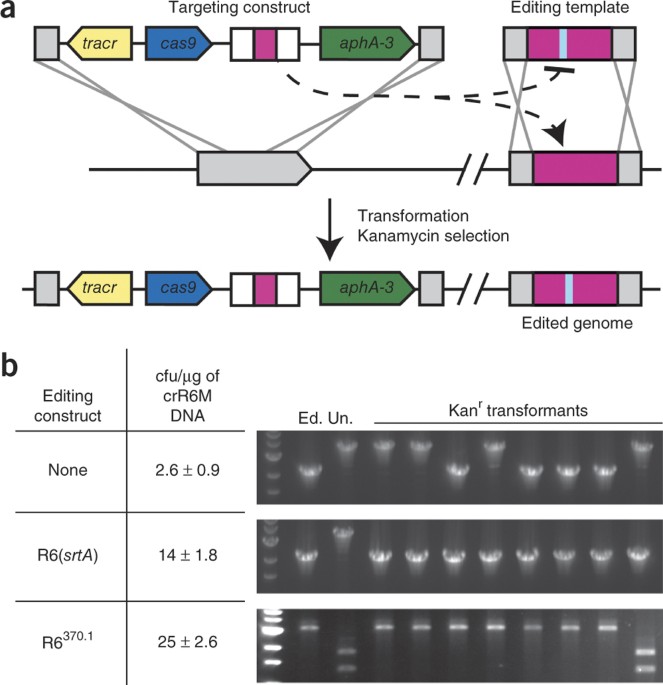
ABSTRACT Here we use the clustered, regularly interspaced, short palindromic repeats (CRISPR)–associated Cas9 endonuclease complexed with dual-RNAs to introduce precise mutations in the
genomes of _Streptococcus pneumoniae_ and _Escherichia coli_. The approach relies on dual-RNA:Cas9-directed cleavage at the targeted genomic site to kill unmutated cells and circumvents the
need for selectable markers or counter-selection systems. We reprogram dual-RNA:Cas9 specificity by changing the sequence of short CRISPR RNA (crRNA) to make single- and multinucleotide
changes carried on editing templates. Simultaneous use of two crRNAs enables multiplex mutagenesis. In _S. pneumoniae_, nearly 100% of cells that were recovered using our approach contained
the desired mutation, and in _E. coli_, 65% that were recovered contained the mutation, when the approach was used in combination with recombineering. We exhaustively analyze dual-RNA:Cas9
target requirements to define the range of targetable sequences and show strategies for editing sites that do not meet these requirements, suggesting the versatility of this technique for
bacterial genome engineering. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your
institution Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access
to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read
our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS A VERSATILE GENETIC ENGINEERING TOOLKIT FOR _E. COLI_ BASED ON CRISPR-PRIME EDITING Article Open access 01
September 2021 SYSTEMATICALLY ATTENUATING DNA TARGETING ENABLES CRISPR-DRIVEN EDITING IN BACTERIA Article Open access 08 February 2023 MAKE-OR-BREAK PRIME EDITING FOR GENOME ENGINEERING IN
_STREPTOCOCCUS PNEUMONIAE_ Article Open access 23 April 2025 ACCESSION CODES PRIMARY ACCESSIONS NCBI REFERENCE SEQUENCE * KC112384 REFERENCES * Urnov, F.D., Rebar, E.J., Holmes, M.C., Zhang,
H.S. & Gregory, P.D. Genome editing with engineered zinc finger nucleases. _Nat. Rev. Genet._ 11, 636–646 (2010). Article CAS PubMed Google Scholar * Bogdanove, A.J. & Voytas,
D.F. TAL effectors: customizable proteins for DNA targeting. _Science_ 333, 1843–1846 (2011). Article CAS PubMed Google Scholar * Stoddard, B.L. Homing endonuclease structure and
function. _Q. Rev. Biophys._ 38, 49–95 (2005). Article CAS PubMed Google Scholar * Bae, T. & Schneewind, O. Allelic replacement in _Staphylococcus aureus_ with inducible
counter-selection. _Plasmid_ 55, 58–63 (2006). Article CAS PubMed Google Scholar * Sung, C.K., Li, H., Claverys, J.P. & Morrison, D.A. An _rpsL_ cassette, janus, for gene replacement
through negative selection in _Streptococcus pneumoniae_. _Appl. Environ. Microbiol._ 67, 5190–5196 (2001). Article CAS PubMed PubMed Central Google Scholar * Sharan, S.K., Thomason,
L.C., Kuznetsov, S.G. & Court, D.L. Recombineering: a homologous recombination-based method of genetic engineering. _Nat. Protoc._ 4, 206–223 (2009). Article CAS PubMed PubMed Central
Google Scholar * Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. _Science_ 337, 816–821 (2012). CAS PubMed PubMed Central Google
Scholar * Deveau, H., Garneau, J.E. & Moineau, S. CRISPR-Cas system and its role in phage-bacteria interactions. _Annu. Rev. Microbiol._ 64, 475–493 (2010). Article CAS PubMed Google
Scholar * Horvath, P. & Barrangou, R. CRISPR-Cas, the immune system of bacteria and archaea. _Science_ 327, 167–170 (2010). Article CAS PubMed Google Scholar * Terns, M.P. &
Terns, R.M. CRISPR-based adaptive immune systems. _Curr. Opin. Microbiol._ 14, 321–327 (2011). Article CAS PubMed PubMed Central Google Scholar * van der Oost, J., Jore, M.M., Westra,
E.R., Lundgren, M. & Brouns, S.J. CRISPR-based adaptive and heritable immunity in prokaryotes. _Trends Biochem. Sci._ 34, 401–407 (2009). Article CAS PubMed Google Scholar * Brouns,
S.J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. _Science_ 321, 960–964 (2008). Article CAS PubMed PubMed Central Google Scholar * Carte, J., Wang, R., Li, H.,
Terns, R.M. & Terns, M.P. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. _Genes Dev._ 22, 3489–3496 (2008). Article CAS PubMed PubMed
Central Google Scholar * Deltcheva, E. et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. _Nature_ 471, 602–607 (2011). Article CAS PubMed PubMed Central
Google Scholar * Hatoum-Aslan, A., Maniv, I. & Marraffini, L.A. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism
anchored at the precursor processing site. _Proc. Natl. Acad. Sci. USA_ 108, 21218–21222 (2011). Article CAS PubMed PubMed Central Google Scholar * Haurwitz, R.E., Jinek, M.,
Wiedenheft, B., Zhou, K. & Doudna, J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. _Science_ 329, 1355–1358 (2010). Article CAS PubMed PubMed Central
Google Scholar * Deveau, H. et al. Phage response to CRISPR-encoded resistance in _Streptococcus thermophilus_. _J. Bacteriol._ 190, 1390–1400 (2008). Article CAS PubMed Google Scholar
* Gasiunas, G., Barrangou, R., Horvath, P. & Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. _Proc. Natl. Acad. Sci.
USA_ 109, E2579–E2586 (2012). Article CAS PubMed PubMed Central Google Scholar * Makarova, K.S., Aravind, L., Wolf, Y.I. & Koonin, E.V. Unification of Cas protein families and a
simple scenario for the origin and evolution of CRISPR-Cas systems. _Biol. Direct_ 6, 38 (2011). Article CAS PubMed PubMed Central Google Scholar * Barrangou, R. RNA-mediated
programmable DNA cleavage. _Nat. Biotechnol._ 30, 836–838 (2012). Article CAS PubMed Google Scholar * Brouns, S.J. Molecular biology. A Swiss army knife of immunity. _Science_ 337,
808–809 (2012). Article CAS PubMed Google Scholar * Carroll, D. A CRISPR approach to gene targeting. _Mol. Ther._ 20, 1658–1660 (2012). Article CAS PubMed PubMed Central Google
Scholar * Bikard, D., Hatoum-Aslan, A., Mucida, D. & Marraffini, L.A. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial
infection. _Cell Host Microbe_ 12, 177–186 (2012). Article CAS PubMed Google Scholar * Sapranauskas, R. et al. The _Streptococcus thermophilus_ CRISPR-Cas system provides immunity in
_Escherichia coli_. _Nucleic Acids Res._ 39, 9275–9282 (2011). Article CAS PubMed PubMed Central Google Scholar * Semenova, E. et al. Interference by clustered regularly interspaced
short palindromic repeat (CRISPR) RNA is governed by a seed sequence. _Proc. Natl. Acad. Sci. USA_ 108, 10098–10103 (2011). Article CAS PubMed PubMed Central Google Scholar *
Wiedenheft, B. et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. _Proc. Natl. Acad. Sci. USA_ 108, 10092–10097 (2011).
Article CAS PubMed PubMed Central Google Scholar * Zahner, D. & Hakenbeck, R. The _Streptococcus pneumoniae_ beta-galactosidase is a surface protein. _J. Bacteriol._ 182, 5919–5921
(2000). Article CAS PubMed PubMed Central Google Scholar * Marraffini, L.A., Dedent, A.C. & Schneewind, O. Sortases and the art of anchoring proteins to the envelopes of
gram-positive bacteria. _Microbiol. Mol. Biol. Rev._ 70, 192–221 (2006). Article CAS PubMed PubMed Central Google Scholar * Motamedi, M.R., Szigety, S.K. & Rosenberg, S.M.
Double-strand-break repair recombination in _Escherichia coli_: physical evidence for a DNA replication mechanism _in vivo_. _Genes Dev._ 13, 2889–2903 (1999). Article CAS PubMed PubMed
Central Google Scholar * Hosaka, T. et al. The novel mutation K87E in ribosomal protein S12 enhances protein synthesis activity during the late growth phase in _Escherichia coli_. _Mol.
Genet. Genomics_ 271, 317–324 (2004). Article CAS PubMed Google Scholar * Costantino, N. & Court, D.L. Enhanced levels of lambda Red-mediated recombinants in mismatch repair mutants.
_Proc. Natl. Acad. Sci. USA_ 100, 15748–15753 (2003). Article CAS PubMed PubMed Central Google Scholar * Edgar, R. & Qimron, U. The _Escherichia coli_ CRISPR system protects from
lambda lysogenization, lysogens, and prophage induction. _J. Bacteriol._ 192, 6291–6294 (2010). Article CAS PubMed PubMed Central Google Scholar * Marraffini, L.A. & Sontheimer,
E.J. Self versus non-self discrimination during CRISPR RNA-directed immunity. _Nature_ 463, 568–571 (2010). Article CAS PubMed PubMed Central Google Scholar * Fischer, S. et al. An
archaeal immune system can detect multiple Protospacer Adjacent Motifs (PAMs) to target invader DNA. _J. Biol. Chem._ 287, 33351–33363 (2012). Article CAS PubMed PubMed Central Google
Scholar * Gudbergsdottir, S. et al. Dynamic properties of the _Sulfolobus_ CRISPR-Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers.
_Mol. Microbiol._ 79, 35–49 (2011). Article CAS PubMed PubMed Central Google Scholar * Wang, H.H. et al. Genome-scale promoter engineering by coselection MAGE. _Nat. Methods_ 9, 591–593
(2012). Article PubMed PubMed Central Google Scholar * Cong, L. et al. Multiplex genome engineering using CRISPR-Cas systems. _Science_ doi:10.1126/science.1231143 (3 January 2013). *
Mali, P. et al. RNA-guided human genome engineering via Cas9. _Science_ doi:10.1126/science.1232033 (3 January 2013). * Cho, S.W., Kim, S., Kim, J.M. & Kim, J.-S. Targeted genome
engineering in human cells with the Cas9 RNA-guided endonuclease. _Nat. Biotechnol._ advance online publication, doi:10.1038/nbt.2507 (29 January 2013). * Hwang, W.Y. et al. Efficient genome
editing in zebrafish using a CRISPR-Cas system. _Nat. Biotechnol._ advance online publication, doi:10.1038/nbt.2501 (29 January 2013). * Hoskins, J. et al. Genome of the bacterium
_Streptococcus pneumoniae_ strain R6. _J. Bacteriol._ 183, 5709–5717 (2001). Article CAS PubMed PubMed Central Google Scholar * Havarstein, L.S., Coomaraswamy, G. & Morrison, D.A.
An unmodified heptadecapeptide pheromone induces competence for genetic transformation in _Streptococcus pneumoniae_. _Proc. Natl. Acad. Sci. USA_ 92, 11140–11144 (1995). Article CAS
PubMed PubMed Central Google Scholar * Horinouchi, S. & Weisblum, B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance.
_J. Bacteriol._ 150, 815–825 (1982). CAS PubMed PubMed Central Google Scholar * Horton, R.M. In vitro recombination and mutagenesis of DNA: SOEing Together tailor-made genes. _Methods
Mol. Biol._ 15, 251–261 (1993). CAS PubMed Google Scholar * Podbielski, A., Spellerberg, B., Woischnik, M., Pohl, B. & Lutticken, R. Novel series of plasmid vectors for gene
inactivation and expression analysis in group A streptococci (GAS). _Gene_ 177, 137–147 (1996). Article CAS PubMed Google Scholar * Husmann, L.K., Scott, J.R., Lindahl, G. &
Stenberg, L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of _Streptococcus pyogenes_. _Infect. Immun._ 63, 345–348 (1995). CAS
PubMed PubMed Central Google Scholar * Gibson, D.G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. _Nat. Methods_ 6, 343–345 (2009). Article CAS PubMed
Google Scholar Download references ACKNOWLEDGEMENTS We thank V. Fischetti and C. Euler for plasmid pLZ12spec, D. Court for the HME63 strain, J. Kern for plasmid pKD46, the Rockefeller
University Genomic Resource Center for technical support and J. Roberts for the MG1655 strain. D.B. is supported by a Harvey L. Karp Discovery Award and the Bettencourt Schuller Foundation.
D.C. is supported by the Medical Scientist Training Program. F.Z. is supported by a US National Institutes of Health (NIH) Director's Pioneer Award (DP1MH100706), Transformative R01,
the Keck, McKnight, Gates, Damon Runyon, Searle Scholars, Klingenstein, and Simons Foundations, Bob Metcalfe, Mike Boylan and Jane Pauley. L.A.M. is supported by the Searle Scholars Program,
the Rita Allen Scholars Program, a Irma T. Hirschl Award and a NIH Director's New Innovator Award (1DP2AI104556-01). AUTHOR INFORMATION Author notes * Wenyan Jiang and David Bikard:
These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Laboratory of Bacteriology, The Rockefeller University, New York, New York, USA Wenyan Jiang, David Bikard &
Luciano A Marraffini * Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA David Cox & Feng Zhang * Department of Brain and Cognitive Science and Department of Biological
Engineering, McGovern Institute for Brain Research, MIT, Cambridge, Massachusetts, USA David Cox & Feng Zhang Authors * Wenyan Jiang View author publications You can also search for this
author inPubMed Google Scholar * David Bikard View author publications You can also search for this author inPubMed Google Scholar * David Cox View author publications You can also search
for this author inPubMed Google Scholar * Feng Zhang View author publications You can also search for this author inPubMed Google Scholar * Luciano A Marraffini View author publications You
can also search for this author inPubMed Google Scholar CONTRIBUTIONS W.J., D.B. and L.A.M. designed the experiments; W.J., D.B. and D.C. performed experiments; W.J., D.B., F.Z. and L.A.M.
wrote the paper. CORRESPONDING AUTHORS Correspondence to David Bikard or Luciano A Marraffini. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Discussion, Supplementary Figures 1–11 and Supplementary Tables 1–3 (PDF 2790 kb) SUPPLEMENTARY DATA Supplementary Data
(XLSX 36 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Jiang, W., Bikard, D., Cox, D. _et al._ RNA-guided editing of bacterial genomes using
CRISPR-Cas systems. _Nat Biotechnol_ 31, 233–239 (2013). https://doi.org/10.1038/nbt.2508 Download citation * Received: 29 November 2012 * Accepted: 16 January 2013 * Published: 29 January
2013 * Issue Date: March 2013 * DOI: https://doi.org/10.1038/nbt.2508 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry,
a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative





:max_bytes(150000):strip_icc():focal(585x519:587x521)/salley-carson-bachelor-in-paradise-101122-1211d1c7abd04d92b28a42cb324326d5.jpg)