- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Sarcopenia is defined as a progressive decrease in skeletal muscle mass and strength due to ageing, and is associated with adverse outcomes such as falls, fractures, and physical disability,
leading to increased healthcare expenditures and mortality risks. Although sarcopenia has recently become a major challenge, there are currently no effective interventions or drugs marketed
for sarcopenia. Therefore, new biomarkers for early diagnosis and targeted drugs for the treatment of sarcopenia are urgently needed. In the present research, transcriptomics and proteomics
analyses were combined and experiments at the transcriptional and protein levels were conducted to identify key genes and molecular mechanisms underlying sarcopenia in
senescence-accelerated mouse prone 8 (SAM-P8) mice, followed by molecular docking to predict targeted drugs. The combined omics analysis identified 8 key genes, while the experiments
verified that only CD9 significantly decreased in sarcopenia. The gene set enrichment analysis (GSEA) results suggested that CD9 involved in ATP biosynthesis, mitochondrial biogenesis, and
oxidative phosphorylation. Besides, dapoxetine, levomilnacipran, and milnacipran were predicted to target CD9 through molecular docking. Our study reported for the first time that CD9 is a
novel potential biomarker of sarcopenia, and targeting CD9 may be a new idea for the development of therapeutic drugs for sarcopenia.
Sarcopenia was first proposed in 1989 by Irwin Rosenberg1,2. And now, the Global Leadership Initiative in Sarcopenia (GLIS) defines sarcopenia as the age-related loss in muscle mass and
strength/function3. It is conservatively estimated that 10%-16% of elderly individuals worldwide suffer from sarcopenia4. In China, the prevalence of sarcopenia among the elderly population
is even higher (20.7%), and varies by sex and age; that is, the prevalence among women (21.6%) is greater than that among men (19.2%), and the prevalence among people aged ≥ 80 years (45.4%)
is the highest, followed by people aged 70–79 years (27.2%) and 60–69 years (15.7%)5. In addition, the muscle mass decreases at an annual rate of 1–2% approximately after the age of 50, and
the muscle strength decreases by 1.5% between the ages of 50 and 60 years and by 3% thereafter6. The occurrence of sarcopenia leads to instability, an increased risk of falls and
consequently an increased demand for medical and social care. Indeed, sarcopenia was found to be closely associated with all-cause mortality7,8.
However, the pathogenesis mechanism of sarcopenia remains unclear, and studies suggest that it is associated with ageing, lifestyle changes, physical activity, nutritional levels, and
neurological factors9,10,11. These factors contribute to skeletal muscle inflammation, oxidative stress, mitochondrial dysfunction, and disorders of protein metabolism12,13,14, eventually
leading to sarcopenia. Given the enormous socioeconomic burden that sarcopenia can impose, preventive measures and effective treatments are urgently needed. At present, interventions for
sarcopenia are limited, including exercise therapy and nutritional support15,16,17,18,19, and patients benefit little, especially for patients who are unsuitable for exercise or are
bedridden. Although several medications have been shown to be potentially effective in treating sarcopenia, such as formoterol20, allopurinol21, and myostatin-specific antibodies22, no drugs
have been marketed to date23. As a result, new methods for the rapid and efficient early diagnosis and treatment of sarcopenia are urgently needed, including novel biomarkers, underlying
mechanisms, and targeted drugs.
Naturally, ageing mice are the preferred choice for sarcopenia research, but their application has been hampered by their long rearing time and high cost. Fortunately, animal models of
accelerated ageing shorten the time required to wait for ageing. The senescence-accelerated mouse (SAM) is one of the most common ageing models developed by Takeda et al.24, and is
considered a "fast-forward" option for ageing studies. The line consists of 18 different strains, 11 of which are SAM-prone (SAM-P, susceptible to ageing) and 7 of which are SAM-resistant
(SAM-R, resistant to ageing)25. Compared with SAM-R1 mice, SAM-P8 mice have significantly lower skeletal muscle weight and contractility, and more obvious atrophy of type II muscle fibres,
indicating the significant characteristics of accelerated skeletal muscle ageing26, thus becoming a common animal model for the study of sarcopenia27,28.
The main process of gene expression includes transcription and protein synthesis, where mRNA is the intermediate of gene expression, and protein is the executive of gene function.
Transcriptomics and proteomics elucidate the changes in related genes and molecular mechanisms involved in the occurrence, development and prognosis of diseases from the perspectives of gene
transcription and protein synthesis, respectively29. As a result, the combined analysis of transcriptomics and proteomics has become an important means to discover and study the biomarkers
of diseases from the the source30,31.
In this study, a combined transcriptomics and proteomics analysis strategy was employed to screen key genes and possible signalling pathways that exhibit significant changes throughout the
pathological process of sarcopenia in SAM-P8 mice, aiming to identify novel biomarkers and possible regulatory mechanisms for the early recognition of and interventions for sarcopenia.
Furthermore, biomarkers for targeted drugs are predicted to provide new directions for the development of targeted drugs for sarcopenia.
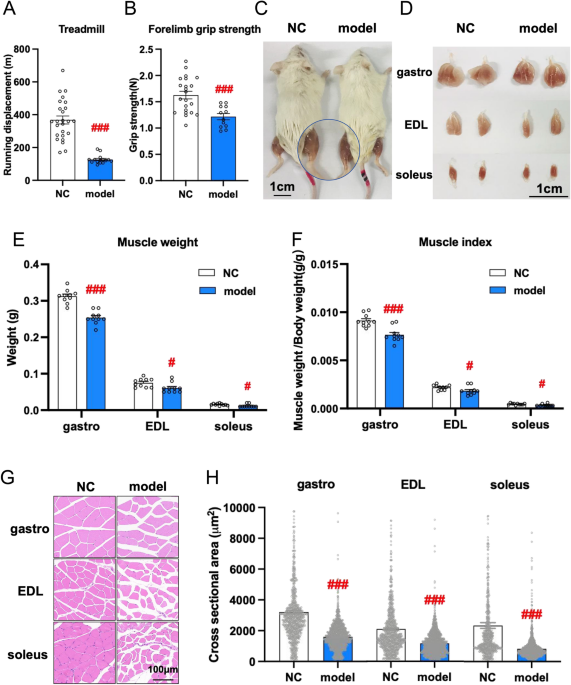
Initially, the treadmill test and forelimb grip strength test were performed, and the results revealed that muscular endurance (Fig. 1A) and explosive power (Fig. 1B) were dramatically lower
in 28-week-old SAM-P8 mice (model group) than in SAM-R1 mice (NC group) (### p value










