- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Analysis of the widely used ITS region is confounded by the presence of intragenomic variants (IGVs). In _Symbiodinium_, the algal symbionts of reef building corals, deep-sequencing
analyses are used to characterise communities within corals, yet these analyses largely overlook IGVs. Here we consider that distinct ITS2 sequences could represent IGVs rather than
distinct symbiont types and argue that symbionts can be distinguished by their proportional composition of IGVs, described as their ITS2 metahaplotype. Using our metahaplotype approach on
Minimum Entropy Decomposition (MED) analysis of ITS2 sequences from the corals _Acropora downingi_, _Cyphastrea microphthalma_ and _Playgyra daedalea_, we show the dominance of a single
species-specific _Symbiodinium_ C3 variant within each coral species. We confirm the presence of these species-specific symbionts using the psbA non-coding region. Our findings highlight the
importance of accounting for IGVs in ITS2 analyses and demonstrate their capacity to resolve biological patterns that would otherwise be overlooked. SIMILAR CONTENT BEING VIEWED BY OTHERS
HOST-SYMBIONT COEVOLUTION, CRYPTIC STRUCTURE, AND BLEACHING SUSCEPTIBILITY, IN A CORAL SPECIES COMPLEX (SCLERACTINIA; PORITIDAE) Article Open access 12 October 2020 CRYPTIC DIVERSITY OF
SHALLOW AND MESOPHOTIC _STEPHANOCOENIA INTERSEPTA_ CORALS ACROSS FLORIDA KEYS NATIONAL MARINE SANCTUARY Article 27 June 2024 RESOLVING CRYPTIC SPECIES COMPLEXES IN MARINE PROTISTS:
PHYLOGENETIC HAPLOTYPE NETWORKS MEET GLOBAL DNA METABARCODING DATASETS Article Open access 15 February 2021 MAIN The taxonomy of diverse microbial groups (for example, fungi, green algae and
dinoflagellates) is frequently distinguished using the ITS region of rDNA (LaJeunesse, 2001; Mai and Coleman, 1997; Schoch et al., 2012; Stern et al., 2012). However, there are multiple
copies of rDNA present in genomes, with differing forms known as intragenomic variants (IGV). Such IGVs can confound ITS-based analyses (Álvarez and Wendel, 2003; Thornhill et al., 2007;
Stat et al., 2011; Stern et al., 2012), but in the absence of more suitable markers, the ITS region remains a dominant means of characterising diverse microbial communities. The ITS2 region
underpins the taxonomy of the dinoflagellate _Symbiodinium_ (LaJeunesse, 2005), a symbiont widely associated with cnidarians, most notably reef building corals. _Symbiodinium_ is fundamental
to the coral holobiont, contributing >90% of the energy budget and influencing corals’ capacity to respond to environmental stress (Muscatine et al., 1984; Berkelmans and Van Oppen
2006). Deep sequencing of _Symbiodinium_ communities typically use the ITS2 region and are processed using OTU or phylotyping approaches (for example, Arif et al., 2014; Cunning et al.,
2015). These approaches either incorporate IGVs within OTUs or do not consider them in analyses. Instead of considering IGVs as incidental, here we show that the composition of ITS2 IGVs can
be used to differentiate distinct taxonomic sub-groups, revealing biological patterns consistent with a highly polymorphic marker. We compared symbiont communities among three coral species
(_Acropora downingii_, _Cyphastrea microphthalma_ and _Platygyra deadalea_) focusing on clade C as it is the most numerically dominant clade among these species at our sampling location
(Hume et al., 2015; Supplementary Table 1). We assessed the _Symbiodinium_ ITS2 sequence composition using an amplicon sequencing approach (see Supplementary Methods; _N_=13–15 ind./coral
sp.). Briefly, amplification of ITS2 and psbA non-coding region (psbAncr) was performed with modified Sym_Var primers (Hume et al., 2015). Normalised libraries were then sequenced on the
MiSeq Platform (Illumina) using the MiSeq v3 600-cycle kit. Demultiplexing, quality filtering, adapter removal, chimera removal and rarefaction (Supplementary Figure 1) were performed prior
to analysis with the Minimum Entropy Decomposition (MED) pipeline (Eren et al., 2015). MED nodes were analysed using _vegan_ (in R) and permutation multivariate analysis of variance
(_adonis_ in _vegan_). We compared our IGV approach with the OTU approach processed using the _mothur_ pipeline for _Symbiodinium_ (Schloss et al., 2009; Arif et al., 2014). Symbiont
communities from each species were initially analysed using the traditional OTU-based approach, with a 97% cutoff; this showed the presence of a single OTU, identified as a C3-type symbiont,
accounting for 100% of all ITS2 sequences. In essence, the OTU approach characterised all coral species as hosting the same host-generalist C3 symbiont type. As oligotyping (including
minimum entropy decomposition, MED) has been shown to reveal biologically relevant patterns masked by OTU approaches (Eren et al., 2014, Eren et al., 2015), we then applied MED analyses to
these symbiont communities. MED is ideally suited to analysing the extreme abundance of rare ITS2 variants in _Symbiodinium_ (Arif et al., 2014) by identifying the biologically informative
positions and therefore removing methodological and biological noise. Despite collapsing the data into distinct nodes, MED analyses retain the patterns observed in haplotype-based analyses
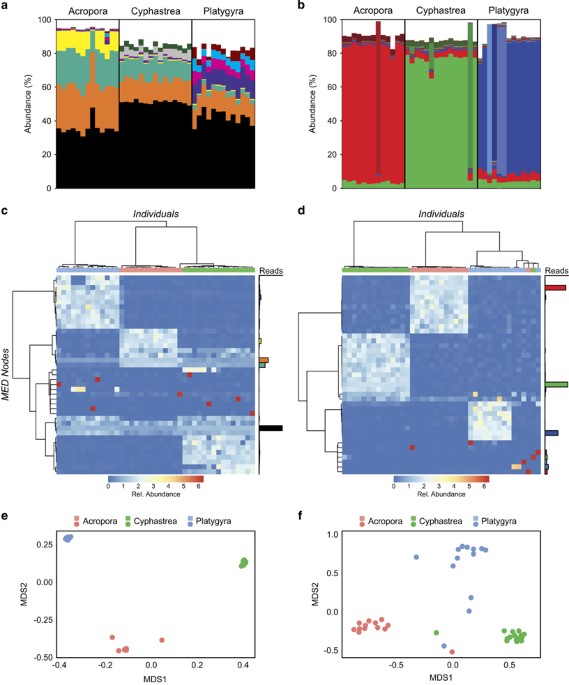
(Supplementary Figures 2 and 3). In contrast to the OTU approach, MED analysis revealed 44 distinct MED nodes (sequences grouped together exclusively by informative positions), characterised
by sequence variants of ITS2 types C3 and C3 Gulf (Supplementary Figure 4). The most abundant ITS2 nodes were common to all coral species, with the most abundant node accounting for 30–50%
of ITS2 sequences in all three coral species. Although the relative proportion of nodes varied among species, they were remarkably consistent within species. Species-specific grouping was
confirmed by multivariate analysis (Adonis, ITS2, F=213.52, _r_2=0.918, _P_=0.000999). Without an additional marker, interpretation of these data is complex as each MED node could either
represent a distinct symbiont or an intragenomic variant (IGV). There are two biologically distinct scenarios in which these data could be interpreted. These coral species could host a mixed
community of symbionts (for example, Sampayo et al., 2007), kept in relatively consistent proportions within species, although the most abundant symbiont taxa would be shared across all
coral species. Alternatively, each coral species could host a single symbiont taxon, which is distinguished only by the species-specific composition of its IGVs, herein referred to as an
‘ITS2 metahaplotype’. Here we will describe these two scenarios using the terms ‘mixed community’ and ‘single taxon’ (an unresolved taxonomic unit), respectively. In order to distinguish
between these scenarios, we performed MED analysis on psbAncr, a marker that provides high phylogenetic resolution in _Symbiodinium_ and although multi-copy, is typically present as a single
dominant sequence with low abundances of IGVs (LaJeunesse and Thornhill, 2011). Under the mixed community scenario, we would predict a mixture of psbAncr MED nodes in each individual coral.
For example, in _A. downingi_, we observed four abundant ITS2 MED nodes and if each node represents a separate symbiont, we should expect to see four correspondingly abundant psbAncr MED
nodes. Furthermore, the abundant nodes should be shared across coral species. Our analyses shows this is not the case. Instead, each individual coral was dominated by a single psbAncr MED
node, which is indicative of the presence of a single symbiont type (Figure 1d). In agreement with the ITS2 data, the psbAncr MED node composition results in species-specific clustering
(Figures 1e and f—Adonis, psbAncr, F=38.51, _r_2=0.670, _P_=0.000999). Overall, these results support the single taxon scenario: that each coral species hosts a single species-specific
symbiont characterised by its unique metahaplotype. These observations highlight the important role that IGVs could play in improving the taxonomic characterisation of _Symbiodinium_ and
other microbial groups, by providing insights into unique species-specific symbiont signatures that would have gone unnoticed by traditional ITS2 approaches. _Symbiodinium_ C3 was initially
considered a widespread generalist (LaJeunesse, 2005). However, our observation of diverse variants, which are coral host species-specific, within C3 support previous work that shows that
there is substantial cryptic diversity that is currently unresolved (Thornhill et al., 2014). This has important implications for our understanding of specificity and diversity of
coral–algal symbioses, and approaches that make use of IGVs provide the opportunity to shed light on these relationships. We have demonstrated the applicability of the metahaplotype approach
for _Symbiodinium_ clade C and in principle, it should be broadly applicable to other clades. Combining ITS2 and psbAncr amplicon sequencing analyses provides a framework whereby mixed ITS2
communities found elsewhere (for example, Sampayo et al., 2007) can be tested to identify whether monotypic communities and host specificity are found in other regions and species. This
will provide a greater appreciation of the different symbiotic strategies employed by corals and help identify the environmental and biological drivers behind the single taxon and mixed
community approaches. The power of such IGV approaches is not limited to coral symbionts, and can be applied to the diverse range of microbial organisms that are currently classified by
traditional ITS2 methods (for example, Schoch et al., 2012; Stern et al., 2012), particularly in metagenomics studies where IGVs could inflate diversity estimates. Our analyses demonstrate
the applicability of our MED-based metahaplotype approach to characterizing relatively simple communities but it could be transferred to more complex communities. However, analysis of mixed
communities would greatly benefit from the development of approaches that could identify individual metahaplotypes within a heterogeneous dataset. Transferring the metahaplotype approach to
mixed communities would be a worthwhile endeavor as we clearly show that incorporating ITS2 IGVs into analyses of microbial communities reveals fundamental ecological processes that would
have been, otherwise, overlooked. REFERENCES * Álvarez I, Wendel JF . (2003). Ribosomal ITS sequences and plant phylogenetic inference. _Mol Phylogenet Evol_ 29: 417–434. Article Google
Scholar * Arif C, Daniels C, Bayer T, Banguera‐Hinestroza E, Barbrook A, Howe CJ _et al_. (2014). Assessing Symbiodinium diversity in scleractinian corals via next‐generation
sequencing‐based genotyping of the ITS2 rDNA region. _Mol Ecol_ 23: 4418–4433. Article CAS Google Scholar * Berkelmans R, Van Oppen MJ . (2006). The role of zooxanthellae in the thermal
tolerance of corals: a ‘nugget of hope’ for coral reefs in an era of climate change. _Proc R Soc B Biol Sci_ 273: 2305–2312. Article Google Scholar * Cunning R, Yost DM, Guarinello ML,
Putnam HM, Gates RD . (2015). Variability of Symbiodinium communities in waters, sediments, and corals of thermally distinct reef pools in American Samoa. _PLoS One_ 10: e0145099. Article
Google Scholar * Eren AM, Borisy GG, Huse SM, Welch JLM . (2014). Oligotyping analysis of the human oral microbiome. _Proc Natl Acad Sci USA_ 111: E2875–E2884. Article CAS Google Scholar
* Eren AM, Morrison HG, Lescault PJ, Reveillaud J, Vineis JH, Sogin ML . (2015). Minimum entropy decomposition: Unsupervised oligotyping for sensitive partitioning of high-throughput
marker gene sequences. _ISME J_ 9: 968–979. Article CAS Google Scholar * Hume B, D'Angelo C, Smith E, Stevens J, Burt J, Wiedenmann J . (2015). Symbiodinium thermophilum sp. nov., a
thermotolerant symbiotic alga prevalent in corals of the world's hottest sea, the Persian/Arabian Gulf. _Sci Rep_ 5: 8562. Article CAS Google Scholar * LaJeunesse TC . (2001).
Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: in search of a ‘species’ level marker. _J Phycol_ 37:
866–880. Article CAS Google Scholar * LaJeunesse TC . (2005). ‘Species’ radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition.
_Mol Biol Evol_ 22: 570–581. Article CAS Google Scholar * LaJeunesse TC, Thornhill DJ . (2011). Improved resolution of reef-coral endosymbiont (Symbiodinium) species diversity, ecology,
and evolution through psbA non-coding region genotyping. _PLoS ONE_ 6: e29013. Article CAS Google Scholar * Mai JC, Coleman AW . (1997). The internal transcribed spacer 2 exhibits a
common secondary structure in green algae and flowering plants. _J Mol Evol_ 44: 258–271. Article CAS Google Scholar * Muscatine L, Falkowski P, Porter J, Dubinsky Z . (1984). Fate of
photosynthetic fixed carbon in light-and shade-adapted colonies of the symbiotic coral Stylophora pistillata. _Proc R Soc Lond B Biol Sci_ 222: 181–202. Article CAS Google Scholar *
Sampayo E, Franceschinis L, Hoegh-Guldberg O, Dove S . (2007). Niche partitioning of closely related symbiotic dinoflagellates. _Mol Ecol_ 16: 3721. Article CAS Google Scholar * Schloss
PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB _et al_. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing
microbial communities. _Appl Environ Microbiol_ 75: 7537–7541. Article CAS Google Scholar * Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA _et al_. (2012). Nuclear
ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. _Proc Natl Acad Sci USA_ 109: 6241–6246. Article CAS Google Scholar * Stat M, Bird CE,
Pochon X, Chasqui L, Chauka LJ, Concepcion GT _et al_. (2011). Variation in Symbiodinium ITS2 sequence assemblages among coral colonies. _PLoS ONE_ 6: e15854. Article CAS Google Scholar *
Stern RF, Andersen RA, Jameson I, Küpper FC, Coffroth M-A, Vaulot D _et al_. (2012). Evaluating the ribosomal internal transcribed spacer (ITS) as a candidate dinoflagellate barcode marker.
_PLoS ONE_ 7: e42780. Article CAS Google Scholar * Thornhill DJ, Lajeunesse TC, Santos SR . (2007). Measuring rDNA diversity in eukaryotic microbial systems: how intragenomic variation,
pseudogenes, and PCR artifacts confound biodiversity estimates. _Mol Ecol_ 16: 5326–5340. Article CAS Google Scholar * Thornhill DJ, Lewis AM, Wham DC, LaJeunesse TC . (2014).
Host‐specialist lineages dominate the adaptive radiation of reef coral endosymbionts. _Evolution_ 68: 352–367. Article CAS Google Scholar Download references ACKNOWLEDGEMENTS We thank the
New York University Abu Dhabi Institute for funding and the Environment Agency Abu Dhabi for permitting. This work was supported by the New York University Core Sequencing and
Bioinformatics Groups. Data used in this study are deposited in the Dryad Digital Repository. (http://dx.doi.org/10.5061/dryad.h6s54). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Center
for Genomics and Systems Biology, New York University Abu Dhabi, Abu Dhabi, UAE Edward G Smith, Remi N Ketchum & John A Burt Authors * Edward G Smith View author publications You can
also search for this author inPubMed Google Scholar * Remi N Ketchum View author publications You can also search for this author inPubMed Google Scholar * John A Burt View author
publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Edward G Smith. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
conflict of interest. ADDITIONAL INFORMATION Supplementary Information accompanies this paper on ISME J website SUPPLEMENTARY INFORMATION SUPPLEMENTARY METHODS (DOCX 22 KB) SUPPLEMENTARY
FIGURE 1 (JPG 152 KB) SUPPLEMENTARY FIGURE 2 (JPG 60 KB) SUPPLEMENTARY FIGURE 3 (JPG 414 KB) SUPPLEMENTARY FIGURE 4 (JPG 43 KB) SUPPLEMENTARY FIGURE 5 (JPG 63 KB) SUPPLEMENTARY FIGURE 6 (JPG
78 KB) SUPPLEMENTARY TABLE 1 (XLSX 11 KB) SUPPLEMENTARY TABLE 2 (DOCX 16 KB) SUPPLEMENTARY TABLE 3 (XLSX 19 KB) SUPPLEMENTARY INFORMATION (DOCX 16 KB) RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Smith, E., Ketchum, R. & Burt, J. Host specificity of _Symbiodinium_ variants revealed by an ITS2 metahaplotype approach. _ISME J_ 11,
1500–1503 (2017). https://doi.org/10.1038/ismej.2016.206 Download citation * Received: 15 June 2016 * Revised: 20 October 2016 * Accepted: 25 November 2016 * Published: 17 February 2017 *
Issue Date: June 2017 * DOI: https://doi.org/10.1038/ismej.2016.206 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative