- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Anion exchanger 1 (AE1), a member of the solute carrier (SLC) family, is the primary bicarbonate transporter in erythrocytes, regulating pH levels and CO2 transport between lungs
and tissues. Previous studies characterized its role in erythrocyte structure and provided insight into transport regulation. However, key questions remain regarding substrate binding and
transport, mechanisms of drug inhibition and modulation by membrane components. Here we present seven cryo-EM structures in apo, bicarbonate-bound and inhibitor-bound states. These, combined
with uptake and computational studies, reveal important molecular features of substrate recognition and transport, and illuminate sterol binding sites, to elucidate distinct inhibitory
mechanisms of research chemicals and prescription drugs. We further probe the substrate binding site via structure-based ligand screening, identifying an AE1 inhibitor. Together, our
findings provide insight into mechanisms of solute carrier transport and inhibition. Access through your institution Buy or subscribe This is a preview of subscription content, access via
your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days
cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THE LIPID REGULATION OF HUMAN ANION EXCHANGER 2
Article Open access 26 January 2024 STRUCTURAL INSIGHTS INTO HUMAN ORGANIC CATION TRANSPORTER 1 TRANSPORT AND INHIBITION Article Open access 15 March 2024 MECHANISM OF ANION EXCHANGE AND
SMALL-MOLECULE INHIBITION OF PENDRIN Article Open access 06 January 2024 DATA AVAILABILITY Density maps and structure coordinates have been deposited in the Electron Microscopy Data Bank
(EMDB) and the Protein Data Bank (PDB): AE1-apo (EMD-26165 and PDB 7TY4), AE1–bicarbonate (EMD-26168 and PDB 7TY7), AE1–DIDS (EMD-41082 and PDB 8T6V), AE1–H2DIDS (EMD-26167 and PDB 7TY6),
AE1–DEPC (EMD-26171 and PDB 7TYA), AE1–dipyridamole (EMD-41081 and PDB 8T6U), AE1–NIF (EMD-26169 and PDB 7TY8). Source data are provided with this paper. REFERENCES * Elgsaeter, A., Stokke,
B. T., Mikkelsen, A. & Branton, D. The molecular basis of erythrocyte shape. _Science_ 234, 1217–1223 (1986). Article CAS PubMed Google Scholar * Poole, J. The Diego blood group
system–an update. _Immunohematology_ 15, 135–143 (1999). Article CAS PubMed Google Scholar * Levine, P., Robinson, E. A., Layrisse, M., Arends, T. & Domingues Sisco, R. The Diego
blood factor. _Nature_ 177, 40–41 (1956). Article CAS PubMed Google Scholar * Reithmeier, R. A. et al. Band 3, the human red cell chloride/bicarbonate anion exchanger (AE1, SLC4A1), in a
structural context. _Biochim. Biophys. Acta_ 1858, 1507–1532 (2016). Article CAS PubMed Google Scholar * Jennings, M. L. Cell physiology and molecular mechanism of anion transport by
erythrocyte band 3/AE1. _Am. J. Physiol. Cell Physiol._ 321, C1028–C1059 (2021). Article CAS PubMed PubMed Central Google Scholar * Kodippili, G. C. et al. Analysis of the mobilities of
band 3 populations associated with ankyrin protein and junctional complexes in intact murine erythrocytes. _J. Biol. Chem._ 287, 4129–4138 (2012). Article CAS PubMed Google Scholar *
Thornell, I. M. & Bevensee, M. O. Regulators of _Slc4_ bicarbonate transporter activity. _Front. Physiol._ 6, 166 (2015). Article PubMed PubMed Central Google Scholar * Vallese, F.
et al. Architecture of the human erythrocyte ankyrin-1 complex. _Nat. Struct. Mol. Biol._ 29, 706–718 (2022). Article CAS PubMed PubMed Central Google Scholar * Xia, X., Liu, S. &
Zhou, Z. H. Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane. _Nat. Struct. Mol. Biol._ 29, 698–705 (2022). Article CAS PubMed PubMed Central
Google Scholar * Falke, J. J. & Chan, S. I. Molecular mechanisms of band 3 inhibitors. 1. Transport site inhibitors. _Biochemistry_ 25, 7888–7894 (1986). Article CAS PubMed Google
Scholar * Falke, J. J. & Chan, S. I. Molecular mechanisms of band 3 inhibitors. 2. Channel blockers. _Biochemistry_ 25, 7895–7898 (1986). Article CAS PubMed Google Scholar * Falke,
J. J. & Chan, S. I. Molecular mechanisms of band 3 inhibitors. 3. Translocation inhibitors. _Biochemistry_ 25, 7899–7906 (1986). Article CAS PubMed Google Scholar * Galanter, W. L.
& Labotka, R. J. The binding of nitrate to the human anion exchange protein (AE1) studied with 14N nuclear magnetic resonance. _Biochim. Biophys. Acta_ 1079, 146–151 (1991). Article CAS
PubMed Google Scholar * Arakawa, T. et al. Crystal structure of the anion exchanger domain of human erythrocyte band 3. _Science_ 350, 680–684 (2015). Article CAS PubMed Google
Scholar * Huynh, K. W. et al. CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1. _Nat. Commun._ 9, 900 (2018). Article PubMed PubMed Central Google Scholar
* Wang, W. et al. Cryo-EM structure of the sodium-driven chloride/bicarbonate exchanger NDCBE. _Nat. Commun._ 12, 5690 (2021). Article CAS PubMed PubMed Central Google Scholar *
Casey, J. R., Pirraglia, C. A. & Reithmeier, R. A. Enzymatic deglycosylation of human Band 3, the anion transport protein of the erythrocyte membrane. Effect on protein structure and
transport properties. _J. Biol. Chem._ 267, 11940–11948 (1992). Article CAS PubMed Google Scholar * Zhang, D., Kiyatkin, A., Bolin, J. T. & Low, P. S. Crystallographic structure and
functional interpretation of the cytoplasmic domain of erythrocyte membrane band 3. _Blood_ 96, 2925–2933 (2000). Article CAS PubMed Google Scholar * Figueroa, D. The Diego blood group
system: a review. _Immunohematology_ 29, 73–81 (2013). Article PubMed Google Scholar * Daniels, G. L. et al. Blood group terminology 2004: from the International Society of Blood
Transfusion committee on terminology for red cell surface antigens. _Vox Sang._ 87, 304–316 (2004). Article CAS PubMed Google Scholar * Schubert, D. & Boss, K. Band 3
protein–cholesterol interactions in erythrocyte membranes. Possible role in anion transport and dependency on membrane phospholipid. _FEBS Lett._ 150, 4–8 (1982). Article CAS PubMed
Google Scholar * Kalli, A. C. & Reithmeier, R. A. F. Interaction of the human erythrocyte Band 3 anion exchanger 1 (AE1, SLC4A1) with lipids and glycophorin A: molecular organization of
the Wright (Wr) blood group antigen. _PLoS Comput. Biol._ 14, e1006284 (2018). Article PubMed PubMed Central Google Scholar * Gregg, V. A. & Reithmeier, R. A. Effect of cholesterol
on phosphate uptake by human red blood cells. _FEBS Lett._ 157, 159–164 (1983). Article CAS PubMed Google Scholar * Zhu, Q. & Casey, J. R. The substrate anion selectivity filter in
the human erythrocyte Cl−/HCO3− exchange protein, AE1. _J. Biol. Chem._ 279, 23565–23573 (2004). Article CAS PubMed Google Scholar * Lu, F. et al. Structure and mechanism of the uracil
transporter UraA. _Nature_ 472, 243–246 (2011). Article CAS PubMed Google Scholar * Case, D. A. et al. AMBER 2020 (Univ. California, San Francisco, 2020). * Guarnieri, F. & Mezei, M.
Simulated annealing of chemical potential: a general procedure for locating bound waters. Application to the study of the differential hydration propensities of the major and minor grooves
of DNA. _J. Am. Chem. Soc._ 118, 8493–8494 (1996). Article CAS Google Scholar * Knauf, P. A., Law, F. Y., Leung, T. W., Gehret, A. U. & Perez, M. L. Substrate-dependent reversal of
anion transport site orientation in the human red blood cell anion-exchange protein, AE1. _Proc. Natl Acad. Sci. USA_ 99, 10861–10864 (2002). Article CAS PubMed PubMed Central Google
Scholar * Okubo, K., Kang, D., Hamasaki, N. & Jennings, M. L. Red blood cell band 3. Lysine 539 and lysine 851 react with the same H2DIDS
(4,4′-diisothiocyanodihydrostilbene-2,2′-disulfonic acid) molecule. _J. Biol. Chem._ 269, 1918–1926 (1994). Article CAS PubMed Google Scholar * Lu, J. & Boron, W. F. Reversible and
irreversible interactions of DIDS with the human electrogenic Na/HCO3 cotransporter NBCe1-A: role of lysines in the KKMIK motif of TM5. _Am. J. Physiol. Cell Physiol._ 292, C1787–C1798
(2007). Article CAS PubMed Google Scholar * Jin, X. R., Abe, Y., Li, C. Y. & Hamasaki, N. Histidine-834 of human erythrocyte band 3 has an essential role in the conformational
changes that occur during the band 3-mediated anion exchange. _Biochemistry_ 42, 12927–12932 (2003). Article CAS PubMed Google Scholar * Izuhara, K., Okubo, K. & Hamasaki, N.
Conformational change of band 3 protein induced by diethyl pyrocarbonate modification in human erythrocyte ghosts. _Biochemistry_ 28, 4725–4728 (1989). Article CAS PubMed Google Scholar
* Liantonio, A. et al. Niflumic acid inhibits chloride conductance of rat skeletal muscle by directly inhibiting the CLC-1 channel and by increasing intracellular calcium. _Br. J.
Pharmacol._ 150, 235–247 (2007). Article CAS PubMed Google Scholar * Cousin, J. L. & Motais, R. Inhibition of anion permeability by amphiphilic compounds in human red cell: evidence
for an interaction of niflumic acid with the band 3 protein. _J. Membr. Biol._ 46, 125–153 (1979). Article CAS PubMed Google Scholar * Friesner, R. A. et al. Glide: a new approach for
rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. _J. Med. Chem._ 47, 1739–1749 (2004). Article CAS PubMed Google Scholar * Carlsson, J. et al. Ligand
discovery from a dopamine D3 receptor homology model and crystal structure. _Nat. Chem. Biol._ 7, 769–778 (2011). Article CAS PubMed PubMed Central Google Scholar * Maneri, L. R. &
Low, P. S. Structural stability of the erythrocyte anion transporter, band 3, in different lipid environments. A differential scanning calorimetric study. _J. Biol. Chem._ 263, 16170–16178
(1988). Article CAS PubMed Google Scholar * Maneri, L. R. & Low, P. S. Fatty acid composition of lipids which copurify with band 3. _Biochem. Biophys. Res. Commun._ 159, 1012–1019
(1989). Article CAS PubMed Google Scholar * Stewart, A. K. et al. Functional characterization and modified rescue of novel AE1 mutation R730C associated with overhydrated cation leak
stomatocytosis. _Am. J. Physiol. Cell Physiol._ 300, C1034–C1046 (2011). Article CAS PubMed PubMed Central Google Scholar * Jennings, M. L. & Smith, J. S. Anion–proton cotransport
through the human red blood cell band 3 protein. Role of glutamate 681. _J. Biol. Chem._ 267, 13964–13971 (1992). Article CAS PubMed Google Scholar * Yang, E. et al. A Ser725Arg mutation
in Band 3 abolishes transport function and leads to anemia and renal tubular acidosis. _Blood_ 131, 1759–1763 (2018). Article CAS PubMed PubMed Central Google Scholar * Zhekova, H. R.
et al. Identification of multiple substrate binding sites in SLC4 transporters in the outward-facing conformation: insights into the transport mechanism. _J. Biol. Chem._ 296, 100724 (2021).
Article CAS PubMed PubMed Central Google Scholar * Thurtle-Schmidt, B. H. & Stroud, R. M. Structure of Bor1 supports an elevator transport mechanism for SLC4 anion exchangers.
_Proc. Natl Acad. Sci. USA_ 113, 10542–10546 (2016). Article CAS PubMed PubMed Central Google Scholar * Ficici, E., Faraldo-Gomez, J. D., Jennings, M. L. & Forrest, L. R. Asymmetry
of inverted-topology repeats in the AE1 anion exchanger suggests an elevator-like mechanism. _J. Gen. Physiol._ 149, 1149–1164 (2017). Article CAS PubMed PubMed Central Google Scholar *
Kalli, A. C. & Reithmeier, R. A. F. Organization and dynamics of the red blood cell band 3 anion exchanger SLC4A1: insights from molecular dynamics simulations. _Front. Physiol._ 13,
817945 (2022). Article PubMed PubMed Central Google Scholar * Colas, C., Ung, P. M. & Schlessinger, A. SLC transporters: structure, function, and drug discovery. _MedChemComm_ 7,
1069–1081 (2016). Article CAS PubMed Google Scholar * Garaeva, A. A. & Slotboom, D. J. Elevator-type mechanisms of membrane transport. _Biochem. Soc. Trans._ 48, 1227–1241 (2020).
Article CAS PubMed PubMed Central Google Scholar * Gasbjerg, P. K. & Brahm, J. Kinetics of bicarbonate and chloride transport in human red cell membranes. _J. Gen. Physiol._ 97,
321–349 (1991). Article CAS PubMed Google Scholar * Garaeva, A. A., Guskov, A., Slotboom, D. J. & Paulino, C. A one-gate elevator mechanism for the human neutral amino acid
transporter ASCT2. _Nat. Commun._ 10, 3427 (2019). Article PubMed PubMed Central Google Scholar * Gunn, R. B. & Frohlich, O. Asymmetry in the mechanism for anion exchange in human
red blood cell membranes. Evidence for reciprocating sites that react with one transported anion at a time. _J. Gen. Physiol._ 74, 351–374 (1979). Article CAS PubMed Google Scholar *
Bjornsson, T. D. & Mahony, C. Clinical pharmacokinetics of dipyridamole. _Thromb. Res. Suppl._ 4, 93–104 (1983). Article CAS PubMed Google Scholar * Legrum, B. & Passow, H.
Inhibition of inorganic anion transport across the human red blood cell membrane by chloride-dependent association of dipyridamole with a stilbene disulfonate binding site on the band 3
protein. _Biochim. Biophys. Acta_ 979, 193–207 (1989). Article CAS PubMed Google Scholar * Offman, E. et al. Pharmacokinetics and pharmacodynamics of the antiplatelet combination aspirin
(acetylsalicylic acid) plus extended-release dipyridamole are not altered by coadministration with the potent CYP2C19 inhibitor omeprazole. _Am. J. Cardiovasc. Drugs_ 13, 113–120 (2013).
Article CAS PubMed Google Scholar * Saniabadi, A. R. et al. Dipyridamole increases human red blood cell deformability. _Eur. J. Clin. Pharmacol._ 42, 651–654 (1992). Article CAS PubMed
Google Scholar * Gimsa, J. & Ried, C. Do band 3 protein conformational changes mediate shape changes of human erythrocytes? _Mol. Membr. Biol._ 12, 247–254 (1995). Article CAS
PubMed Google Scholar * Wagner, J. R. et al. POVME 3.0: software for mapping binding pocket flexibility. _J. Chem. Theory Comput._ 13, 4584–4592 (2017). Article CAS PubMed PubMed
Central Google Scholar * Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. _Nat. Methods_ 14, 331–332 (2017). Article
CAS PubMed PubMed Central Google Scholar * Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure
determination. _Nat. Methods_ 14, 290–296 (2017). Article CAS PubMed Google Scholar * Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron
micrographs. _J. Struct. Biol._ 192, 216–221 (2015). Article PubMed PubMed Central Google Scholar * Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive
regularization improves single-particle cryo-EM reconstruction. _Nat. Methods_ 17, 1214–1221 (2020). Article CAS PubMed Google Scholar * Yamashita, K., Palmer, C. M., Burnley, T. &
Murshudov, G. N. Cryo-EM single-particle structure refinement and map calculation using Servalcat. _Acta Crystallogr. D Struct. Biol._ 77, 1282–1291 (2021). Article CAS PubMed PubMed
Central Google Scholar * Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. _Acta Crystallogr. D Struct.
Biol._ 75, 861–877 (2019). Article CAS PubMed PubMed Central Google Scholar * The PyMOL Molecular Graphics System, Version 2.0 (Schrödinger, LLC). * Sanematsu, K. et al. Intracellular
acidification is required for full activation of the sweet taste receptor by miraculin. _Sci. Rep._ 6, 22807 (2016). Article CAS PubMed PubMed Central Google Scholar * Jo, S., Kim, T.,
Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. _J. Comput. Chem._ 29, 1859–1865 (2008). Article CAS PubMed Google Scholar * Mezei, M. Simulaid: a
simulation facilitator and analysis program. _J. Comput. Chem._ 31, 2658–2668 (2010). Article CAS PubMed Google Scholar * Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. &
Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. _J. Comput. Aided Mol. Des._ 27, 221–234 (2013). Article PubMed Google
Scholar * Lyne, P. D., Lamb, M. L. & Saeh, J. C. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring.
_J. Med. Chem._ 49, 4805–4808 (2006). Article CAS PubMed Google Scholar * Sterling, T. & Irwin, J. J. ZINC 15—ligand discovery for everyone. _J. Chem. Inf. Model_ 55, 2324–2337
(2015). Article CAS PubMed PubMed Central Google Scholar * Jorgensen, W. L., Maxwell, D. S. & Tirado-Rives, J. Development and testing of the OPLS all-atom force field on
conformational energetics and properties of organic liquids. _J. Am. Chem. Soc._ 118, 11225–11236 (1996). Article CAS Google Scholar * Friesner, R. A. et al. Extra precision glide:
docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. _J. Med. Chem._ 49, 6177–6196 (2006). Article CAS PubMed Google Scholar * Schlessinger,
A. et al. Structure-based discovery of prescription drugs that interact with the norepinephrine transporter, NET. _Proc. Natl Acad. Sci. USA_ 108, 15810–15815 (2011). Article CAS PubMed
PubMed Central Google Scholar * Adams, D. J. Grand canonical ensemble Monte Carlo for a Lennard–Jones Fluid. _Mol. Phys._ 29, 307–311 (1975). Article CAS Google Scholar Download
references ACKNOWLEDGEMENTS This work was supported by NIH grant GM133504, a Sloan Research Fellowship in Neuroscience, an Edward Mallinckrodt, Jr. Foundation Grant, a McKnight Foundation
Scholars Award (all to D.W.), NIH T32 Training Grant GM062754 and DA053558 (G.Z.), R01 CA277794 (A.S. and K.H.), NIH grant U01AG046170 (B.Z.), NIH grant RF1AG057440 (B.Z.), NIH grant
R01AG068030 (B.Z.) and NIH grants R01DK073681, R01DK067555 and R01DK061659 (R.O.). Some of this work was performed at the National Center for cryo-EM Access and Training (NCCAT) and the
Simons Electron Microscopy Center located at the New York Structural Biology Center, supported by the NIH Common Fund Transformative High Resolution Cryo-Electron Microscopy program (U24
GM129539,) and by grants from the Simons Foundation (SF349247) and NY State Assembly. We further acknowledge cryo-EM resources at the National Resource for Automated Molecular Microscopy
located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR and the NIH National Institute of General Medical Sciences (GM103310)
with additional support from Agouron Institute (F00316) and NIH (OD019994). For additional data collection we are also grateful to staff at the Laboratory for BioMolecular Structure (LBMS),
which is supported by the DOE Office of Biological and Environmental Research (KP160711). This work was supported in part through the computational resources and staff expertise provided by
Scientific Computing at the Icahn School of Medicine at Mount Sinai. We also thank J. F. Fay for help with initial data processing. AUTHOR INFORMATION Author notes * These authors
contributed equally: Michael J. Capper, Shifan Yang, Alexander C. Stone. AUTHORS AND AFFILIATIONS * Department of Pharmacological Sciences, Icahn School of Medicine at Mount Sinai, New York,
NY, USA Michael J. Capper, Yamuna Kalyani Mathiharan, Raul Habib, Keino Hutchinson, Yihan Zhao, Avner Schlessinger, Mihaly Mezei, Roman Osman, Bin Zhang & Daniel Wacker * Department of
Genetics and Genomic Sciences, Icahn School of Medicine at Mount Sinai, New York, NY, USA Shifan Yang, Alexander C. Stone, Sezen Vatansever, Yihan Zhao & Bin Zhang * Mount Sinai Center
for Transformative Disease Modeling, Icahn School of Medicine at Mount Sinai, New York, NY, USA Shifan Yang, Alexander C. Stone, Sezen Vatansever & Bin Zhang * Department of
Neuroscience, Icahn School of Medicine at Mount Sinai, New York, NY, USA Gregory Zilberg, Yamuna Kalyani Mathiharan, Bin Zhang & Daniel Wacker * Department of Artificial Intelligence and
Human Health, Icahn School of Medicine at Mount Sinai, New York, NY, USA Avner Schlessinger & Bin Zhang * Icahn Genomics Institute, Icahn School of Medicine at Mount Sinai, New York,
NY, USA Bin Zhang Authors * Michael J. Capper View author publications You can also search for this author inPubMed Google Scholar * Shifan Yang View author publications You can also search
for this author inPubMed Google Scholar * Alexander C. Stone View author publications You can also search for this author inPubMed Google Scholar * Sezen Vatansever View author publications
You can also search for this author inPubMed Google Scholar * Gregory Zilberg View author publications You can also search for this author inPubMed Google Scholar * Yamuna Kalyani Mathiharan
View author publications You can also search for this author inPubMed Google Scholar * Raul Habib View author publications You can also search for this author inPubMed Google Scholar *
Keino Hutchinson View author publications You can also search for this author inPubMed Google Scholar * Yihan Zhao View author publications You can also search for this author inPubMed
Google Scholar * Avner Schlessinger View author publications You can also search for this author inPubMed Google Scholar * Mihaly Mezei View author publications You can also search for this
author inPubMed Google Scholar * Roman Osman View author publications You can also search for this author inPubMed Google Scholar * Bin Zhang View author publications You can also search for
this author inPubMed Google Scholar * Daniel Wacker View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.J.C. designed experiments, expressed
and purified protein for grid freezing, collected data, refined structures and edited the manuscript. S.Y. and A.C.S. purified protein, prepared samples for grid freezing, established and
performed functional assays and edited the manuscript. S.V. performed molecular docking calculations with help from Y.Z., and helped analyze the structures. G.Z. prepared grids for structure
determination and assisted with data collection, processing and refinement. Y.K.M. helped with data processing and structure refinement. R.H. helped establish protein expression and
purification. K.H. performed volume calculations. A.S. supervised docking and volume calculation experiments and helped write the paper. R.O. performed molecular simulations and SACP
analysis of substrate binding with help from M.M. B.Z. contributed to the study design and supervised computational studies. D.W. designed experiments, analyzed the data, supervised the
overall project and management and wrote the manuscript. CORRESPONDING AUTHOR Correspondence to Daniel Wacker. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Structural & Molecular Biology_ thanks Reinhart A. F. Reithmeier and the other, anonymous, reviewer(s) for their contribution to
the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Katarzyna Ciazynska, in collaboration with the _Nature Structural & Molecular Biology_ team.
Peer reviewer reports are available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
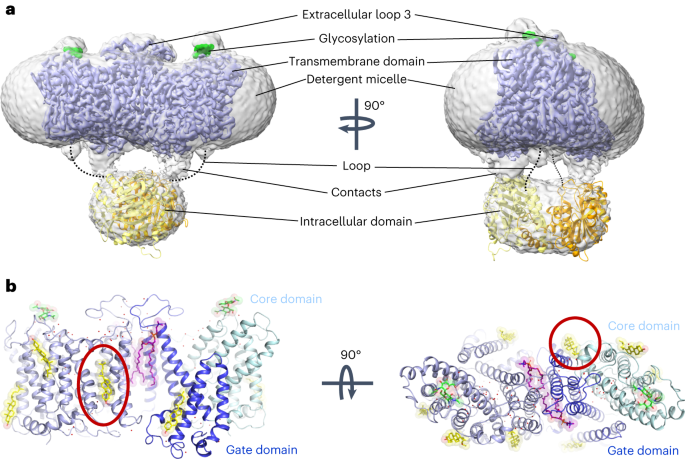
affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 CRYO-EM STRUCTURE DETERMINATION OF AE1 COMPLEXES AS EXEMPLIFIED BY AE1-DIDS COMPLEX. A, Analytical size exclusion chromatography and SDS-PAGE
show monodisperse and pure protein of AE1-DIDS. Data were collected on 300 keV Krios, a representative micrograph is shown, and processed in cryoSPARC: particles were picked from motion
corrected micrographs, subjected to 2D classification (representative classes are shown), followed by ab initio model building and 3D classification. After multiple rounds of 3D
classification, the final particle stack was subjected to local CTF refinement followed by local refinement of the masked AE1 membrane domain with imposed C2 symmetry. Final map was obtained
with GS-FSC indicating a resolution of 2.95 Å (AE1-DIDS) applying the 0.143 cutoff. Viewing direction distribution analysis (cryoSPARC) indicates sufficient coverage. An initial model was
built in PHENIX, and then further refined in ServalCat for the generation of final maps and coordinates of mdAE1. Calculations in cryoSPARC indicate local resolutions of up to 2.5 Å around
substrate and inhibitor binding sites. Viewing direction analysis indicates isotropic distribution of views in final particle stack. B, Superposition with other AE1 structures (AE1, PDB ID:
7TW2, dark red; AE1-Glycophorin complex, PDB ID: 7UZ3, lime green)7,8. Root mean square deviations of 0.997 Å (7TW2) and 0.372 Å (7UZ3) highlight similarity of protein conformation.
Glycosylation modifications, cholesterols, lipids, waters, PIP2, and Glycophorin are shown in green, yellow, purple, red, grey, and teal, respectively. C, Cryo-EM structures allowed us to
assign the complete extracellular surface including all Diego antigens (see Extended Data Table 2) with sidechains shown as spheres with yellow carbon atoms. Source data EXTENDED DATA FIG. 2
CRYO-EM STRUCTURE DETERMINATION OF APO AE1 AND AE1-BICARBONATE COMPLEXES. A, B, workflow of Apo AE1 (A) and AE1-Bicarbonate (B) data processing, showing representative 2D classes, followed
by 2D classification, ab initio modeling and several rounds of 3D classification, with representative 3D classes shown. After multiple rounds of 3D classification, final particle stacks were
subjected to CTF refinement followed by local refinement of the masked AE1 membrane domain with imposed C2 symmetry. GS-FSC calculations indicate global resolutions of 2.99 Å (Apo AE1) and
3.37 (AE1-Bicarbonate) applying the 0.143 cutoff. Viewing direction distribution analysis (cryoSPARC) indicates sufficient coverage. Initial models were built in PHENIX, and then further
refined in ServalCat for the generation of final maps and coordinates. Calculations in cryoSPARC indicate local resolutions of lower than 3 Å around substrate and inhibitor binding sites.
Source data EXTENDED DATA FIG. 3 CRYO-EM STRUCTURE DETERMINATION OF AE1-H2DIDS AND AE1-DEPC COMPLEXES. A, B, workflow of AE1-H2DIDS (A) and AE1-DEPC (B) data processing, showing
representative 2D classes, followed by 2D classification, ab initio modeling and several rounds of 3D classification, with representative 3D classes shown. After multiple rounds of 3D
classification, final particle stacks were subjected to CTF refinement followed by local refinement of the masked AE1 membrane domain with imposed C2 symmetry. GS-FSC calculations indicate
global resolutions of 2.98 Å (AE1-H2DIDS) and 3.07 (AE1-DEPC) applying the 0.143 cutoff. Viewing direction distribution analysis (cryoSPARC) indicates sufficient coverage. Initial models
were built in PHENIX, and then further refined in ServalCat for the generation of final maps and coordinates. Calculations in cryoSPARC indicate local resolutions of lower than 3 Å around
substrate and inhibitor binding sites. Source data EXTENDED DATA FIG. 4 CRYO-EM STRUCTURE DETERMINATION OF AE1-DIPYRIDAMOLE AND AE1-NIF COMPLEXES. A, B, workflow of AE1-Dipyridamole (A) and
AE1-NIF (B) data processing, showing representative 2D classes, followed by 2D classification, ab initio modeling and several rounds of 3D classification, with representative 3D classes
shown. After multiple rounds of 3D classification, final particle stacks were subjected to CTF refinement followed by local refinement of the masked AE1 membrane domain with imposed C2
symmetry. GS-FSC calculations indicate global resolutions of 3.13 Å (AE1-Dipyridamole) and 3.18 (AE1-NIF) applying the 0.143 cutoff. Viewing direction distribution analysis (cryoSPARC)
indicates sufficient coverage. Initial models were built in PHENIX, and then further refined in ServalCat for the generation of final maps and coordinates. Calculations in cryoSPARC indicate
local resolutions of lower than 3 Å around substrate and inhibitor binding sites. Source data EXTENDED DATA FIG. 5 SUBSTRATE-BINDING IN SLC4 AND SLC26 TRANSPORTERS. A, Cryo-EM density of
bicarbonate and binding site residues (marine mesh) using a contour level of 6σ. B, Binding site of uracil in the UraA-Uracil structure (PDB ID: 3QE7)24. C, D, Carbonate and sodium ions in
the rabbit NDCBE (SLC4A8) cryo-EM structure (PDB ID: 7RTM, EMD-24683)15, with clashes between CO32− and T538, as well as Na+ and the backbone shown as red dotted lines. Ionic interactions
and hydrogen bonds are shown as grey dotted lines. E, SACP simulations allow to calculate bicarbonate affinity to AE1 anion binding site (see methods). In some structures (~5%) we have
identified a second bicarbonate binding site in the putative exit channel. F, Surface display shows putative anion exit channel leading from the bicarbonate binding site to the cytoplasmic
site. Channel extends below the plane of section. Source data EXTENDED DATA FIG. 6 STRUCTURE AND DENSITY OF AE1-BOUND INHIBITORS. A, Chemical structures of different AE1 inhibitors and
ligands used in this study. B, Cryo-EM density of DIDS (magenta) and key residues at a contour level of 6σ. C, H2DIDS (magenta) binding mode. D, Cryo-EM density of H2DIDS (magenta) and key
residues at a contour level of 5σ. E, F, DEPC modifications of K539 and K851 (E) and corresponding cryo-EM densities (F) shown at a contour level of 8σ. G, H, Binding pocket of apo structure
(G) and cryo-EM densities of residues (H) at a contour level of 6σ shown for comparison. I, Cryo-EM density of Dipyridamole (magenta) and key lysines at a contour level of 6σ. AE1’s gate
and core domains are shown in tv blue and palecyan, respectively, and cryo-EM densities are shown as marine colored mesh. EXTENDED DATA FIG. 7 BINDING OF NIF TO AE1. A-E, Close up of NIF
binding site (A) and validation by molecular docking (B), with the best scoring docking pose shown (ΔG binding score: -52.33). C, Cryo-EM density of NIF and NIF binding site residues shown
at a contour level of 3.5σ. D, Overlay of Apo (salmon) and NIF-bound AE1 (light blue) reveals subtle conformational rearrangements required for NIF binding. E, Calculation of NIF binding
pocket volume and surface in POVME355. AE1’s gate and core domains are shown in tv blue and palecyan, respectively, and cryo-EM densities are shown as marine colored mesh. EXTENDED DATA FIG.
8 EXPERIMENTAL TESTING OF COMPOUND 22 (CMPD 22) ANALOGS. A, Inhibitory activity of 24 commercially available analogs of Cmpd 22 at 20 µM. Uptake experiments were performed with 2–6
technical repeats and are averaged from 3 independent experiments (n = 3). Data are represented as mean±s.e.m. B, Chemical structure of Cmpd 22 and selected analogs. C, D, Concentration
response experiments show that Cmpd 22-J, Cmpd 22-R, and Cmpd 22-D have comparable inhibitory potencies as Cmpd 22 (C), while Cmpd 22-P and Cmpd 22-C appear to not inhibit AE1 (D).
Concentration response uptake experiments were performed in triplicates and are averaged from 4 independent experiments (n = 4). Apparent potencies are calculated as apparent IC50 (mean) and
error bars denote 95% confidence intervals. All uptake data have been normalized to Cmpd 22. Source data SUPPLEMENTARY INFORMATION REPORTING SUMMARY PEER REVIEW FILE SOURCE DATA SOURCE DATA
FIG. 4 AND EXTENDED DATA FIGS. 1–5 AND 8 AE1 uptake source data. SOURCE DATA EXTENDED DATA FIG. 1 Size-exclusion chromatogram source data, FSC curve source data and uncropped SDS–PAGE
image. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or
other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Capper, M.J., Yang, S., Stone, A.C. _et al._ Substrate binding and inhibition of the anion exchanger 1 transporter. _Nat Struct Mol Biol_ 30,
1495–1504 (2023). https://doi.org/10.1038/s41594-023-01085-6 Download citation * Received: 14 December 2022 * Accepted: 28 July 2023 * Published: 07 September 2023 * Issue Date: October
2023 * DOI: https://doi.org/10.1038/s41594-023-01085-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable
link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative