- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT TAZ promotes growth, development and tumorigenesis by regulating the expression of target genes. However, the manner in which TAZ orchestrates the transcriptional responses is
poorly defined. Here we demonstrate that TAZ forms nuclear condensates through liquid–liquid phase separation to compartmentalize its DNA-binding cofactor TEAD4, coactivators BRD4 and MED1,
and the transcription elongation factor CDK9 for transcription. TAZ forms phase-separated droplets in vitro and liquid-like nuclear condensates in vivo, and this ability is negatively
regulated by Hippo signalling through LATS-mediated phosphorylation and is mediated by the coiled-coil (CC) domain. Deletion of the TAZ CC domain or substitution with the YAP CC domain
prevents the phase separation of TAZ and its ability to induce the expression of TAZ-specific target genes. Thus, we identify a mechanism of transcriptional activation by TAZ and demonstrate
that pathway-specific transcription factors also engage the phase-separation mechanism for efficient and specific transcriptional activation. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get
Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $209.00 per year only
$17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS TAF14 RECOGNIZES A COMMON
MOTIF IN TRANSCRIPTIONAL MACHINERIES AND FACILITATES THEIR CLUSTERING BY PHASE SEPARATION Article Open access 21 August 2020 NUCLEATED TRANSCRIPTIONAL CONDENSATES AMPLIFY GENE EXPRESSION
Article 14 September 2020 A CHAPERONE-LIKE FUNCTION OF FUS ENSURES TAZ CONDENSATE DYNAMICS AND TRANSCRIPTIONAL ACTIVATION Article 03 January 2024 DATA AVAILABILITY Source data for Figs. 1–4
and 6–8 and Extended Data Figs. 1, 3, 4 and 8 are available online. The RNA-seq data are available in the Gene Expression Omnibus (GEO) with the accession number GSE142474. All other data
supporting the findings of this study are available from the corresponding author on reasonable request. REFERENCES * Pan, D. The Hippo signaling pathway in development and cancer. _Dev.
Cell_ 19, 491–505 (2010). Article CAS PubMed PubMed Central Google Scholar * Ma, S., Meng, Z., Chen, R. & Guan, K. L. The Hippo pathway: biology and pathophysiology. _Annu. Rev.
Biochem._ 88, 577–604 (2018). Article CAS Google Scholar * Tapon, N. & Harvey, K. F. The Hippo pathway—from top to bottom and everything in between. _Semin. Cell Dev. Biol._ 23,
768–769 (2012). Article PubMed Google Scholar * Badouel, C. & McNeill, H. SnapShot: the Hippo signaling pathway. _Cell_ 145, 484 (2011). Article CAS PubMed Google Scholar *
Schroeder, M. C. & Halder, G. Regulation of the Hippo pathway by cell architecture and mechanical signals. _Semin. Cell Dev. Biol._ 23, 803–811 (2012). Article CAS PubMed Google
Scholar * Yu, F. X. & Guan, K. L. The Hippo pathway: regulators and regulations. _Genes Dev._ 27, 355–371 (2013). Article CAS PubMed PubMed Central Google Scholar * Dong, J. X. et
al. Elucidation of a universal size-control mechanism in _Drosophila_ and mammals. _Cell_ 130, 1120–1133 (2007). Article CAS PubMed PubMed Central Google Scholar * Ota, M. & Sasaki,
H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. _Development_ 135, 4059–4069 (2008). Article CAS PubMed
Google Scholar * Enderle, L. & McNeill, H. Hippo gains weight: added insights and complexity to pathway control. _Sci. Signal._ 6, re7 (2013). * Shreberk-Shaked, M. & Oren, M. New
insights into YAP/TAZ nucleo-cytoplasmic shuttling: new cancer therapeutic opportunities? _Mol. Oncol._ 13, 1335–1341 (2019). PubMed PubMed Central Google Scholar * Galli, G. G. et al.
YAP drives growth by controlling transcriptional pause release from dynamic enhancers. _Mol. Cell_ 60, 328–337 (2015). Article CAS PubMed PubMed Central Google Scholar * Zanconato, F.
et al. Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. _Nat. Med._ 24, 1599–1610 (2018). Article CAS PubMed PubMed Central Google Scholar * Boija, A. et
al. Transcription factors activate genes through the phase-separation capacity of their activation domains. _Cell_ 175, 1842–1855 (2018). Article CAS PubMed Google Scholar * Chan, S. W.
et al. A role for TAZ in migration, invasion, and tumorigenesis of breast cancer cells. _Cancer Res._ 68, 2592–2598 (2008). Article CAS PubMed Google Scholar * Cordenonsi, M. et al. The
Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. _Cell_ 147, 759–772 (2011). Article CAS PubMed Google Scholar * Plouffe, S. W. et al. The Hippo
pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. _J. Biol. Chem._ 293, 11230–11240 (2018). Article CAS PubMed PubMed Central Google Scholar
* Xin, M. et al. Hippo pathway effector Yap promotes cardiac regeneration. _Proc. Natl Acad. Sci. USA_ 110, 13839–13844 (2013). Article PubMed PubMed Central Google Scholar * Nishioka,
N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. _Dev. Cell_ 16, 398–410 (2009). Article CAS
PubMed Google Scholar * Morin-Kensicki, E. M. et al. Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65.
_Mol. Cell. Biol._ 26, 77–87 (2006). Article CAS PubMed PubMed Central Google Scholar * Hossain, Z. et al. Glomerulocystic kidney disease in mice with a targeted inactivation of Wwtr1.
_Proc. Natl Acad. Sci. USA_ 104, 1631–1636 (2007). Article CAS PubMed PubMed Central Google Scholar * Passaniti, A., Brusgard, J. L., Qiao, Y. T., Sudol, M. & Finch-Edmondson, M.
Roles of RUNX in Hippo pathway signaling. _Adv. Exp. Med. Biol._ 962, 435–448 (2017). Article CAS PubMed PubMed Central Google Scholar * Zhao, B. et al. TEAD mediates YAP-dependent gene
induction and growth control. _Gene Dev._ 22, 1962–1971 (2008). Article CAS PubMed PubMed Central Google Scholar * Zhao, B., Tumaneng, K. & Guan, K. L. The Hippo pathway in organ
size control, tissue regeneration and stem cell self-renewal. _Nat. Cell Biol._ 13, 877–883 (2011). Article CAS PubMed PubMed Central Google Scholar * Alberti, S., Gladfelter, A. &
Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. _Cell_ 176, 419–434 (2019). Article CAS PubMed PubMed Central Google
Scholar * Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. _Science_ 361, eaar3958 (2018). Article CAS PubMed PubMed Central
Google Scholar * Lu, H., Liu, R. & Zhou, Q. Balanced between order and disorder: a new phase in transcription elongation control and beyond. _Transcription_ 10, 157–163 (2019). Article
CAS PubMed PubMed Central Google Scholar * Kaan, H. Y. K. et al. Crystal structure of TAZ-TEAD complex reveals a distinct interaction mode from that of YAP-TEAD complex. _Sci. Rep._ 7,
2035 (2017). Article CAS PubMed PubMed Central Google Scholar * Piccolo, S., Dupont, S. & Cordenonsi, M. The biology of Yap/Taz: Hippo signaling and beyond. _Physiol. Rev._ 94,
1287–1312 (2014). Article CAS PubMed Google Scholar * Hyman, A. A., Weber, C. A. & Juelicher, F. Liquid-liquid phase separation in biology. _Annu. Rev. Cell Dev. Biol._ 30, 39–58
(2014). Article CAS PubMed Google Scholar * Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. _Trends Cell Biol._ 28, 420–435 (2018). Article CAS PubMed
PubMed Central Google Scholar * Cai, D. et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. _Nat. Cell Biol._ 21, 1578–1589 (2019). Article
CAS PubMed PubMed Central Google Scholar * Henis, Y. I., Rotblat, B. & Kloog, Y. FRAP beam-size analysis to measure palmitoylation-dependent membrane association dynamics and
microdomain partitioning of Ras proteins. _Methods_ 40, 183–190 (2006). Article CAS PubMed Google Scholar * Eisenberg, S., Giehl, K., Henis, Y. I. & Ehrlich, M. Differential
interference of chlorpromazine with the membrane interactions of oncogenic K-Ras and its effects on cell growth. _J. Biol. Chem._ 283, 27279–27288 (2008). Article CAS PubMed Google
Scholar * Shvartsman, D. E. et al. Src kinase activity and SH2 domain regulate the dynamics of Src association with lipid and protein targets. _J. Cell Biol._ 178, 675–686 (2007). Article
CAS PubMed PubMed Central Google Scholar * Wolfenson, H. et al. A role for the juxtamembrane cytoplasm in the molecular dynamics of focal adhesions. _PLoS ONE_ 4, e4304 (2009). Article
CAS PubMed PubMed Central Google Scholar * Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. _Cell_
163, 123–133 (2015). Article CAS PubMed PubMed Central Google Scholar * Nott, T. J. et al. Phase transition of a disordered nuage protein generates environmentally responsive
membraneless organelles. _Mol. Cell_ 57, 936–947 (2015). Article CAS PubMed PubMed Central Google Scholar * Fan, R., Kim, N. G. & Gumbiner, B. M. Regulation of Hippo pathway by
mitogenic growth factors via phosphoinositide 3-kinase and phosphoinositide-dependent kinase-1. _Proc. Natl Acad. Sci. USA_ 110, 2569–2574 (2013). Article PubMed PubMed Central Google
Scholar * Kim, N. G. & Gumbiner, B. M. Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. _J. Cell Biol._ 210, 503–515 (2015). Article CAS PubMed PubMed
Central Google Scholar * Totaro, A., Panciera, T. & Piccolo, S. YAP/TAZ upstream signals and downstream responses. _Nat. Cell Biol._ 20, 888–899 (2018). Article CAS PubMed PubMed
Central Google Scholar * Yu, F. X. et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. _Cell_ 150, 780–791 (2012). Article CAS PubMed PubMed Central
Google Scholar * Panciera, T., Azzolin, L., Cordenonsi, M. & Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. _Nat. Rev. Mol. Cell Biol._ 18, 758–770 (2017). Article
CAS PubMed PubMed Central Google Scholar * Hergovich, A. The roles of NDR protein kinases in Hippo signalling. _Genes_ 7, 21 (2016). Article CAS PubMed Central Google Scholar *
Chan, S. W. et al. TEADs mediate nuclear retention of TAZ to promote oncogenic transformation. _J. Biol. Chem._ 284, 14347–14358 (2009). Article CAS PubMed PubMed Central Google Scholar
* Noland, C. L. et al. Palmitoylation of TEAD transcription factors is required for their stability and function in Hippo pathway signaling. _Structure_ 24, 179–186 (2016). Article CAS
PubMed Google Scholar * Zhang, H. et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. _J. Biol. Chem._ 284, 13355–13362
(2009). Article CAS PubMed PubMed Central Google Scholar * Rausch, V. et al. The Hippo pathway regulates caveolae expression and mediates flow response via caveolae. _Curr. Biol._ 29,
242–255 (2019). Article CAS PubMed PubMed Central Google Scholar * Misra, J. R. & Irvine, K. D. The Hippo signaling network and its biological functions. _Annu. Rev. Genet._ 52,
65–87 (2018). Article CAS PubMed PubMed Central Google Scholar * Zhang, L., Yue, T. & Jiang, J. Hippo signaling pathway and organ size control. _Fly_ 3, 68–73 (2009). Article
PubMed Google Scholar * Gill, M. K. et al. A feed forward loop enforces YAP/TAZ signaling during tumorigenesis. _Nat. Commun._ 9, 3510 (2018). Article CAS PubMed PubMed Central Google
Scholar * Richardson, H. E. & Portela, M. Tissue growth and tumorigenesis in _Drosophila_: cell polarity and the Hippo pathway. _Curr Opin. Cell Biol._ 48, 1–9 (2017). Article CAS
PubMed Google Scholar * Zeng, M. et al. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. _Cell_ 166, 1163–1175 (2016). Article
CAS PubMed PubMed Central Google Scholar * Fang, X. et al. _Arabidopsis_ FLL2 promotes liquid-liquid phase separation of polyadenylation complexes. _Nature_ 569, 265–269 (2019).
Article CAS PubMed PubMed Central Google Scholar * Varelas, X. et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. _Nat. Cell
Biol._ 10, 837–848 (2008). Article CAS PubMed Google Scholar * Zhang, J. M. et al. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo
pathway. _Nat. Cell Biol._ 11, 1444–1450 (2009). Article CAS PubMed PubMed Central Google Scholar * Kwon, I. et al. Phosphorylation-regulated binding of rna polymerase ii to fibrous
polymers of low-complexity domains. _Cell_ 155, 1049–1060 (2013). Article CAS PubMed PubMed Central Google Scholar * Kato, M. et al. Cell-free formation of RNA granules: low complexity
sequence domains form dynamic fibers within hydrogels. _Cell_ 149, 753–767 (2012). Article CAS PubMed PubMed Central Google Scholar * Hagenbeek, T. J. et al. The Hippo pathway effector
TAZ induces TEAD-dependent liver inflammation and tumors. _Sci. Signal._ 11, eaaj1757 (2018). Article CAS PubMed Google Scholar * Lu, H. et al. Phase-separation mechanism for C-terminal
hyperphosphorylation of RNA polymerase II. _Nature_ 558, 318–323 (2018). Article CAS PubMed PubMed Central Google Scholar * Plouffe, S. W. et al. Characterization of Hippo pathway
components by gene inactivation. _Mol. Cell_ 64, 993–1008 (2016). Article CAS PubMed PubMed Central Google Scholar * Zhu, Q. et al. SnoN antagonizes the Hippo kinase complex to promote
TAZ signaling during breast carcinogenesis. _Dev. Cell_ 37, 399–412 (2016). Article CAS PubMed PubMed Central Google Scholar * Eisenberg, S. et al. Raft protein clustering alters N-Ras
membrane interactions and activation pattern. _Mol. Cell. Biol_ 31, 3938–3952 (2011). Article CAS PubMed PubMed Central Google Scholar * Efron, B. & Tibshirani, R. in _An
Introduction to Bootstrap_ (eds Cox, D. R. et al.) 124–130 (Chapman & Hall, 1993). * Rashidian, J. et al. Ski regulates Hippo and TAZ signaling to suppress breast cancer progression.
_Sci. Signal._ 8, ra14 (2015). Article CAS PubMed PubMed Central Google Scholar * Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. _Bioinformatics_ 29, 15–21 (2013). Article
CAS PubMed Google Scholar * Anders, S., Pyl, P. T. & Huber, W. HTSeq–a Python framework to work with high-throughput sequencing data. _Bioinformatics_ 31, 166–169 (2015). Article
CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank K.-L. Guan and A. Mauviel for providing cDNAs of components of the Hippo pathway and H. Sasaki for the
8xGT-IIC-δ51LucII construct; J. He for technical assistance and Q. Zhu for helpful suggestions, discussions and help with experimental procedures; and D. Schichnes and S. Ruzin at the CNR
biological imaging facility at the University of California, Berkeley for assistance with microscopy. This study was supported by DOD/US Army Medical Research And Materiel Command
W81XWH-15-1-0068 (to K.L. and Q.Z.), a Tel Aviv University-University of California Berkeley collaborative research grant (to Y.I.H. and K.L.), and NIH R01AI41757 (to Q.Z.). Y.I.H. is an
incumbent of the Zalman Weinberg Chair in Cell Biology. Y.L. is supported by the Berkeley Scholars program, and T.W. was supported by the China Scholarship Council. AUTHOR INFORMATION Author
notes * These authors contributed equally: Yi Lu, Tiantian Wu. AUTHORS AND AFFILIATIONS * Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA, USA Yi
Lu, Tiantian Wu, Huasong Lu, Qiang Zhou & Kunxin Luo * State Key Laboratory of Cellular Stress Biology, School of Life Sciences, Xiamen University, Xiamen, China Tiantian Wu * Department
of Neurobiology, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv, Israel Orit Gutman & Yoav I. Henis Authors * Yi Lu View author publications You can also search
for this author inPubMed Google Scholar * Tiantian Wu View author publications You can also search for this author inPubMed Google Scholar * Orit Gutman View author publications You can also
search for this author inPubMed Google Scholar * Huasong Lu View author publications You can also search for this author inPubMed Google Scholar * Qiang Zhou View author publications You
can also search for this author inPubMed Google Scholar * Yoav I. Henis View author publications You can also search for this author inPubMed Google Scholar * Kunxin Luo View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Y.L., T.W. and K.L. designed the research. Y.L. performed in vivo experiments. T.W. performed in vitro
experiments. Y.I.H. designed and O.G. performed FRAP experiments. Y.L., T.W., H.L., Y.I.H., Q.Z. and K.L. analysed data and wrote the paper. K.L. conceived and directed the project. All of
the authors discussed the results and commented on the manuscript. CORRESPONDING AUTHOR Correspondence to Kunxin Luo. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA
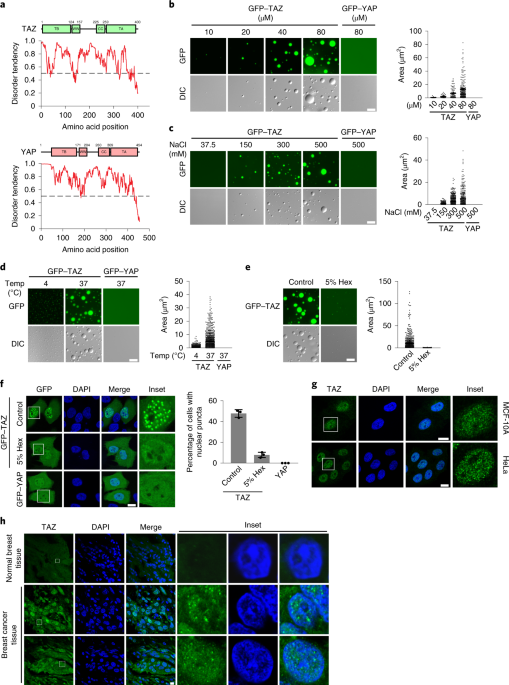
EXTENDED DATA FIG. 1 REGULATION OF TAZ DROPLET FORMATION _IN VITRO_ AND NUCLEAR PUNCTA FORMATION _IN VIVO_. A, GFP-TAZ purified from _E. coil_ were analysed by SDS-PAGE and visualized by
Coomassie blue staining. B. 50 μM GFP-TAZ were heated-inactivated (5 min at 95 °C and immediately put on ice for 5 min) or treated with 100 μg/ml Proteinase K for 30 min at 40 °C, and then
subjected to droplet formation assay _in vitro_ in the presence of 500 mM NaCl at room temperature. C, Ectopically expressed GFP-TAZ was expressed at a lower level than endogenous TAZ in
MCF-10A cells as shown by western blotting. GAPDH was used as a loading control. D, Flag-TAZ formed nuclear puncta when transfected into the MCF-10A cells, as detected by immunofluorescence
staining with anti-Flag. Scale bar, 10 μm. Experiments in A–D were repeated independently three times with similar results. Unprocessed blots are provided in Unprocessed Blots Extended Data
Fig. 1. Source data EXTENDED DATA FIG. 2 YAP DOES NOT FORM DROPLETS _IN VITRO_ AND _IN VIVO_ IN THE ABSENCE OF CROWDING AGENTS. A, GFP-YAP purified from _E. coil_ were analyzed by SDS-PAGE
and visualized by Coomassie blue staining. B, GFP-YAP at varying concentrations was subjected to the droplet formation assay at room temperature and in the presence of 500 mM NaCl. C, 50 μM
GFP-YAP was subjected to the droplet formation assay at room temperature in the presence of indicated salt concentrations. D, 50 μM GFP-YAP was subjected to droplet formation in the presence
of 150 mM NaCl at 4 °C or 37 °C. E, Two YAP isoforms, GFP-YAP1–1β or GFP-YAP1–2α, did not form droplets (50 μM protein, 500 mM NaCl and room temperature). aa, amino acids. F, 50 μM GFP-YAP
formed droplets in the presence of 10% PEG-8000, Ficoll or Dextran but not 10% glycerol or sucrose. Droplet formation assay was performed in the presence of 500 mM NaCl at room temperature.
G, 50 μM GFP-YAP did not form droplets in the presence of BSA at varying concentrations. H, GFP-YAP did not form nuclear puncta in both HeLa cells and 293T cells. Scale bars, 10 μm.
Experiments in A–H were repeated independently three times with similar results. EXTENDED DATA FIG. 3 THE CC AND WW DOMAINS ARE REQUIRED FOR TAZ TO FORM NUCLEAR PUNCTA. A, Domain structure
of TAZ and TAZ truncations. The numbers above indicate the position of amino acid residues. B, Bacterially purified GFP-TAZ, ∆TB, ∆WW, ∆CC, and ∆WW+∆CC proteins were analyzed by SDS-PAGE and
detected by Coomasssie blue staining. C, Localization of GFP-TAZ and various mutants in HeLa cells. D, Localization of GFP-TAZ and various TAZ/YAP chimera in HeLa cells. Scale bars, 10 μm.
E, A GST pull-down assay was performed by incubating immobilized GST fusion proteins with lysates of cells expressing HA-tagged WT or mutant TAZ, and the associated TAZ proteins were
detected by western blotting with anti-HA (upper). GST fusion proteins were assessed by western blotting with anti-GST, and HA-TAZ proteins in the cell lysates were measured by western
blotting (lower). Experiments in B–E were repeated independently three times with similar results. Unprocessed blots are provided in Unprocessed Blots Extended Data Fig. 3. Source data
EXTENDED DATA FIG. 4 TAZ CC DOMAIN ENHANCES YAP PHASE SEPARATION IN THE PRESENCE OF PEG. A, Domain structure of YAP chimera. B, Substitution of the YAP CC and WW domains with that of TAZ is
not sufficient to enable YAP to undergo LLPS in MCF10A cells in the absence of PEG. C, Coomasssie blue staining of various recombinant proteins purified from _E. coil_. D, 25 μM bacterially
purified GFP-YAP chimera proteins were subjected to droplet formation assay in the presence of 10% PEG-8000. Quantification of the droplets is on the right. Scale bar, 10 μm. Data shown as
the mean ± s.e.m. Statistical significance was evaluated using One-way ANOVA with Krusk-Wallis test. Droplets in n = 3 fields in each group were quantified. E, The TAZ CC and WW domains
enhanced LLPS by GFP-YAP in transfected MCF10A cells in the presence of PEG as shown by confocal microscopy. Scale bar, 10 μm. Quantification of the percentage of cells that displayed
nuclear puncta is shown on the right. Data shown as the mean ± s.e.m.. _P_ value was determined by unpaired two-tailed Student’s _t_-test. 80 transfected cells in each group were quantified.
n = 3 biologically independent samples. Experiments in B, C, E were repeated independently three times with similar results. Experiments in D were repeated twice with similar results.
Statistical source data for D, E, are provided in Statistical Source Date Extended Data Fig. 4. Source data EXTENDED DATA FIG. 5 HIPPO SIGNALING NEGATIVELY REGULATES TAZ PHASE SEPARATION IN
HELA CELLS. TAZ localization was examined by immunofluorescence staining with anti-TAZ (green) in HeLa cells that have been subjected to the following treatments: A, Serum-starved HeLa cells
were treated with 1 μM LPA or 50 ng/ml EGF for 1 h. B, Serum-starved HeLa cells were seeded on fibronectin-coated coverslips for 10 min or 2 h in serum-free medium. C, HeLa cells were grown
on fibronectin-coated polyacrylamide hydrogels of 1 kPa and 40 kPa stiffness. D, HeLa cells were treated with 1 μg/ml Latrunculin B for 1 h. Alexa Fluor 555-conjugated phalloidin (Red)
staining was performed to detect F-actin in b-d. Scale bar, 10 µm. Experiments in A–D were repeated independently three times with similar results. EXTENDED DATA FIG. 6 LATS2 REGULATES TAZ
LLPS AND RECRUITMENT OF TEAD4 AND BRD4. A, MCF-10A cells transfected with GFP-TAZ-S89A and Flag-LATS2 were subjected to immunofluorescence staining with anti-Flag (Red). Scale bar, 10 µm. B,
MCF-10A cells stably expressing siLATS1/2 were transfected with GFP-TAZ and Flag-TEAD4. TEAD localization at high cell density was detected by immunofluorescence staining with anti-Flag
(Red). Scale bar, 10 μm. C, MCF-10A cells stably expressing siLATS1/2 were transfected with GFP-TAZ. Endogenous BRD4 localization was examined by immunofluorescence staining with anti-BRD4
(Red). Scale bar, 10 μm. All experiments were repeated independently three times with similar results. EXTENDED DATA FIG. 7 TAZ NUCLEAR CONDENSATES DO NOT CO-LOCALIZE WITH THE PML BODIES,
CAJAL BODIES OR NUCLEOLI. The PML nuclear bodies, Cajal Bodies and nucleoli in MCF-10A cells expressing GFP-TAZ (green) were detected by immunofluorescence staining with antibodies targeting
PML, Coilin and Fibrillarin, respectively (red). Scale bar, 10 μm. Experiments were repeated independently three times with similar results. EXTENDED DATA FIG. 8 TAZ MUTANTS LACKING THE CC
DOMAIN STILL BIND TO LAST2 AND TEAD4. A, HA-tagged WT and mutant TAZ were co-transfected into 293T cells with Flag-LATS2. TAZ proteins associated with LATS2 were isolated by
immunoprecipitation with anti-Flag and detected by western blotting with anti-HA antibodies (upper panels). The abundance of these proteins in the cell lysates was assessed by western
blotting (lower panels). GAPDH was used as a loading control. B, Interaction of various TAZ mutants with Flag-TEAD4 was analyzed by co-IP assay as described in A. C, Interaction of various
TAZ/YAP chimera with LATS2 was analyzed by co-IP as described in A. All experiments were repeated independently three times with similar results. Unprocessed blots are provided in
Unprocessed Blots Extended Data Fig. 8. Source data SUPPLEMENTARY INFORMATION REPORTING SUMMARY SOURCE DATA SOURCE DATA FIG. 1 Statistical source data. SOURCE DATA FIG. 2 Statistical source
data. SOURCE DATA FIG. 3 Unprocessed blots. SOURCE DATA FIG. 3 Statistical source data. SOURCE DATA FIG. 4 Unprocessed blots. SOURCE DATA FIG. 4 Statistical source data. SOURCE DATA FIG. 6
Unprocessed blots. SOURCE DATA FIG. 6 Statistical source data. SOURCE DATA FIG. 7 Unprocessed blots. SOURCE DATA FIG. 8 Unprocessed blots. SOURCE DATA FIG. 8 Statistical source data. SOURCE
DATA EXTENDED DATA FIG. 1 Unprocessed blots. SOURCE DATA EXTENDED DATA FIG. 3 Unprocessed blots. SOURCE DATA EXTENDED DATA FIG. 4 Statistical source data. SOURCE DATA EXTENDED DATA FIG. 8
Unprocessed blots. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Lu, Y., Wu, T., Gutman, O. _et al._ Phase separation of TAZ compartmentalizes the
transcription machinery to promote gene expression. _Nat Cell Biol_ 22, 453–464 (2020). https://doi.org/10.1038/s41556-020-0485-0 Download citation * Received: 28 May 2019 * Accepted: 14
February 2020 * Published: 23 March 2020 * Issue Date: April 2020 * DOI: https://doi.org/10.1038/s41556-020-0485-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to
read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative