- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Given the ongoing SARS-CoV-2 pandemic, identification of immunogenic targets against the coronavirus spike glycoprotein will provide crucial advances towards the development of
sensitive diagnostic tools and potential vaccine candidate targets. In this study, using pools of overlapping linear B-cell peptides, we report two IgG immunodominant regions on SARS-CoV-2
spike glycoprotein that are recognised by sera from COVID-19 convalescent patients. Notably, one is specific to SARS-CoV-2, which is located in close proximity to the receptor binding
domain. The other region, which is localised at the fusion peptide, could potentially function as a pan-SARS target. Functionally, antibody depletion assays demonstrate that antibodies
targeting these immunodominant regions significantly alter virus neutralisation capacities. Taken together, identification and validation of these neutralising B-cell epitopes will provide
insights towards the design of diagnostics and vaccine candidates against this high priority coronavirus. SIMILAR CONTENT BEING VIEWED BY OTHERS THE SARS-COV-2 SPIKE RESIDUES 616/644 AND
1138/1169 DELINEATE TWO ANTIBODY EPITOPES IN COVID-19 MRNA COMIRNATY VACCINE (PFIZER/BIONTECH) Article Open access 09 April 2022 EPITOPE MAPPING OF SARS-COV-2 SPIKE PROTEIN USING
NATURALLY-ACQUIRED IMMUNE RESPONSES TO DEVELOP MONOCLONAL ANTIBODIES Article Open access 09 May 2025 A SARS-COV-2 NEUTRALIZING ANTIBODY WITH EXTENSIVE SPIKE BINDING COVERAGE AND MODIFIED FOR
OPTIMAL THERAPEUTIC OUTCOMES Article Open access 11 May 2021 INTRODUCTION In December 2019, a cluster of pneumonia cases of unknown aetiology was reported in the city of Wuhan in the
province of Hubei. The previously unidentified pathogen, which induces symptoms resembling an infection by the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV), was later identified
as a novel coronavirus, SARS-CoV-21. To date, there are more than four million laboratory-confirmed cases of human Coronavirus Disease 2019 (COVID-19), with over 280,000 deaths across 212
countries and territories (For up to date information consult https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/). After being declared a pandemic by World
Health Organization (WHO) on 11th March 2020, there is a compelling need to understand and develop effective therapeutic interventions against SARS-CoV-2. SARS-CoV-2 uses the spike (S)
glycoprotein to bind to the angiotensin-converting enzyme 2 (ACE2) receptor with a better affinity than SARS-CoV S glycoprotein for entry2. Thus, blocking the binding to ACE2, or blocking
host protease cleavage of S glycoprotein to release the fusion peptide is an efficient strategy to prevent coronavirus entry3,4,5. Several studies have assessed the immunogenicity of
structural domains of recombinant SARS-CoV-2 S glycoprotein6,7. At the time of writing, findings on SARS-CoV-2 linear epitopes remain mostly limited to bioinformatics prediction of human B-
and T-cell epitopes using SARS-CoV as a model8,9,10, and one recent pre-print described the use of a microarray of overlapping peptides to assess linear epitopes in 10 COVID-19 patients11.
Five regions on the S glycoprotein of SARS-CoV (residues 274–306, 510–586, 587–628, 784–803, and 870-893) were predicted to be associated with a robust immune response8, whereas other
studies reported candidate epitopes9,10 that require validation with patient samples. In this study, we report the antibody profiles of COVID-19 patients and the identification of two
immunodominant linear B-cell epitopes on the S glycoprotein of SARS-CoV-2. Interestingly, using S glycoprotein pseudotyped lentiviruses, we demonstrate that antibodies recognising these two
linear epitopes account for a high proportion of the anti-spike response. These epitopes can potentially be used in the design of more sensitive serological assays for epidemiological or
vaccine efficiency assessments. RESULTS A SPIKE PSEUDOTYPED LENTIVIRUS ASSAY FOR VIRUS NEUTRALISATION To investigate whether a biosafety level (BSL) 2 approved pseudotyped lentivirus
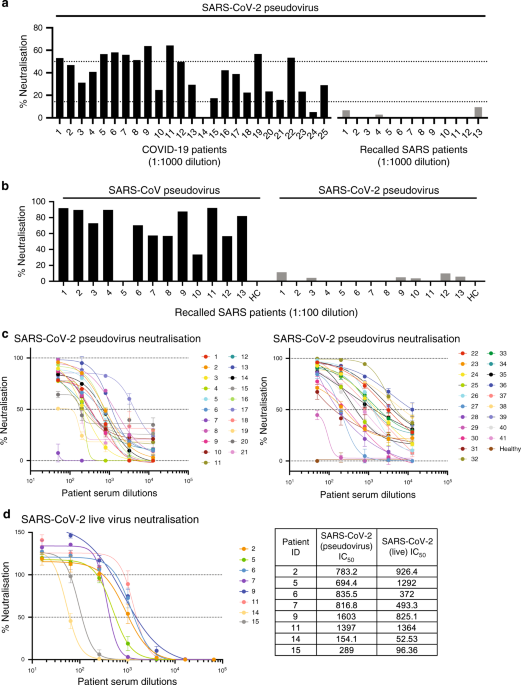
expressing SARS-CoV-2 S glycoprotein tagged with a luciferase reporter could detect neutralising antibodies, we performed an initial screen at 1:1000 dilution using sera from 25 convalescent
COVID-19 patients and from 13 SARS patients recalled in January–February 2020 as controls (Fig. 1a). Majority of the COVID-19 patients’ sera were able to neutralise >50% of SARS-CoV-2
pseudovirus entry, whereas recalled SARS patients did not show neutralisation. To validate the absence of neutralisation from the 13 recalled SARS patients, we assessed their neutralisation
capacity at a lower dilution of 1:100 against the pseudotyped lentivirus expressing SARS-CoV S or SARS-CoV-2 S glycoproteins (Fig. 1b). The results indicate that these recalled SARS patients
still possess antibodies specific to SARS-CoV albeit at low levels, making them an appropriate control group for subsequent linear B-cell epitope mapping. Next, this assay was used to
determine the IC50 values of anti-SARS-CoV-2 S-neutralising antibodies from the sera of 41 convalescent COVID-19 patients (Fig. 1c). To further validate that this safer pseudotyped
lentivirus assay is representative of live SARS-CoV-2 virus neutralisation, we performed antibody neutralisation titrations for eight patients under BSL3 conditions. IC50 values obtained
were comparable, validating the lentivirus assay (Fig. 1d). Six patients (2, 5, 6, 7, 8, 9) with sufficient amount of sera and good neutralising capacity were then selected for further
characterisation. Notably, sera from these patients showed similar IC50 values ranging from 694 to 836, except for patient 9, who showed the strongest neutralising activity with an IC50
value of 1603 (Fig. 1c, Supplementary Table 4). TWO SPECIFIC LINEAR EPITOPES ON THE SARS-COV-2 S PROTEIN We next assessed the linear antigenic targets from sera of the six selected COVID-19
patients and five recalled SARS patients using a linear B-cell peptide library spanning the entire S glycoprotein of either SARS-CoV-2 or SARS-CoV, in pools of five overlapping peptides
(Fig. 2a, Supplementary Fig. 1). Interestingly, two distinct peptide pools from SARS-CoV-2 S library, pools S14 and S21, were strongly detected by sera from COVID-19 patients (Fig. 2a) and
not by recalled SARS patients or healthy control serum (Supplementary Fig. 1a). Two COVID-19 patients could detect SARS-CoV S library pool S51, which partially overlaps with SARS-CoV-2 pool
S21 (Fig. 2a, Supplementary Fig. 1b). This region encompasses the fusion peptide, which is highly conserved among coronaviruses12,13, suggesting a potential pan-SARS epitope at this
location. Further assessment of individual peptides within pools S14 and S21 narrowed down the specific region of interest to peptides S14P5 and S21P2, respectively (Fig. 2b). Recognition of
S14P5 and S21P2 was stronger for the peptides of SARS-CoV-2 than on the corresponding SARS-CoV peptides (Fig. 2c). The use of these peptides as potential detection epitopes for serology
assessment was further validated with 41 COVID-19 patients and 28 healthy donors (collected before the pandemic). Detection for both S14P5 and S21P2 was consistently and significantly higher
in COVID-19 patients (Fig. 2d). More importantly, the level of antibodies targeting these two specific peptides determined by enzyme-linked immunosorbent assay (ELISA) significantly
correlated with sera neutralising IC50 values (Fig. 2e), suggesting that antibodies directed at these epitopes could neutralise SARS-CoV-2. ANTIBODIES AGAINST S14P5 AND S21P2 CAN NEUTRALISE
SARS-COV-2 Using a recently published structure of SARS-CoV-2 S glycoprotein in the prefusion conformation, S14P5 was shown to localise in close proximity to the receptor binding domain
(RBD) (Fig. 3a), whereas S21P2 covers part of the fusion peptide (Fig. 3b). To assess the importance of these regions in controlling SARS-CoV-2 infection, antibody depletion assays were
performed against S14P5 and S21P2 (Fig. 3c). Depletion efficiency and specificity were validated by ELISA, and results showed that only antibodies against the respective peptides were
depleted (Fig. 3d). Interestingly, sera depleted for antibodies targeting either peptides S14P5, S21P2, or S14P5 + S21P2 significantly reduced the ability to neutralise SARS-CoV-2
pseudovirus infection, as compared with the non-depleted sera controls (Fig. 3e). Taken together, these results demonstrated that antibodies targeting these two linear epitopes account for a
significant fraction of the anti-S-neutralising response. DISCUSSION In this study, we identified two immunodominant linear B-cell epitopes, S14P5 and S21P2, on the SARS-CoV-2 S
glycoprotein, and further assessed the functional capacity of COVID-19 patient sera antibodies against these regions using a pseudotyped lentivirus assay. This assay uses safer
third-generation lentiviruses, which will greatly benefit the scientific community in allowing rapid and safer assessments and characterisation of neutralising antibody titres in patient
blood, and potential monoclonal antibodies (mAbs). Depletion assays functionally validated the positive correlation between antibody levels against these epitopes and neutralisation titres
against SARS-CoV-2 pseudotyped lentiviruses. Future studies will be needed to fully understand the role and neutralisation capacity of antibodies targeting these regions. Peptide S14P5 is
localised in close proximity to the RBD. As such, it is plausible that antibodies binding to this region may sterically hinder binding to the ACE2 receptor, thereby abolishing virus
infection14. Another possibility could be an allosteric effect on ACE2 binding. Supporting our results, the partial sequence of peptide S14P5 was computationally predicted to be
immunogenic8,10. Peptide S21P2 partially overlaps with an epitope identified in a recent pre-print11 and contains a part of the fusion peptide sequence (Fig. 3b). As such, alterations to
this region may potentially affect virus fusion. Indeed, targeting the SARS-CoV and MERS-CoV fusion peptide region was demonstrated to neutralise coronavirus infection with a pan-coronavirus
fusion inhibitor peptide15. Although our findings showed a robust IgG response against the two identified linear epitopes, it is plausible that they represent a small proportion of the
total anti-S antibody response6,7. Nevertheless, antibody depletion assays against S14P5 or S21P2 led to >20% reduction in pseudotyped lentivirus neutralisation, validating that
antibodies targeting these linear S regions are important for neutralising SARS-CoV-2 infection. Surprisingly, depletion of antibodies directed against both S14P5 and S21P2 did not
significantly decrease the neutralisation as compared with single depletions, suggesting that neutralisation at these regions is not synergistic. Future studies involve the isolation of mAbs
targeting these linear epitopes to allow proper quantification and comparison of peptide-specific IgG titres with antibodies directed against the RBD domain, or to other conformational
epitopes. It would be interesting to also assess the level of persistence of these antibodies against linear and other conformational epitopes. Interestingly, IgG levels against each peptide
correlated positively with the patient neutralisation IC50 values, suggesting that quantitative serological assays against these peptides could be used as a proxy for virus exposure status
as well as protection levels. However, this will require validation with other patient cohorts. Notably, the two identified epitopes present a low-to-moderate rate to impact mutations, which
would minimise the possibility of false negatives in serological assays (Supplementary Table 5)16. Together, these results will be essential to guide the design and evaluation of efficient
and specific serological assays against linear epitopes, as well as help prioritise vaccine target designs during this unprecedented crisis. METHODS ETHICS STATEMENT Written informed consent
was obtained from participants in accordance with the tenets of the Declaration of Helsinki. For COVID-19 serum/plasma collection “A Multi-centred Prospective Study to Detect Novel
Pathogens and Characterize Emerging Infections (The PROTECT study group)”, a domain specific review board (DSRB) evaluated the study design and protocol, which was approved under study
number 2012/00917. Serum/plasma collection of recalled SARS patients “Comparison of host immune responses to coronavirus infections” was approved by DSRB under study number 2020/00091. Sera
from healthy volunteers “Study of blood cell subsets and their products in models of infection, inflammation and immune regulation” was approved under study number 2017/2806. PATIENT SERUM
AND PLASMA FRACTIONS Serum was collected in BD Vacutainer SST II Advance tubes (Fisher Scientific, #12927696). After clotting, serum was separated using centrifugation for 10 minutes at 1000
rcf, and aliquoted before storing at −80 °C. Patient serum was heat-inactivated for 30 minutes at 56 °C before usage for this study. Plasma fraction was harvested after 20 minutes
centrifugation at 1700 rcf of blood collected in BD Vacutainer CPT tubes (BD, #362753). Plasma samples were treated by solvent/detergent treatment with a final concentration of 1% Triton
X-100 (Thermo Fisher Scientific, #28314) for virus inactivation at RT for 2 hours17. Information on selected patients is provided in Supplementary Table 1. Patient demographics and clinical
characteristics are described in Supplementary Table 2. LINEAR PEPTIDE LIBRARY The sequences used for the design of biotinylated linear peptides of the S glycoprotein of SARS-CoV and
SARS-CoV-2 are under GenBank accession numbers NC_004718.3 and MN908947.3. Preliminary epitope screening was used with a library of peptides (Mimotopes, Mulgrave, VIC, Australia) consisting
of 18‐mer overlapping sequences. Peptides were used individually or as pooled sets. Five to eight peptides were combined to form one pooled peptide set. Lyophilised individual peptides were
dissolved in 200 μL of DMSO (Sigma‐Aldrich, #D8418-100ML) to obtain a stock solution. PEPTIDE-BASED ELISA B-cell linear library ELISA was performed in a similar manner to a previously
established peptide-based screen18. In brief, streptavidin‐coated plates (Thermo Fisher Scientific, #15125) were blocked with 0.1% PBST (0.1% v/v Tween‐20, Sigma-Aldrich, #P1379-500ML, in
PBS, Gibco, #20012-043) containing 1% w/v sodium caseinate (Sigma‐Aldrich, #C8654-500G, lot BCBP6469) and 1% w/v bovine serum albumin (BSA; Sigma‐Aldrich, #A7030-500G, lot SLBW5033)
overnight at 4 °C, before addition of pooled or single biotinylated peptides at 1:1000 dilution in 0.1% PBST. Heat‐inactivated patient serum samples were added at 1:1000 dilution in 0.1%
PBST. Horseradish peroxidase-conjugated goat anti-human IgG (H + L) antibody (Jackson ImmunoResearch, #109-035-088, lot 139159) prepared in 10% blocking buffer was used for detection of
peptide‐bound antibodies. In total, 100 μL of TMB substrate (Sigma‐Aldrich, #T8665, lot SLCB5343) was used for a 5 minute development and was stopped by addition of 100 μL of 0.16 M sulfuric
acid prepared from 95% to 97% Sulfuric Acid stock solution (Merck, #1.00731.1000), prior to absorbance measurements. Absorbance was measured with the following parameters: 450 nm minus 690
nm (bandwidth of 9 nm) in five flashes after a 10 second shaking at 1 mm amplitude on an Infinite M200 plate reader (Tecan, firmware V_2.02_11/06). PEPTIDES S14P5 AND S21P2 ELISA FOR 41
COVID-19 PATIENTS Owing to the limitation of available serum samples, ELISA was performed with 1% Triton X-100 (Thermo Fisher Scientific, #28314) treated plasma fractions. ELISA was
performed in similar conditions as described above with the following modifications. Nunc Maxisorp flat-bottom 96-well plates (Thermo Fisher Scientific, #442404) were coated overnight with
50 μL per well of 0.5 μg/mL of NeutrAvidin protein (Thermo Fisher Scientific, #31050). Blocking was performed for 1 hour with 0.01% polyvinyl alcohol (PVA; Sigma-Aldrich, #341584) in 0.1%
PBST (blocking buffer) prepared from stock of 0.5% PVA w/v in distilled H2O. Peptide coating was performed at 1:2000 dilution for 1 hour. Secondary antibody was incubated for 1 hour in
blocking buffer at 1:1000 dilution. Development was performed with 50 μL of TMB and stopped with 50 μL of 0.16 M sulfuric acid. PEPTIDE AFFINITY DEPLETION OF POOLED SERA Using principles
similar to previous work19,20, we performed affinity depletion as follows. Selected synthetic biotinylated peptides were added at 1:1000 dilution in 0.1% PBST to pre-blocked
streptavidin-coated plates and incubated at room temperature for 1 hour. Plates were washed three times with 0.1% PBST followed by PBS wash to remove traces of Tween-20. Pooled patient sera
were prepared at a dilution of 1:100 in Dulbecco’s Modified Eagle’s Medium (DMEM; HyClone, #SH30243.01, lot AE29431634), and 50 μL was added to each well and incubated for 20 minutes at room
temperature for adsorption. The unbound fraction was collected after 24 rounds of adsorption. ELISA analysis was performed as described above but at 1:2000 dilution to assess the levels of
peptide-specific antibodies before and after affinity depletion. Adsorbed samples were then mixed with lentiviruses pseudotyped with SARS-CoV-2 S protein as described below. Selected peptide
sequences are given in Supplementary Table 3. CELL LINES AND CELL CULTURE The human embryonic kidney epithelial cell 293T (ATCC, CRL-3216) and VERO E6 C1008 (ATCC CRL-1586 were cultured in
DMEM (Hyclone, #SH30022.01) supplemented with 10% heat-inactivated foetal bovine serum (FBS; Gibco, #10270-106). A stable cell line expressing human ACE2, CHO-ACE2 (a kind gift from
Professor Yee-Joo Tan, Department of Microbiology, NUS & IMCB, A*STAR, Singapore)21 was maintained in DMEM supplemented with 10% heat-inactivated FBS, 1% MEM non-essential amino acids
solution (Gibco, #11140-050) and 0.5 mg/mL of Geneticin Selective Antibiotic (Gibco, #10131-027). Every 2–3 days, cells were passaged by dissociating the cells with StemPro Accutase Cell
Dissociation Reagent (Gibco, #A1110501). ACE2 surface expression on CHO-ACE2 cells was verified using anti-human ACE2 AF647 (Santa Cruz Biotech, #sc-390851, lot B0320). Cells were routinely
tested for mycoplasma contamination. SARS-COV-2 AND SARS-COV PSEUDOTYPED LENTIVIRUS PRODUCTION Based on the third-generation lentivirus system, pseudotyped viral particles expressing
SARS-CoV or SARS-CoV-2 S proteins were produced by reverse transfection of 30 × 106 of 293 T cells with 12 µg pMDLg/pRRE (Addgene, #12251), 6 µg pRSV-Rev (Addgene, #12253), 12 µg
pTT5LnX-coV-SP (SARS-CoV-2 spike) or pXJ3’-S (SARS-CoV spike, a kind gift from Professor Yee-Joo Tan, Department of Microbiology, NUS & IMCB, A*STAR, Singapore)22 and 24 µg
pHIV-Luc-ZsGreen (Addgene, #39196) using Lipofectamine 2000 transfection reagent (Invitrogen, #11668-019) and cultured in a 37 °C incubator for 3 days. Viral supernatant was harvested, spun
down by centrifugation to remove cell debris and filtered through a 0.45 µm filter unit (Sartorius, #16555). Lenti-X p24 rapid titre kit (Takara Bio, #632200) was used to quantify the viral
titres following the manufacturer’s instructions. PSEUDOTYPED LENTIVIRUS NEUTRALISATION ASSAY CHO-ACE2 cells were seeded at a density of 2.5 × 104 cells in 100 µL of complete medium without
Geneticin in 96-well Flat Clear Bottom Black Polystyrene TC-treated Microplates (Corning, #3904). After heat-inactivation at 56 °C for 30 minutes, serially diluted patient sera were
incubated in a 96-well flat-bottom cell culture plate (Costar, #3596) with an equal volume of pseudotyped virus (12 ng of p24) at the final volume of 50 μL at 37 °C for 1 h, and the mixture
was added to the monolayer of pre-seeded CHO-ACE2 cells. After 1 hour of pseudotyped viral infection at 37 °C, 150 µL of culture medium was added to each well and the cells were further
incubated for another 48 h. Upon removal of culture medium, cells were washed twice with sterile PBS, and then lysed in 20 µL of 1 × Passive lysis buffer (Promega, #E1941) with gentle
shaking at 400 rpm at 37 °C for 30 minutes. Luciferase activity was then assessed using Luciferase Assay System (Promega, #E1510) on a Promega GloMax Luminometer. LIVE SARS-COV-2
NEUTRALISATION ASSAY IN BSL3 Using a 96-well opaque (white) plate, 25 µL of 100 TCID50 of SARS-CoV-2 isolated from a naso-pharengeal swab of a patient in Singapore23, was incubated with 25
µL of the indicated sera dilution for 1 hour at 37 °C with 5% CO2. After incubation, 50 µL of 4 × 105 cells/ml (VERO E6 C1008) was added into each well. The plate was subsequently incubated
for 4 days at 37 °C with 5% CO2. Cell viability was then determined using Viral ToxGlo Assay (Promega, #G8941). In brief, 100 µL of the reagent was added into each well and incubated for 10
minutes at room temperature prior to measurement of luminescence readout using microplate reader (Tecan). DATA VISUALISATION AND STATISTICAL ANALYSIS Structural data of SARS-CoV-2 S protein
was retrieved from Protein Databank (PDB ID: 6VSB) in homotrimeric prefusion conformation and visualised using PyMOL (Schrodinger, version 2.2.0). Data were analysed using Excel for Mac
16.16.8 and GraphPad Prism for macOS version 8.4.1. Statistical tests are indicated in the figure legends. IC50 values of individual patients were calculated using the [Inhibitor] vs
response variable slope four parameter of GraphPad Prism, with negative values forced to zero. Correlation between pseudovirus IC50 and OD values were analysed using the non-parametric
Spearman correlation and straight line non-linear regression robust fit functions of Prism with data from Supplementary Table 4. For the neutralisation assay with non-depleted and depleted
pooled serum, one-sample _t_ test for each experiment was perform to assess if the values were significantly different from 100. REPORTING SUMMARY Further information on research design is
available in the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY The source data underlying Figs. 1–3 and Supplementary Fig. 1 are provided as a Source Data file.
Other data can be obtained upon reasonable request to the corresponding author. Source data are provided with this paper. REFERENCES * Cohen, J. & Normile, D. New SARS-like virus in
China triggers alarm. _Science_ 367, 234–235 (2020). ADS CAS PubMed Google Scholar * Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. _Science_
367, 1260–1263 (2020). ADS CAS PubMed PubMed Central Google Scholar * Walls, A. C. et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. _Cell_,
https://doi.org/10.1016/j.cell.2020.02.058 (2020). PubMed PubMed Central Google Scholar * Letko, M., Marzi, A. & Munster, V. Functional assessment of cell entry and receptor usage for
SARS-CoV-2 and other lineage B betacoronaviruses. _Nat. Microbiol._ https://doi.org/10.1038/s41564-020-0688-y (2020). CAS PubMed Google Scholar * Hoffmann, M. et al. SARS-CoV-2 Cell
entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. _Cell_, https://doi.org/10.1016/j.cell.2020.02.052 (2020). PubMed PubMed Central Google Scholar
* Lv, H. et al. Cross-reactive antibody response between SARS-CoV-2 and SARS-CoV infections. _Cell Rep_. In press. https://doi.org/10.1016/j.celrep.2020.107725 (2020). * Wu, F. et al.
Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. Available at SSRN: https://doi.org/10.2139/ssrn.3566211 (2020). * Grifoni, A. et
al. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. _Cell Host Microbe_, https://doi.org/10.1016/j.chom.2020.03.002 (2020).
Google Scholar * Zheng, M. & Song, L. Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. _Cell Mol. Immunol_,
https://doi.org/10.1038/s41423-020-0385-z (2020). CAS Google Scholar * Ahmed, S. F., Quadeer, A. A. & McKay, M. R. Preliminary identification of potential vaccine targets for the
COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. _Viruses_ 12, https://doi.org/10.3390/v12030254 (2020). CAS PubMed Central Google Scholar * Wang, H. et al.
SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. Preprint at: https://doi.org/10.1101/2020.03.26.994756 (2020). * Madu, I. G., Roth, S. L.,
Belouzard, S. & Whittaker, G. R. Characterisation of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a
viral fusion peptide. _J. Virol._ 83, 7411–7421 (2009). CAS PubMed PubMed Central Google Scholar * Alsaadi, E. A. J., Neuman, B. W. & Jones, I. M. A. Fusion peptide in the spike
protein of MERS coronavirus. _Viruses_ 11, 825 (2019). CAS PubMed Central Google Scholar * Tian, X. et al. Potent binding of 2019 novel coronavirus spike protein by a SARS
coronavirus-specific human monoclonal antibody. _Emerg. Microbes Infect._ 9, 382–385 (2020). CAS PubMed PubMed Central Google Scholar * Xia, S. et al. A pan-coronavirus fusion inhibitor
targeting the HR1 domain of human coronavirus spike. _Sci. Adv._ 5, eaav4580 (2019). ADS CAS PubMed PubMed Central Google Scholar * Zhao, W. M. et al. The 2019 novel coronavirus
resource. _Yi Chuan_ 42, 212–221 (2020). PubMed Google Scholar * Darnell, M. E. & Taylor, D. R. Evaluation of inactivation methods for severe acute respiratory syndrome coronavirus in
noncellular blood products. _Transfusion_ 46, 1770–1777 (2006). PubMed PubMed Central Google Scholar * Amrun, S. N. et al. Novel differential linear B-cell epitopes to identify Zika and
dengue virus infections in patients. _Clin. Transl. Immunol._ 8, e1066 (2019). Google Scholar * Kam, Y. W. et al. Early neutralizing IgG response to Chikungunya virus in infected patients
targets a dominant linear epitope on the E2 glycoprotein. _EMBO Mol. Med._ 4, 330–343 (2012). CAS PubMed PubMed Central Google Scholar * Lee, C. Y.-P. et al. Type I interferon shapes the
quantity and quality of the anti-Zika virus antibody response. _Clin. Transl. Immunol._ 9, e1126 (2020). Google Scholar * Lip, K. M. et al. Monoclonal antibodies targeting the HR2 domain
and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. _J. Virol._ 80, 941–950 (2006). CAS PubMed
PubMed Central Google Scholar * Ng, O. W. et al. Substitution at aspartic acid 1128 in the SARS coronavirus spike glycoprotein mediates escape from a S2 domain-targeting neutralizing
monoclonal antibody. _PLoS ONE_ 9, e102415 (2014). ADS PubMed PubMed Central Google Scholar * Young, B. E., et al. Epidemiologic Features and Clinical Course of Patients Infected With
SARS-CoV-2 in Singapore. _JAMA_. https://doi.org/10.1001/jama.2020.3204 (2020). CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank Professor Yee-Joo
Tan (Department of Microbiology, NUS; Institute of Molecular and Cell Biology—IMCB, A*STAR) who kindly provided CHO-ACE2 cells and pXJ3’-S plasmid. We acknowledge the excellent technical
work in the BSL3 facility of Wong Pui San (Wong P.S.) and Chye De Ho (Chye D.H.). We also like to thank the study participants who donated their blood samples to this project, and the
healthcare workers caring for COVID-19 patients. This work was supported by core research grants provided to Singapore Immunology Network by the Biomedical Research Council (BMRC), and by
the A*ccelerate GAP-funded project (ACCL/19-GAP064-R20H-H) from the Agency of Science, Technology and Research (A*STAR). Subject recruitment and sample collection were funded by the National
Medical Research Council (NMRC) COVID-19 Research fund (COVID19RF-001). AUTHOR INFORMATION Author notes * These authors contributed equally: Chek Meng Poh, Guillaume Carissimo, Bei Wang.
AUTHORS AND AFFILIATIONS * Singapore Immunology Network, Agency of Science, Technology and Research, Immunos, Biopolis, Singapore, 138648, Singapore Chek Meng Poh, Guillaume Carissimo, Bei
Wang, Siti Naqiah Amrun, Cheryl Yi-Pin Lee, Rhonda Sin-Ling Chee, Siew-Wai Fong, Nicholas Kim-Wah Yeo, Wen-Hsin Lee, Anthony Torres-Ruesta, Cheng-I Wang, Laurent Renia & Lisa F. P. Ng *
Department of Biological Science, National University of Singapore, Singapore, Singapore Siew-Wai Fong * Department of Biochemistry, Yong Loo Lin School of Medicine, National University of
Singapore, 8 Medical Drive, Singapore, 117596, Singapore Anthony Torres-Ruesta & Lisa F. P. Ng * National Centre for Infectious Diseases, 16 Jalan Tan Tock Seng, Singapore, 308442,
Singapore Yee-Sin Leo, Mark I-Cheng Chen, Barnaby Edward Young & David C. Lye * Department of Infectious Diseases, Tan Tock Seng Hospital, 11 Jalan Tan Tock Seng, Singapore, 308433,
Singapore Yee-Sin Leo, Louis Yi Ann Chai, Barnaby Edward Young & David C. Lye * Lee Kong Chian School of Medicine, Nanyang Technological University, 11 Mandalay Road, Singapore, 308232,
Singapore Yee-Sin Leo, Barnaby Edward Young & David C. Lye * Yong Loo Lin School of Medicine, National University of Singapore and National University Health System, 10 Medical Drive,
Singapore, 117597, Singapore Yee-Sin Leo & David C. Lye * Saw Swee Hock School of Public Health, National University of Singapore and National University Health System, 12 Science Drive
2, #10-01, Singapore, 117549, Singapore Mark I-Cheng Chen * Department of Infectious Diseases, Changi General Hospital, 2 Simei Street 3, Singapore, 529889, Singapore Seow-Yen Tan *
Department of Medicine, National University Hospital, 5 Lower Kent Ridge Road, Singapore, 119074, Singapore Louis Yi Ann Chai * Department of Infectious Diseases, Singapore General Hospital,
31 Third Hospital Ave, #03-03 Bowyer Block C, Singapore, 168753, Singapore Shirin Kalimuddin & Siew-Yee Thien * Emerging Infectious Disease Program, Duke-NUS Medical School, 8 College
Road, Singapore, 169857, Singapore Shirin Kalimuddin * Biological Defence Program, DSO National Laboratories, 27 Medical Drive, Singapore, 117510, Singapore Shirley Seah Gek Kheng &
Brendon John Hanson * Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, 8 West Derby Street, Liverpool, L7 3EA, United Kingdom Lisa F. P. Ng
Authors * Chek Meng Poh View author publications You can also search for this author inPubMed Google Scholar * Guillaume Carissimo View author publications You can also search for this
author inPubMed Google Scholar * Bei Wang View author publications You can also search for this author inPubMed Google Scholar * Siti Naqiah Amrun View author publications You can also
search for this author inPubMed Google Scholar * Cheryl Yi-Pin Lee View author publications You can also search for this author inPubMed Google Scholar * Rhonda Sin-Ling Chee View author
publications You can also search for this author inPubMed Google Scholar * Siew-Wai Fong View author publications You can also search for this author inPubMed Google Scholar * Nicholas
Kim-Wah Yeo View author publications You can also search for this author inPubMed Google Scholar * Wen-Hsin Lee View author publications You can also search for this author inPubMed Google
Scholar * Anthony Torres-Ruesta View author publications You can also search for this author inPubMed Google Scholar * Yee-Sin Leo View author publications You can also search for this
author inPubMed Google Scholar * Mark I-Cheng Chen View author publications You can also search for this author inPubMed Google Scholar * Seow-Yen Tan View author publications You can also
search for this author inPubMed Google Scholar * Louis Yi Ann Chai View author publications You can also search for this author inPubMed Google Scholar * Shirin Kalimuddin View author
publications You can also search for this author inPubMed Google Scholar * Shirley Seah Gek Kheng View author publications You can also search for this author inPubMed Google Scholar *
Siew-Yee Thien View author publications You can also search for this author inPubMed Google Scholar * Barnaby Edward Young View author publications You can also search for this author
inPubMed Google Scholar * David C. Lye View author publications You can also search for this author inPubMed Google Scholar * Brendon John Hanson View author publications You can also search
for this author inPubMed Google Scholar * Cheng-I Wang View author publications You can also search for this author inPubMed Google Scholar * Laurent Renia View author publications You can
also search for this author inPubMed Google Scholar * Lisa F. P. Ng View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS C.M.P., G.C., S.N.A.,
C.Y.P.L. conceptualised, designed, acquired, analysed, interpreted the data and wrote the manuscript. B.W. acquired, analysed, interpreted the data, and wrote the manuscript. R.S.L.C.,
S.W.F., N.K.W.Y., S.W.H.L., S.S.G.K., and A.T.R. acquired and analysed the data. Y.S.L., M.I.C.C., S.Y. Thien, L.Y.A.C., S.K., S.Y. Tan, B.E.Y., and D.C.L. designed and supervised sample
collection. B.J.H., C.I.W., L.R., L.F.P.N. conceptualised, designed, analysed, and wrote the manuscript. All authors revised and approved the final version of the manuscript. CORRESPONDING
AUTHOR Correspondence to Lisa F. P. Ng. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature
Communications_ thanks the anonymous reviewers for their contribution to the peer review of this work. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims
in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE REPORTING SUMMARY SOURCE DATA SOURCE DATA RIGHTS AND PERMISSIONS OPEN
ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format,
as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third
party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the
article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright
holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Poh, C.M., Carissimo, G., Wang, B.
_et al._ Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. _Nat Commun_ 11, 2806 (2020).
https://doi.org/10.1038/s41467-020-16638-2 Download citation * Received: 04 May 2020 * Accepted: 17 May 2020 * Published: 01 June 2020 * DOI: https://doi.org/10.1038/s41467-020-16638-2 SHARE
THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative