- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
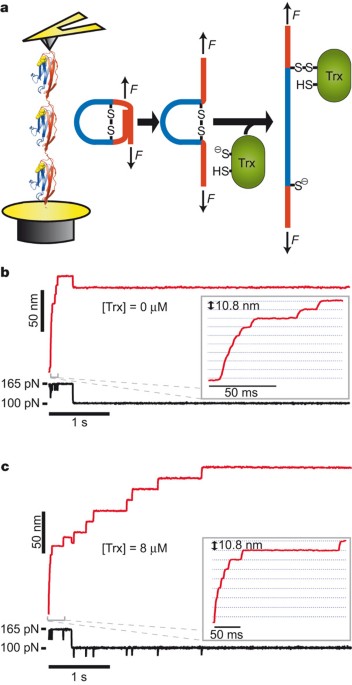
ABSTRACT Thioredoxins are enzymes that catalyse disulphide bond reduction in all living organisms1. Although catalysis is thought to proceed through a substitution nucleophilic bimolecular
(SN2) reaction1,2, the role of the enzyme in modulating this chemical reaction is unknown. Here, using single-molecule force-clamp spectroscopy3,4, we investigate the catalytic mechanism of
_Escherichia coli_ thioredoxin (Trx). We applied mechanical force in the range of 25–600 pN to a disulphide bond substrate and monitored the reduction of these bonds by individual enzymes.
We detected two alternative forms of the catalytic reaction, the first requiring a reorientation of the substrate disulphide bond, causing a shortening of the substrate polypeptide by 0.79 ±
0.09 Å (± s.e.m.), and the second elongating the substrate disulphide bond by 0.17 ± 0.02 Å (± s.e.m.). These results support the view that the Trx active site regulates the geometry of the
participating sulphur atoms with sub-ångström precision to achieve efficient catalysis. Our results indicate that substrate conformational changes may be important in the regulation of Trx
activity under conditions of oxidative stress and mechanical injury, such as those experienced in cardiovascular disease5,6. Furthermore, single-molecule atomic force microscopy techniques,
as shown here, can probe dynamic rearrangements within an enzyme’s active site during catalysis that cannot be resolved with any other current structural biological technique. Access through
your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 51
print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject
to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT
BEING VIEWED BY OTHERS SINGLE-MOLECULE MAGNETIC TWEEZERS TO PROBE THE EQUILIBRIUM DYNAMICS OF INDIVIDUAL PROTEINS AT PHYSIOLOGICALLY RELEVANT FORCES AND TIMESCALES Article 11 March 2024 THE
ROLE OF SINGLE-PROTEIN ELASTICITY IN MECHANOBIOLOGY Article 24 October 2022 FABRICATION OF ELECTRON TUNNELING PROBES FOR MEASURING SINGLE-PROTEIN CONDUCTANCE Article 07 July 2023 REFERENCES
* Holmgren, A. Thioredoxin. _Annu. Rev. Biochem._ 54, 237–271 (1985) Article CAS Google Scholar * Holmgren, A. Thioredoxin structure and mechanism: conformational changes on oxidation of
the active-site sulfhydryls to a disulfide. _Structure_ 3, 239–243 (1995) Article CAS Google Scholar * Schlierf, M., Li, H. & Fernandez, J. M. The unfolding kinetics of ubiquitin
captured with single-molecule force-clamp techniques. _Proc. Natl Acad. Sci. USA_ 101, 7299–7304 (2004) Article ADS CAS Google Scholar * Wiita, A. P., Ainavarapu, S. R. K., Huang, H. H.
& Fernandez, J. M. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. _Proc. Natl Acad. Sci. USA_ 103, 7222–7227 (2006) Article ADS
CAS Google Scholar * Paravicini, T. M. & Touyz, R. M. Redox signaling in hypertension. _Cardiovasc. Res._ 71, 247–258 (2006) Article CAS Google Scholar * World, C. J., Yamawaki,
H. & Berk, B. C. Thioredoxin in the cardiovascular system. _J. Mol. Med._ 84, 997–1003 (2006) Article CAS Google Scholar * Kraut, D. A., Carroll, K. S. & Herschlag, D. Challenges
in enzyme mechanism and energetics. _Annu. Rev. Biochem._ 72, 517–571 (2003) Article CAS Google Scholar * Hammes-Schiffer, S. & Benkovic, S. J. Relating protein motion to catalysis.
_Annu. Rev. Biochem._ 75, 519–541 (2006) Article CAS Google Scholar * Carrion-Vazquez, M. et al. Mechanical and chemical unfolding of a single protein: a comparison. _Proc. Natl Acad.
Sci. USA_ 96, 3694–3699 (1999) Article ADS CAS Google Scholar * Grandbois, M., Beyer, M., Rief, M., Clausen-Schaumann, H. & Gaub, H. E. How strong is a covalent bond? _Science_ 283,
1727–1730 (1999) Article ADS CAS Google Scholar * Ainavarapu, S. R. et al. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. _Biophys. J._
92, 225–233 (2007) Article ADS CAS Google Scholar * Abbondanzieri, E. A., Greenleaf, W. J., Shaevitz, J. W., Landick, R. & Block, S. M. Direct observation of base-pair stepping by
RNA polymerase. _Nature_ 438, 460–465 (2005) Article ADS CAS Google Scholar * Holmgren, A. Reduction of disulfides by thioredoxin. Exceptional reactivity of insulin and suggested
functions of thioredoxin in mechanism of hormone action. _J. Biol. Chem._ 254, 9113–9119 (1979) CAS PubMed Google Scholar * Krause, G., Lundstrom, J., Barea, J. L., Pueyo de la Cuesta, C.
& Holmgren, A. Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in _Escherichia coli_ thioredoxin. _J. Biol. Chem._ 266, 9494–9500 (1991) CAS
PubMed Google Scholar * Bell, G. I. Models for the specific adhesion of cells to cells. _Science_ 200, 618–627 (1978) Article ADS CAS Google Scholar * Qin, J., Clore, G. M. &
Gronenborn, A. M. The high-resolution three-dimensional solution structures of the oxidized and reduced states of human thioredoxin. _Structure_ 2, 503–522 (1994) Article CAS Google
Scholar * Eklund, H., Gleason, F. K. & Holmgren, A. Structural and functional relations among thioredoxins of different species. _Proteins_ 11, 13–28 (1991) Article CAS Google Scholar
* Qin, J., Clore, G. M., Kennedy, W. P., Huth, J. R. & Gronenborn, A. M. Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from
the transcription factor NFκB. _Structure_ 3, 289–297 (1995) Article CAS Google Scholar * Qin, J., Clore, G. M., Kennedy, W. P., Kuszewski, J. & Gronenborn, A. M. The solution
structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. _Structure_ 4, 613–620 (1996) Article CAS Google Scholar * Rosenfield, R. E.,
Parthasarathy, R. & Dunitz, J. D. Directional preferences of nonbonded atomic contacts with divalent sulfur. 1. Electrophiles and nucleophiles. _J. Am. Chem. Soc._ 99, 4860–4862 (1977)
Article CAS Google Scholar * Pappas, J. A. Theoretical studies of reactions of sulfur–sulfur bond. 1. General heterolytic mechanisms. _J. Am. Chem. Soc._ 99, 2926–2930 (1977) Article CAS
Google Scholar * Fernandes, P. A. & Ramos, M. J. Theoretical insights into the mechanism for thiol/disulfide exchange. _Chem. Eur. J._ 10, 257–266 (2004) Article CAS Google Scholar
* Foloppe, N. & Nilsson, L. The glutaredoxin -C-P-Y-C- motif: influence of peripheral residues. _Structure_ 12, 289–300 (2004) CAS PubMed Google Scholar * Grosberg, A. Y. &
Khokhlov, A. R. _Statistical Physics of Macromolecules_ (AIP, New York, 1994) Google Scholar * Tao, L. et al. Cardioprotective effects of thioredoxin in myocardial ischemia and reperfusion:
role of S-nitrosation. _Proc. Natl Acad. Sci. USA_ 101, 11471–11476 (2004) Article ADS Google Scholar * Perez-Jimenez, R., Godoy-Ruiz, R., Ibarra-Molero, B. & Sanchez-Ruiz, J. M. The
effect of charge-introduction mutations on _E. _ _coli_ thioredoxin stability. _Biophys. Chem._ 115, 105–107 (2005) Article CAS Google Scholar * Holmgren, A. & Reichard, P.
Thioredoxin 2: cleavage with cyanogen bromide. _Eur. J. Biochem._ 2, 187–196 (1967) Article CAS Google Scholar * Holmgren, A. & Slaby, I. Thioredoxin-C': mechanism of noncovalent
complementation and reactions of the refolded complex and the active site containing fragment with thioredoxin reductase. _Biochemistry_ 18, 5591–5599 (1979) Article CAS Google Scholar *
Ren, X., Bjornstedt, M., Shen, B., Ericson, M. L. & Holmgren, A. Mutagenesis of structural half-cystine residues in human thioredoxin and effects on the regulation of activity by
selenodiglutathione. _Biochemistry_ 32, 9701–9708 (1993) Article CAS Google Scholar * Fernandez, J. M. & Li, H. Force-clamp spectroscopy monitors the folding trajectory of a single
protein. _Science_ 303, 1674–1678 (2004) Article ADS CAS Google Scholar * Oberhauser, A. F., Hansma, P. K., Carrion-Vazquez, M. & Fernandez, J. M. Stepwise unfolding of titin under
force-clamp atomic force microscopy. _Proc. Natl Acad. Sci. USA_ 98, 468–472 (2001) Article ADS CAS Google Scholar * Sigworth, F. J. & Neher, E. Single Na+ channel currents observed
in cultured rat muscle cells. _Nature_ 287, 447–449 (1980) Article ADS CAS Google Scholar * Aldrich, R. W., Corey, D. P. & Stevens, C. F. A reinterpretation of mammalian sodium
channel gating based on single channel recording. _Nature_ 306, 436–441 (1983) Article ADS CAS Google Scholar * Efron, B. _The Jackknife, the Bootstrap, and Other Resampling Plans_
(SIAM, Philadelphia, 1982) Book Google Scholar * Taylor, J. R. _An Introduction to Error Analysis_ 2nd edn 261–293 (Univ. Science Books, Sausalito, 1997) Google Scholar * Press, W. H.,
Teukolsky, S. A., Vetterling, W. T. & Flannery, B. P. _Numerical Recipes in C_ (Cambridge Univ. Press, Cambridge, UK, 1992) MATH Google Scholar * Lindahl, E., Hess, B. & van der
Spoel, D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. _J. Mol. Model._ 7, 306–317 (2001) Article CAS Google Scholar * Jorgensen, W. L. & Swenson, C. J.
Optimized intermolecular potential functions for amides and peptides. Structure and properties of liquid amides. _J. Am. Chem. Soc._ 107, 569–578 (1985) Article CAS Google Scholar * Hess,
B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: a linear constraint solver for molecular simulations. _J. Comput. Chem._ 18, 1463–1472 (1997) Article CAS Google
Scholar * Miyamoto, S. & Kollman, P. A. Settle—an analytical version of the shake and rattle algorithm for rigid water models. _J. Comput. Chem._ 13, 952–962 (1992) Article CAS Google
Scholar * Darden, T., York, D. & Pedersen, L. Particle mesh Ewald—an _N_.log(_N_) method for Ewald sums in large systems. _J. Chem. Phys._ 98, 10089–10092 (1993) Article ADS CAS
Google Scholar * Berendsen, H. J. C., Postma, J. P. M., Vangunsteren, W. F., Dinola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. _J. Chem. Phys._ 81,
3684–3690 (1984) Article ADS CAS Google Scholar * Delano, W. L. _Pymol Manual_ 〈http://www.delanoscientific.com〉 (2001) Google Scholar * Improta, S., Politou, A. S. & Pastore, A.
Immunoglobulin-like modules from titin I-band: extensible components of muscle elasticity. _Structure_ 4, 323–337 (1996) Article CAS Google Scholar * Vriend, G. WHAT IF: a molecular
modeling and drug design program. _J. Mol. Graph._ 8, 52–58 (1990) Article CAS Google Scholar * Grubmüller, H., Heymann, B. & Tavan, P. Ligand binding: molecular mechanics calculation
of the streptavidin biotin rupture force. _Science_ 271, 997–999 (1996) Article ADS Google Scholar Download references ACKNOWLEDGEMENTS We thank D. Rodriguez-Larrea for thioredoxin
purification and S. Posy for assistance with structural modelling. This work was supported by NIH grants to J.M.F., an NIH grant to B.J.B., a grant from the Swedish Society for Medical
Research to A.H., and a grant from the Spanish Ministry of Science and Education to J.M.S.-R. F.G. is supported by an ISE Columbia University grant to J.M.F. and B.J.B. A.P.W. is supported
by an NIH Medical Scientist Training Program grant to Columbia University. AUTHOR CONTRIBUTIONS A.P.W., R.P.-J. and J.M.F. designed the experiments. A.P.W. and R.P.-J. performed the
experiments and analysed the data. K.A.W. designed the kinetic model and performed error analysis. F.G. and B.J.B. performed molecular dynamics simulations. A.H. provided TRX. J.M.S.-R.
provided Trx and Trx(P34H). A.P.W., F.G., K.A.W., R.P.-J. and J.M.F. wrote the paper. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Biological Sciences,, Arun P. Wiita, Raul
Perez-Jimenez, Kirstin A. Walther & Julio M. Fernandez * Graduate Program in Neurobiology and Behavior,, Arun P. Wiita * Department of Physics,, Kirstin A. Walther * Department of
Chemistry, Columbia University, New York, New York 10027, USA, Frauke Gräter & B. J. Berne * Department of Medical Biochemistry and Biophysics, Medical Nobel Institute for Biochemistry,
Karolinska Institutet, SE-171 77, Stockholm, Sweden, Arne Holmgren * Departamento de Quimica Fisica, Facultad de Ciencias, Universidad de Granada, 18071, Granada, Spain, Jose M. Sanchez-Ruiz
Authors * Arun P. Wiita View author publications You can also search for this author inPubMed Google Scholar * Raul Perez-Jimenez View author publications You can also search for this
author inPubMed Google Scholar * Kirstin A. Walther View author publications You can also search for this author inPubMed Google Scholar * Frauke Gräter View author publications You can also
search for this author inPubMed Google Scholar * B. J. Berne View author publications You can also search for this author inPubMed Google Scholar * Arne Holmgren View author publications
You can also search for this author inPubMed Google Scholar * Jose M. Sanchez-Ruiz View author publications You can also search for this author inPubMed Google Scholar * Julio M. Fernandez
View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Julio M. Fernandez. ETHICS DECLARATIONS COMPETING INTERESTS The
authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION The file contains Supplementary Results, Supplementary Tables 1-4 and Supplementary
Figures 1-10. This file includes the description of our theoretical model for estimating Δx12 in the Supplementary Results as well as all of the Supplementary Tables, Figures, and associated
Legends referenced in the main text. (PDF 1441 kb) SUPPLEMENTARY SETUP PARAMETERS 1 This file contains the setup parameters for the molecular dynamics (MD) simulations of the Trx:NF-kB
complex. (TXT 3393 kb) SUPPLEMENTARY SETUP PARAMETERS 2 This file contains the setup parameters for the molecular dynamics (MD) simulations of the unbound (apo) Trx enzyme. (TXT 2231 kb)
SUPPLEMENTARY SETUP PARAMETERS 3 This file contains the setup parameters for the molecular dynamics (MD) simulations of the Trx:Ref-1 complex (PDB: 1CQH). (TXT 2574 kb) SUPPLEMENTARY MD
SIMULATIONS The file contains overall parameters for the MD simulations (ZIP 2 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Wiita, A.,
Perez-Jimenez, R., Walther, K. _et al._ Probing the chemistry of thioredoxin catalysis with force. _Nature_ 450, 124–127 (2007). https://doi.org/10.1038/nature06231 Download citation *
Received: 21 May 2007 * Accepted: 07 September 2007 * Issue Date: 01 November 2007 * DOI: https://doi.org/10.1038/nature06231 SHARE THIS ARTICLE Anyone you share the following link with will
be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt
content-sharing initiative