- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Missense variants throughout _ACTA2_, encoding α-smooth muscle actin (αSMA), predispose to adult-onset thoracic aortic disease, but variants disrupting arginine 179 (R179) lead to
smooth muscle dysfunction syndrome characterized by diverse childhood-onset vascular diseases. Here we show that αSMA localizes to the nucleus in wild-type smooth muscle cells (SMCs),
enriches in the nucleus with SMC differentiation, and associates with chromatin remodeling complexes and SMC contractile gene promoters. The _ACTA2_ p.Arg179 αSMA variant shows decreased
nuclear localization. Primary SMCs from _Acta2_SMC-R179C/+ mice are less differentiated than wild-type SMCs in vitro and in vivo and have global changes in chromatin accessibility. Induced
pluripotent stem cells from participants with _ACTA2_ p.Arg179 variants fail to fully differentiate from neuroectodermal progenitor cells to SMCs, and single-cell transcriptomic analyses of
an _ACTA2_ p.Arg179His participant’s aortic tissue show increased SMC plasticity. Thus, nuclear αSMA participates in SMC differentiation, and loss of this nuclear activity occurs with
_ACTA2_ p.Arg179 pathogenic variants. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through
your institution Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS INDUCIBLE PRMT1 ABLATION IN ADULT VASCULAR SMOOTH MUSCLE LEADS TO CONTRACTILE
DYSFUNCTION AND AORTIC DISSECTION Article Open access 11 October 2021 INTERLEUKIN-11 IS IMPORTANT FOR VASCULAR SMOOTH MUSCLE PHENOTYPIC SWITCHING AND AORTIC INFLAMMATION, FIBROSIS AND
REMODELING IN MOUSE MODELS Article Open access 20 October 2020 YY1 DIRECTLY INTERACTS WITH MYOCARDIN TO REPRESS THE TRIAD MYOCARDIN/SRF/CARG BOX-MEDIATED SMOOTH MUSCLE GENE TRANSCRIPTION
DURING SMOOTH MUSCLE PHENOTYPIC MODULATION Article Open access 11 December 2020 DATA AVAILABILITY scRNA-seq datasets generated for this manuscript are available in the GEO under accession
number GSE201091. The ATAC-seq dataset generated for this manuscript has been deposited in the GEO and is available under accession number GSE241055. All reagents and resources applicable to
this study are available from the corresponding authors upon reasonable request. Source data are provided with this paper. CODE AVAILABILITY No novel code or algorithm was generated for
this study. REFERENCES * Guo, D. C. et al. Mutations in smooth muscle alpha-actin (_ACTA2_) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease.
_Am. J. Hum. Genet._ 84, 617–627 (2009). Article CAS PubMed PubMed Central Google Scholar * Guo, D. C. et al. Mutations in smooth muscle alpha-actin (_ACTA2_) lead to thoracic aortic
aneurysms and dissections. _Nat. Genet._ 39, 1488–1493 (2007). Article CAS PubMed Google Scholar * Milewicz, D. M. et al. Altered smooth muscle cell force generation as a driver of
thoracic aortic aneurysms and dissections. _Arterioscler. Thromb. Vasc. Biol._ 37, 26–34 (2017). Article CAS PubMed Google Scholar * Milewicz, D. M. et al. De novo _ACTA2_ mutation
causes a novel syndrome of multisystemic smooth muscle dysfunction. _Am. J. Med. Genet. A_ 152A, 2437–2443 (2010). Article PubMed PubMed Central Google Scholar * Regalado, E. S. et al.
Clinical history and management recommendations of the smooth muscle dysfunction syndrome due to ACTA2 arginine 179 alterations. _Genet. Med._ 20, 1206–1215 (2018). Article CAS PubMed
PubMed Central Google Scholar * Lauer, A. et al. Cerebrovascular disease progression in patients with _ACTA2_ Arg179 pathogenic variants. _Neurology_ 96, e538–e552 (2021). Article CAS
PubMed PubMed Central Google Scholar * Munot, P. et al. A novel distinctive cerebrovascular phenotype is associated with heterozygous Arg179 _ACTA2_ mutations. _Brain_ 135, 2506–2514
(2012). Article PubMed PubMed Central Google Scholar * Georgescu, M. M. et al. The defining pathology of the new clinical and histopathologic entity ACTA2-related cerebrovascular
disease. _Acta Neuropathol. Commun._ 3, 81–87 (2015). Article PubMed PubMed Central Google Scholar * Kelpsch, D. J. & Tootle, T. L. Nuclear actin: from discovery to function. _Anat.
Rec._ 301, 1999–2013 (2018). Article CAS Google Scholar * Xie, X. et al. β‐actin‐dependent global chromatin organization and gene expression programs control cellular identity. _FASEB J._
32, 1296–1314 (2019). Google Scholar * Xie, X., Jankauskas, R., Mazari, A. M. A., Drou, N. & Percipalle, P. β-actin regulates a heterochromatin landscape essential for optimal
induction of neuronal programs during direct reprograming. _PLoS Genet._ 14, e1007846 (2018). Article PubMed PubMed Central Google Scholar * Kumar, A. et al. Actin R256 mono-methylation
is a conserved post-translational modification involved in transcription. _Cell. Rep._ 32, 108172 (2020). Article CAS PubMed PubMed Central Google Scholar * Ilik, I. A. & Aktas, T.
Nuclear speckles: dynamic hubs of gene expression regulation. _FEBS J._ 289, 7234–7245 (2022). Article CAS PubMed Google Scholar * Cheung, C., Bernardo, A. S., Pedersen, R. A. &
Sinha, S. Directed differentiation of embryonic origin-specific vascular smooth muscle subtypes from human pluripotent stem cells. _Nat. Protoc._ 9, 929–938 (2014). Article CAS PubMed
Google Scholar * Owens, G. K., Kumar, M. S. & Wamhoff, B. R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. _Physiol. Rev._ 84, 767–801
(2004). Article CAS PubMed Google Scholar * Kaw, A. et al. Mosaicism for the smooth muscle cell (SMC)-specific knock-in of the _Acta2_ R179C pathogenic variant: implications for gene
editing therapies. _J. Mol. Cell. Cardiol._ 171, 102–104 (2022). Article CAS PubMed Google Scholar * Liedtke, S., Stephan, M. & Kogler, G. Oct4 expression revisited: potential
pitfalls for data misinterpretation in stem cell research. _Biol. Chem._ 389, 845–850 (2008). Article CAS PubMed Google Scholar * Mayor, R. & Theveneau, E. The neural crest.
_Development_ 140, 2247–2251 (2013). Article CAS PubMed Google Scholar * Lu, H., Fagnant, P. M., Krementsova, E. B. & Trybus, K. M. Severe molecular defects exhibited by the R179H
mutation in human vascular smooth muscle alpha-actin. _J. Biol. Chem._ 291, 21729–21739 (2016). Article CAS PubMed PubMed Central Google Scholar * Hinz, B., Gabbiani, G. &
Chaponnier, C. The NH2-terminal peptide of alpha-smooth muscle actin inhibits force generation by the myofibroblast in vitro and in vivo. _J. Cell Biol._ 157, 657–663 (2002). Article CAS
PubMed PubMed Central Google Scholar * Papke, C. L. et al. Smooth muscle hyperplasia due to loss of smooth muscle alpha-actin is driven by activation of focal adhesion kinase, altered p53
localization and increased levels of platelet-derived growth factor receptor-beta. _Hum. Mol. Genet._ 22, 3123–3137 (2013). Article CAS PubMed PubMed Central Google Scholar * Yap, C.,
Mieremet, A., de Vries, C. J. M., Micha, D. & de Waard, V. Six shades of vascular smooth muscle cells illuminated by klf4 (krüppel-like factor 4). _Arter. Thromb. Vasc. Biol._ 41,
2693–2707 (2021). Article CAS Google Scholar * Kato, S. et al. Ectopic expression of Smad7 inhibits transforming growth factor-β responses in vascular smooth muscle cells. _Life Sci._ 69,
2641–2652 (2001). Article PubMed Google Scholar * Hideto, O. et al. Smooth muscle cell phenotype-dependent transcriptional regulation of the alpha-1 integrin gene. _J. Biol. Chem._ 272,
26643–26651 (1997). Article Google Scholar * Turczyńska, K. M. et al. Regulation of smooth muscle dystrophin and synaptopodin 2 expression by actin polymerization and vascular Injury.
_Arter. Thromb. Vasc. Biol._ 35, 1489–1497 (2015). Article Google Scholar * Chen, J. et al. Loss of smooth muscle alpha-actin leads to NF-κB-dependent increased sensitivity to angiotensin
II in smooth muscle cells and aortic enlargement. _Circ. Res._ 120, 1903–1915 (2017). Article CAS PubMed PubMed Central Google Scholar * Schildmeyer, L. A. et al. Impaired vascular
contractility and blood pressure homeostasis in the smooth muscle alpha-actin null mouse. _FASEB J._ 14, 2213–2220 (2000). Article CAS PubMed Google Scholar * Pedroza, A. J. et al.
Single-cell transcriptomic profiling of vascular smooth muscle cell phenotype modulation in Marfan syndrome aortic aneurysm. _Arterioscler. Thromb. Vasc. Biol._ 40, 2195–2211 (2020). Article
CAS PubMed PubMed Central Google Scholar * Wirka, R. C. et al. Atheroprotective roles of smooth muscle cell phenotypic modulation and the TCF21 disease gene as revealed by single-cell
analysis. _Nat. Med._ 25, 1280–1289 (2019). Article CAS PubMed PubMed Central Google Scholar * Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating
single-cell transcriptomic data across different conditions, technologies, and species. _Nat. Biotechnol._ 36, 411–420 (2018). Article CAS PubMed PubMed Central Google Scholar * Stuart,
T. et al. Comprehensive integration of single-cell data. _Cell_ 177, 1888–1902 (2019). Article CAS PubMed PubMed Central Google Scholar * Schwarz, D. et al. Ezh2 is required for neural
crest-derived cartilage and bone formation. _Development_ 141, 867–877 (2014). Article CAS PubMed Google Scholar * Takizawa, H. et al. Neural crest-derived cells possess differentiation
potential to keratinocytes in the process of wound healing. _Biomed. Pharmacother._ 146, 112593 (2022). Article CAS PubMed Google Scholar * Crane, J. F. & Trainor, P. A. Neural
crest stem and progenitor cells. _Annu. Rev. Cell Dev. Biol._ 22, 267–286 (2006). Article CAS PubMed Google Scholar * Pan, H. et al. Single-cell genomics reveals a novel cell state
during smooth muscle cell phenotypic switching and potential therapeutic targets for atherosclerosis in mouse and human. _Circulation_ 142, 2060–2075 (2020). Article CAS PubMed PubMed
Central Google Scholar * Li, Y. et al. Single-cell transcriptome analysis reveals dynamic cell populations and differential gene expression patterns in control and aneurysmal human aortic
tissue. _Circulation_ 142, 1374–1388 (2020). Article CAS PubMed PubMed Central Google Scholar * Mahmood, S. R. et al. β-actin dependent chromatin remodeling mediates compartment level
changes in 3D genome architecture. _Nat. Commun._ 12, 5240 (2021). Article CAS PubMed PubMed Central Google Scholar * Su, I. et al. Polycomb group protein ezh2 controls actin
polymerization and cell signaling. _Cell_ 121, 425–436 (2005). Article CAS PubMed Google Scholar * Gunasekaran, S., Miyagawa, Y. & Miyamoto, K. Actin nucleoskeleton in embryonic
development and cellular differentiation. _Curr. Opin. Cell Biol._ 76, 102100 (2022). Article CAS PubMed Google Scholar * Lino Cardenas, C. L., Briere, L. C., Sweetser, D. A., Lindsay,
M. E. & Musolino, P. L. A seed sequence variant in miR-145-5p causes multisystem smooth muscle dysfunction syndrome. _J. Clin. Invest._ 133, e166497 (2023). Article PubMed PubMed
Central Google Scholar * Wang, Y. et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. _Dev. Cell_ 25, 69–80 (2013).
Article CAS PubMed Google Scholar * Cordes, K. R. et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. _Nature_ 460, 705–710 (2009). Article CAS PubMed PubMed
Central Google Scholar * Guo, D. C. et al. Loss-of-function mutations in _YY1AP1_ lead to Grange syndrome and a fibromuscular dysplasia-like vascular disease. _Am. J. Hum. Genet._ 100,
21–30 (2017). Article CAS PubMed Google Scholar * Zhang, M., Fang, H., Zhou, J. & Herring, B. P. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific
gene expression. _J. Biol. Chem._ 282, 25708–25716 (2007). Article CAS PubMed Google Scholar * Zhou, J. et al. The SWI/SNF chromatin remodeling complex regulates myocardin-induced smooth
muscle-specific gene expression. _Arterioscler. Thromb. Vasc. Biol._ 29, 921–928 (2009). Article CAS PubMed PubMed Central Google Scholar * Bohnsack, M. T., Stuven, T., Kuhn, C.,
Cordes, V. C. & Gorlich, D. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. _Nat. Cell Biol._ 8, 257–263 (2006). Article CAS PubMed Google
Scholar * Le, H. Q. et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. _Nat. Cell Biol._ 18, 864–875 (2016). Article CAS
PubMed Google Scholar * Neely, A. & Bao, X. Nuclei Isolation Staining (NIS) method for imaging chromatin-associated proteins in difficult cell types. _Curr. Protoc. Cell Biol._ 84, e94
(2019). Article PubMed PubMed Central Google Scholar * Cheung, C. et al. Generation of human vascular smooth muscle subtypes provides insight into embryological origin–dependent disease
susceptibility. _Nat. Biotechnol._ 30, 165–173 (2012). Article CAS PubMed PubMed Central Google Scholar * Kwartler, C. S. et al. Vascular smooth muscle cell isolation and culture from
mouse aorta. _Bio Protoc._ 6, e2045 (2016). Google Scholar * McLean, C. Y. et al. GREAT improves functional interpretation of _cis_-regulatory regions. _Nat. Biotechnol._ 28, 495–501
(2010). Article CAS PubMed PubMed Central Google Scholar * Robinson, J. T. et al. Integrative genomics viewer. _Nat. Biotechnol._ 29, 24–26 (2011). Article CAS PubMed PubMed Central
Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by an America Heart Association Merit Award (to D.M.M.), the National Heart, Lung and Blood Institute (RO1
HL146583 to D.M.M.), the Olivia Petrera-Cohen Research Fund (to D.M.M.), the John Ritter Foundation (to D.M.M.), Marylin and Frederick R. Lummis, MD, Fellowship in the Biomedical Sciences
(to A.K.), National Institutes of Health (NIH) TL1TR003169 (to A.K.), NIH UL1TR003167 (to A.K.), NIH F32HL154681 (to A.J.P.), NIH R01HL157949 (to M.P.F.), British Heart Foundation awards
RG/17/5/32936 and FS/18/46/33663 (to S.S.) and American Heart Association grant 20CDA35310689 (to C.S.K.). The funders had no role in study design, data collection and analysis, decision to
publish or preparation of the manuscript. Confocal microscopy was performed at the Center for Advanced Microscopy, Department of Integrative Biology and Pharmacology at McGovern Medical
School, UTHealth. We thank G. Gabbiani and C. Chaponnier from the University of Geneva for their generosity in sharing the SKAfp and SMAfp peptides for this study. AUTHOR INFORMATION Author
notes * Shuangtao Ma Present address: Department of Medicine, Michigan State University, East Lansing, MI, USA AUTHORS AND AFFILIATIONS * Division of Medical Genetics, Department of Internal
Medicine, McGovern Medical School, The University of Texas Health Science Center at Houston, Houston, TX, USA Callie S. Kwartler, Anita Kaw, Pujun Guan, Shuangtao Ma, Xue-yan Duan, Caroline
Kernell, Charis Wang, Jose Emiliano Esparza Pinelo, Mikayla S. Borthwick Bowen, Jiyuan Chen & Dianna M. Milewicz * Department of Cardiothoracic Surgery, Stanford University, Stanford,
CA, USA Albert J. Pedroza & Michael P. Fischbein * Department of Epigenetics and Molecular Carcinogenesis, The University of Texas MD Anderson Cancer Center, Smithville, TX, USA Yuan
Zhong * Wellcome-MRC Cambridge Stem Cell Institute, University of Cambridge, Cambridge, United Kingdom Sanjay Sinha * Institute of Cancer Research, Shenzhen Bay Laboratory, Shenzhen, China
Xuetong Shen Authors * Callie S. Kwartler View author publications You can also search for this author inPubMed Google Scholar * Albert J. Pedroza View author publications You can also
search for this author inPubMed Google Scholar * Anita Kaw View author publications You can also search for this author inPubMed Google Scholar * Pujun Guan View author publications You can
also search for this author inPubMed Google Scholar * Shuangtao Ma View author publications You can also search for this author inPubMed Google Scholar * Xue-yan Duan View author
publications You can also search for this author inPubMed Google Scholar * Caroline Kernell View author publications You can also search for this author inPubMed Google Scholar * Charis Wang
View author publications You can also search for this author inPubMed Google Scholar * Jose Emiliano Esparza Pinelo View author publications You can also search for this author inPubMed
Google Scholar * Mikayla S. Borthwick Bowen View author publications You can also search for this author inPubMed Google Scholar * Jiyuan Chen View author publications You can also search
for this author inPubMed Google Scholar * Yuan Zhong View author publications You can also search for this author inPubMed Google Scholar * Sanjay Sinha View author publications You can also
search for this author inPubMed Google Scholar * Xuetong Shen View author publications You can also search for this author inPubMed Google Scholar * Michael P. Fischbein View author
publications You can also search for this author inPubMed Google Scholar * Dianna M. Milewicz View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS D.M.M. and C.S.K. designed the study. C.S.K. planned the individual experiments. C.S.K., A.K., S.M., X.y.D., C.K., J.E.E.P., M.S.B.B. and J.C. performed the cellular
experiments. A.K. and A.J.P. obtained the sample and analyzed the scRNA-seq on mouse tissue. A.J.P. and M.P.F. obtained the sample and analyzed the scRNA-seq on participant tissue. C.S.K.
and A.J.P. obtained the sample and analyzed the ATAC-seq on cultured SMCs. P.G. performed the integrated analysis combining scRNA-seq and ATAC-seq datasets. Y.Z. and X.S. consulted on
nuclear actin functions and contributed to the design of experiments. S.S. reprogrammed a participant stem cell line and assisted with the stem cell differentiation protocol. D.M.M. and
C.S.K. interpreted the data and drafted the manuscript. D.M.M. and M.P.F. obtained funding for this work. CORRESPONDING AUTHORS Correspondence to Callie S. Kwartler or Dianna M. Milewicz.
ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Cardiovascular Research_ thanks the anonymous reviewers for
their contribution to the peer review of this work. Primary Handling Editor: Vesna Todorovic, in collaboration with the _Nature Cardiovascular Research_ Team. ADDITIONAL INFORMATION
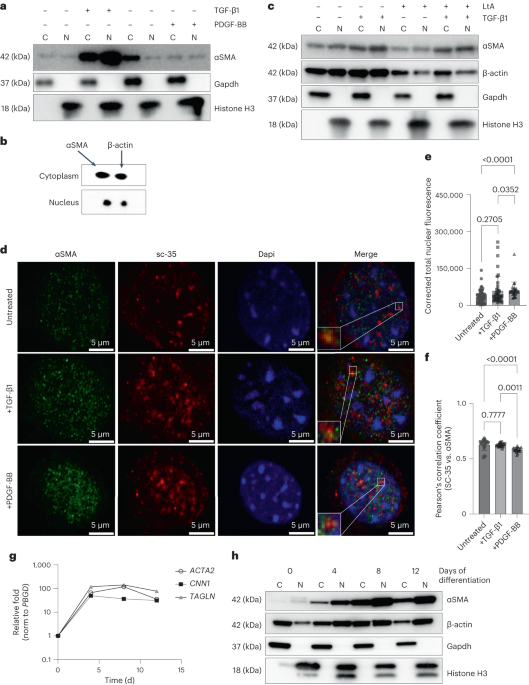
PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 ΑSMA LOCALIZES TO
THE NUCLEUS CONCURRENTLY WITH SMC DIFFERENTIATION. A) Quantitation of immunoblot in Fig. 1A shows that TGFβ1 stimulation for 48 h increases protein levels of αSMA in both the nucleus and
cytosol, while PDGF-BB treatment for 24 h does not affect αSMA nuclear localization (n=3 independent experiments). Significance was assessed by one-way ANOVA followed by Tukey’s multiple
comparisons test. B) Quantitation of the latrunculin (LtA) treatment immunoblot shown in Fig. 1C shows that depolymerization of actins by LtA treatment does not affect nuclear localization
of actin, supporting that the observed nuclear localization is accurate (n = 4 independent experiments). Significance was assessed by Kruskal-Wallis test with Dunn’s multiple comparisons
test. C) Quantitation of the NEPC to SMC differentiation timecourse immunoblot shown in Fig. 1G shows that αSMA levels in both the nucleus and cytosol increase around 15-20 fold during the
differentiation. Over the same timecourse, β-actin levels do not dramatically change, and nuclear accumulation is lower than cytosolic (n = 4 independent experiments using 2 different
control cell lines). All data are presented as mean ± standard deviation. Source data EXTENDED DATA FIG. 2 FORCED NUCLEAR LOCALIZATION OF ACTINS DOES NOT AFFECT MKL1 LEVELS OR LOCALIZATION.
A,B) Immunostaining of NLS-infected SMCs with Mkl1 antibody (A) shows no change in Mkl1 localization or intensity with α-NLS or β-NLS infection, quantified in (B). n = 14 cells infected with
empty vector (EV), 15 cells infected with β-NLS, and 11 cells infected with α-NLS across three independent staining experiments. Significance was assessed by one-way ANOVA with Tukey’s
multiple comparisons test. All data are presented as mean ± standard deviation. Source data EXTENDED DATA FIG. 3 MOUSE _ACTA2_SMC-R179C/+ SMOOTH MUSCLE CELLS HAVE DECREASED NUCLEAR ΑSMA AND
ARE HYPODIFFERENTIATED. A) Quantitation of immunoblot in Fig. 4E shows significant and reproducible decreases in accumulation of contractile proteins in the _Acta2_SMC-R179C/+ cells compared
with WT (n = 4 independent experiments). Significance was assessed by two-way ANOVA followed by Tukey’s multiple comparisons test. B) Quantitation of immunoblot in Fig. 4F shows decreased
nuclear accumulation, of αSMA in the _Acta2_SMC-R179C/+ cells compared with WT. β-actin accumulation to the nucleus was variable across experiments (n = 4 independent experiments).
Significance was assessed by two-way ANOVA followed by Tukey’s multiple comparisons test. C) Quantitation of immunoblot in Fig. 4G shows that LtA treatment further does moderately decrease
nuclear accumulation of β-actin in _Acta2_SMC-R179C/+ cells but does not significantly affect αSMA nuclear accumulation (n = 3 independent experiments). Significance was assessed by two-way
ANOVA followed by Tukey’s multiple comparisons test. D) Co-immunoprecipitation with Brg1 antibody confirms decreased association of both αSMA and β-actin with the Baf chromatin remodeling
complex in in _Acta2_SMC-R179C/+ SMCs which is partially rescued by TGFβ1 treatment (n = 3 independent experiments). All data are presented as mean ± standard deviation. Source data EXTENDED
DATA FIG. 4 DIFFERENTIAL CHROMATIN ACCESSIBILITY ALIGNS WITH TRANSCRIPTIONAL CHANGES IN _ACTA2_SMC-R179C/+ SMCS. A) Distribution of chromatin accessibility peaks around the transcription
start sites of nearby genes for peaks with increased accessibility in WT SMCs. B) Distribution of chromatin accessibility peaks around the transcription start sites of nearby genes for peaks
with increased accessibility in _Acta2_SMC-R179C/+ SMCs. C) Visualization of integrated scRNA-seq and ATAC-seq datasets plots the log2FC in gene expression _in vivo_ on the y-axis and the
log2FC in chromatin accessibility _in vitro_ on the x-axis. Genes indicated by dots inside the red circle have decreased accessibility and decreased expression in _Acta2_SMC-R179C/+ SMCs and
were analyzed by GO enrichment analysis in Fig. 5H. Genes indicated by dots inside the blue circle have increased accessibility and increased expression in _Acta2_SMC-R179C/+ SMCs and were
analyzed by GO enrichment analysis in Fig. 5H. EXTENDED DATA FIG. 5 GENES WITH INCREASED EXPRESSION AND ACCESSIBILITY IN _ACTA2_SMC-R179C/+ SMCS. A) Genome browser view for the _Klf4_ gene
shows chromatin accessibility increased in _Acta2_SMC-R179C/+ SMCs _in vitro_ and transcript levels visualized in UMAP space and quantified shows increased expression in _Acta2_SMC-R179C/+
SMCs _in vivo_. B) Genome browser view for the _Smad7_ gene shows chromatin accessibility increased in _Acta2_SMC-R179C/+ SMCs _in vitro_ and transcript levels visualized in UMAP space and
quantified shows increased expression in _Acta2_SMC-R179C/+ SMCs _in vivo_. P values denote results of Wilcoxon rank sum test with Bonferroni correction for multiple comparisons between all
‘SMC 1’ (5,816 cells) and ‘SMC 2’ (3,229 cells) in the scRNAseq dataset. For all violin plots, line depicts median, hinges depict interquartile range, whiskers depict 1.5 interquartile
ranges above and below the hinges. C) Quantitative RT-PCR validates shRNA-mediated knockdown of _Smad7_ in WT SMCs and shows knockdown of _Smad7_ increases expression of SMC contractile
genes. Graph shows three technical replicates, representative of three independent experiments from three distinct lentiviral infections. Significance was assessed by unpaired, two-tailed
t-test for each gene. All data are presented as mean ± standard deviation. Source data EXTENDED DATA FIG. 6 GENES WITH DECREASED EXPRESSION AND ACCESSIBILITY IN _ACTA2_SMC-R179C/+ SMCS. A)
Genome browser view for the _Itga1_ gene shows chromatin accessibility decreased in _Acta2_SMC-R179C/+ SMCs _in vitro_ and transcript levels visualized in UMAP space and quantified shows
decreased expression in _Acta2_SMC-R179C/+ SMCs _in vivo_. B) Genome browser view for the _Synpo2_ gene shows chromatin accessibility decreased in _Acta2_SMC-R179C/+ SMCs _in vitro_ and
transcript levels visualized in UMAP space and quantified shows decreased expression in _Acta2_SMC-R179C/+ SMCs _in vivo_. P values denote results of Wilcoxon rank sum test with Bonferroni
correction for multiple comparisons between all ‘SMC 1’ (5,816 cells) and ‘SMC 2’ (3,229 cells) in the scRNAseq dataset. For all violin plots, line depicts median, hinges depict
interquartile range, whiskers depict 1.5 interquartile ranges above and below the hinges. EXTENDED DATA FIG. 7 PARTICIPANT-DERIVED _ACTA2_ P.ARG179 SMCS ARE LESS DIFFERENTIATED THAN
CONTROLS. A) Demographic and genotype information for patient and control lines used in this study. B) Quantitation of immunoblot from Fig. 6A reveals significant decreases in contractile
protein accumulation in _ACTA2_ p.R179C patient SMCs compared with control SMCs (n = 4 independent experiments). Significance was assessed by one-way ANOVA with Sidak’s multiple comparisons
test. C-E) Immunoblot showing decreased contractile protein accumulation is a consistent finding across three _ACTA2_ p.R179 patient lines (C), quantified in (D,E). Graph shows biological
replicates- each dot represents one patient or control line. Significance in (D) ( + /- TGFβ1) was assessed by two-way ANOVA with Tukey’s multiple comparisons test. Significance in (E) (no
TGFβ1) was assessed by unpaired, two-tailed t-tests per protein. Grey bars/black dots represent control and green bars/dots represent _ACTA2_ p.R179. All data are presented as mean ±
standard deviation. Source data EXTENDED DATA FIG. 8 PARTICIPANT-DERIVED _ACTA2_ P.ARG179 SMCS HAVE DECREASED NUCLEAR ΑSMA COMPARED WITH CONTROLS. A) Quantitation of immunoblot from Fig. 6C
shows a significant decrease in αSMA nuclear accumulation in patient-derived _ACTA2_ p.R179C SMCs compared with controls. β-actin accumulation to the nucleus was variable and not
significantly altered (n = 3 independent experiments). Significance was assessed by repeated measures one-way ANOVA followed by Tukey’s multiple comparisons test. B,C) Immunoblot showing
decreased nuclear accumulation of αSMA is a consistent phenotype across three _ACTA2_ p.R179 patient lines (B), quantified in (C). Graph shows biological replicates- each dot represents one
patient or control line. Significance was assessed by unpaired, two-tailed t-test for each condition (cytosol, nucleus) for each protein. D) Quantitation of immunoblot from Fig. 6D shows a
significant increase in cytosolic αSMA and decrease in αSMA nuclear accumulation in patient-derived _ACTA2_ p.R179 NEPCs compared with controls. β-actin accumulation to the nucleus was not
significantly altered (n = 3 independent experiments). Significance was assessed by unpaired, two-tailed t-test for each condition (cytosol, nucleus) for each protein. E)
Co-immunoprecipitation with BRG1 antibody confirms decreased association of both αSMA and β-actin with the BAF chromatin remodeling complex in _ACTA2_ p.R179C SMCs. Grey bars/black dots
represent control and green bars/dots represent _ACTA2_ p.R179. All data are presented as mean ± standard deviation. Source data EXTENDED DATA FIG. 9 EXPRESSION OF SKELETAL Α-ACTIN CAN
COMPENSATE FOR LOSS OF NUCLEAR ΑSMA IN _ACTA2_-/- CELLS. A) Immunostaining for αSMA shows partial disruption of αSMA filaments in SMCs with the _ACTA2_ p.R179C pathogenic variant. B)
Subcellular fractionation immunoblot of _Acta2_ KO mouse SMCs shows that sarcomeric actin (encoded by _ACTA1_ and known to be expressed in _Acta2_ KO SMCs as a compensatory mechanism for
loss of αSMA) in these cells can go into the nucleus. C) Quantitative RT-PCR confirms that _ACTA2_ Crispr-edited iPSC-derived SMCs do not have increased expression of _ACTA1_. Graph shows
three technical replicates, results are representative of three independent experiments. All data are presented as mean ± standard deviation. Source data EXTENDED DATA FIG. 10 INTEGRATED
_ACTA2_ P.R179H AND MARFAN SYNDROME (MFS) AORTIC SCRNASEQ DATA. A) Complete dataset following reciprocal PCA data integration demonstrating representation of all major cell clusters in both
datasets. Red arrow indicates SMC cluster selected for downstream analysis. B) Expression plot of _MYH11_ confirming SMC identity for selected cluster. C) Overlaid UMAP plot for _ACTA2_
p.R179H and MFS SMCs demonstrating overrepresented sub-populations in _ACTA2_ patient (black arrows). D) Expression of ‘typical’ SMC modulation markers (_FN1_, _COL1A1_) with gradient of
expression in both patients. E) Expression of representative altered SMC phenotypic trajectory markers in _ACTA2_ patient (see Fig. 7I) not activated in MFS SMCs. F) Activation of
TGFβ1-responsive gene _SERPINE1_ is not observed in ACTA2 p.R179H SMCs as compared to MFS. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary discussion, Tables 1 and 2, Figs.
1–7 and source data and western blots for supplementary figures. REPORTING SUMMARY SUPPLEMENTARY DATA 1 Numerical source data for supplementary figures. SUPPLEMENTARY DATA 2 Image source
data for supplementary figures. SOURCE DATA SOURCE DATA FIG. 1 Unprocessed western blots, statistical source data and original image files. SOURCE DATA FIG. 2 Unprocessed western blots,
statistical source data and original image files. SOURCE DATA FIG. 3 Unprocessed western blots and statistical source data. SOURCE DATA FIG. 4 Unprocessed western blots, statistical source
data and original image files. SOURCE DATA FIG. 6 Unprocessed western blots, statistical source data and original image files. SOURCE DATA EXTENDED DATA FIG./TABLE 1 Statistical source data.
SOURCE DATA EXTENDED DATA FIG./TABLE 2 Statistical source data and original image files. SOURCE DATA EXTENDED DATA FIG./TABLE 3 Unprocessed western blots and statistical source data. SOURCE
DATA EXTENDED DATA FIG./TABLE 5 Statistical source data. SOURCE DATA EXTENDED DATA FIG./TABLE 7 Unprocessed western blots and statistical source data. SOURCE DATA EXTENDED DATA FIG./TABLE 8
Unprocessed western blots and statistical source data. SOURCE DATA EXTENDED DATA FIG./TABLE 9 Unprocessed western blots and statistical source data. RIGHTS AND PERMISSIONS Springer Nature
or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of
the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Kwartler, C.S., Pedroza, A.J., Kaw, A. _et al._ Nuclear smooth muscle α-actin participates in vascular smooth muscle cell differentiation. _Nat Cardiovasc Res_ 2, 937–955 (2023).
https://doi.org/10.1038/s44161-023-00337-4 Download citation * Received: 04 May 2022 * Accepted: 23 August 2023 * Published: 28 September 2023 * Issue Date: October 2023 * DOI:
https://doi.org/10.1038/s44161-023-00337-4 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative