- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Abdominal aortic aneurysms (AAA) is a multifactorial complex disease with life-threatening consequences. While Genome-wide association studies (GWAS) have revealed several single
nucleotide polymorphisms (SNPs) located in the genome of individuals with AAA, the link between SNPs with the associated pathological signals, the influence of risk factors on their
distribution and their combined analysis is not fully understood. We integrated 86 AAA SNPs from GWAS and clinical cohorts from the literature to determine their phenotypical vulnerabilities
and association with AAA risk factors. The SNPs were annotated using snpXplorer AnnotateMe tool to identify their chromosomal position, minor allele frequency, CADD (Combined Annotation
Dependent Depletion), annotation-based pathogenicity score, variant consequence, and their associated gene. Gene enrichment analysis was performed using Gene Ontology and clustered using
REVIGO. The plug-in GeneMANIA in Cytoscape was applied to identify network integration with associated genes and functions. 15 SNPs affecting 20 genes with a CADD score above ten were
identified. AAA SNPs were predominantly located on chromosome 3 and 9. Stop-gained rs5516 SNP obtained high frequency in AAA and associated with proinflammatory and vascular remodeling
phenotypes. SNPs presence positively correlated with hypertension, dyslipidemia and smoking history. GO showed that AAA SNPs and their associated genes could regulate lipid metabolism,
extracellular matrix organization, smooth muscle cell proliferation, and oxidative stress, suggesting that part of these AAA traits could stem from genetic abnormalities. We show a library
of inborn SNPs and associated genes that manifest in AAA. We uncover their pathological signaling trajectories that likely fuel AAA development. SIMILAR CONTENT BEING VIEWED BY OTHERS THE
SNP RS7865618 OF 9P21.3 LOCUS EMERGES AS THE MOST PROMISING MARKER OF CORONARY ARTERY DISEASE IN THE SOUTHERN INDIAN POPULATION Article Open access 09 December 2020 GENOME-WIDE ASSOCIATION
META-ANALYSIS IDENTIFIES RISK LOCI FOR ABDOMINAL AORTIC ANEURYSM AND HIGHLIGHTS PCSK9 AS A THERAPEUTIC TARGET Article Open access 16 October 2023 INTEGRATIVE GENOMIC ANALYSES IDENTIFY
CANDIDATE CAUSAL GENES FOR CALCIFIC AORTIC VALVE STENOSIS INVOLVING TISSUE-SPECIFIC REGULATION Article Open access 18 March 2024 INTRODUCTION Abdominal aortic aneurysms (AAA) are
characterized by localized weakening and dilation of the abdominal aorta1 that results from a cascade of mechanisms including transmural inflammation2, smooth muscle cell (SMC) apoptosis3
and extracellular matrix (ECM) degradation4 that collectively lead to the loss of aortic wall elasticity. AAA often coexists with other cardiovascular diseases5,6,7 and risk factors such as
aging and smoking8. However, the physiopathology of AAA is uniquely distinguished by signaling programs that result in the exaggerated degradation of the core constituents of the elements of
the ECM that lead to life-threatening aortic rupture. SNPs are single base-pair variations that occur in the DNA either within or outside the coding region of genes and have the potential
to interfere at different steps of gene expression depending on their genomic location9. For example, SNPs present in non-coding segments of the genome have been shown to modulate the
efficacy of gene transcription by impeding the accessibility of transcription factors in these response elements10. The presence of SNPs in coding region of genes can give rise to mRNA with
different bases at SNP site which could impact the proper translation of mRNA while the presence of SNPs within a coding sequence can lead to an amino-acid change and protein-misfolding9,11.
Recently, it has been shown that the identification and estimation of variance by all SNPs from GWAS of conventionally unrelated individuals might be a significant determinant of genetic
heritability to particular diseases12,13. Notably, previous genetic studies have reported that AAA occurrence can be heritable within families, reaching up to a 20% increase in AAA
susceptibility within first-degree relatives14,15,16. Notably, a population-based twin study estimated the heritability of AAA to be as strong as between 70 and 77%17. However, while the
inheritance of SNPs in AAA has not been directly studied in family cohorts, several gene polymorphisms such as _COL3A1_, _MYH11_, and _TGFBR2_ were also identified to be transmitted within a
familial lineage of AAA18. Despite this evidence pointing to the familial risk associated with AAA, there is a paucity of information linking these genetic signatures and their underlying
mechanistic significance in the development of AAA. GWAS have facilitated the accessibility of disease-specific SNPs19. Identification of SNPs as predictive markers for disease risk has been
used for Alzheimer’s20, migraines21, and coronary artery disease22. In AAA, GWAS of the Million Veteran Program has examined the genetic associations of genetic variants in a subset of AAA
independent of family history23. As such, identification of AAA-specific SNPs would be beneficial to further characterize the pathogenic pathway associated with each SNP and refine our
understanding of the knowledge gap between genomic inheritance and the molecular trajectory that lead to the degradation of the aortic wall that occurs during AAA. In the current study, we
performed an integrative analysis combining the available GWAS catalogs in AAA along with non-GWAS gene association studies published in the literature. We identified 86 SNPs related to AAA
and its associated risk factors. We curated this result using multiple bioinformatic analyses to expose their potential phenotypical signals via which they could contribute to AAA
development. METHODS LITERATURE SEARCH Identification of studies was conducted through a literature search of PubMed, GWAS central, and GWAS registry. The search query used to retrieve
potentially eligible studies from PubMed was “abdominal aortic aneurysm AND genetics”, “abdominal aortic aneurysm AND SNPs”, and “abdominal aortic aneurysm AND GWAS”. In addition, we
searched for other possible SNPs outside of GWAS central and GWAS catalog by considering AAA candidate gene association studies with detailed clinical characteristics, reporting data on the
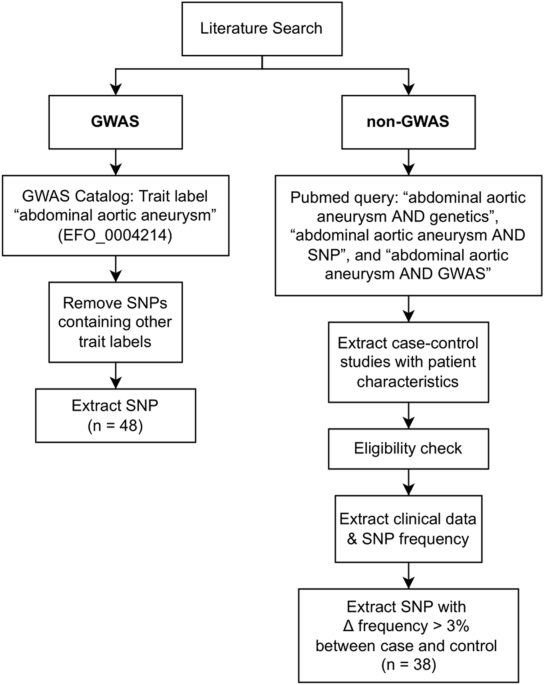
associations between AAA and different SNPs. We only included published full-text articles until April 2022. The flow diagram of the study inclusion is described in Fig. 1. SELECTION
CRITERIA AND QUALITY ASSESSMENT GWAS studies were included if they were listed in the GWAS directory with the trait label “abdominal aortic aneurysm” (EFO_0004214), with no other labels.
Non-GWAS studies were considered eligible if they evaluated the association between AAA and genetic polymorphism through effect measures of odds ratio (OR), with 95% confidence interval
(CI); if they reported allele and minor allele frequency (MAF) of population groups; and if controls were in Hardy–Weinberg equilibrium. We excluded the studies with data published only in
abstract form, studies where MAF in controls were lower than 1% (rare variants), with sample size fewer than 10 cases or controls, studies that were rebutted by others and studies that
combined data with other cardiovascular diseases. systematic reviews and animal in vivo studies were also not included in our analysis. We based our selection criteria on the principles
proposed by the Human Genome Epidemiology Network (HuGeNet) for the meta-analysis of molecular association studies20. The following clinical data were extracted from eligible AAA candidate
gene association studies when available: age, gender percentage, aortic diameter (mm), smoking history (past or current), hypertension, diabetes, coronary artery disease (CAD), peripheral
artery disease (PAD), and dyslipidemia (or hypercholesterolemia). SNPs frequency and genotype data of case and control groups were also extracted. SNPS-GENE ANNOTATION AND PATHWAY ENRICHMENT
ANALYSIS The SNPs used in Gene-set Enrichment Analysis were obtained from GWAS database with the trait label “abdominal aortic aneurysm” (EFO_0004214) and from other studies where allele
frequency in AAA was reported to be 3% more than in controls. The SNPs were annotated using snpXplorer AnnotateMe tool24 with the following settings: SNPs-gene annotation or Gene-set
Enrichment Analysis, GRCh38, GTEx tissue (Blood, Aorta, Coronary) and GO:BP. For each SNP input, SNPs-gene annotation provided the following information: chromosomal position; MAF;
CADD-annotation based pathogenicity score (CADD v1.6), variant consequence, affected gene; GTEx based eQTL (expression quantitative-trait-loci) and sQTL (splicing quantitative-trait-loci);
closest affected gene. Input SNPs approximate affected genes based on position within the genome, expression in GTEx tissue, or their direct coding gene. Gene enrichment analysis was
performed on AAA-related genes using Gene Ontology (GO) terms as the gene-set source. Clustering of the enriched GO terms was performed using REVIGO25, and annotated terms were selected
through a semantic similarity matrix and a dynamic cut tree algorithm for term-based clustering24. The plug-in GeneMANIA in Cytoscape was used as an analytical method to provide an
association network integration to predict gene function and gene–gene interaction of the SNPs-associated gene in this study26,27. GeneMANIA weighted each functional genomic dataset
according to its predictive value according to its gene query while suggesting more genes with similar domain structure or physical interaction27. Gene interaction was plotted via Cytoscape.
A Mann–Whitney U test was performed using R software28 on patient clinical data. Comparisons of age, aortic diameter, male percentage, smoking history (past and current), hypertension,
dyslipidemia, diabetes, CAD, and PAD were performed if data was available. Data were plotted using R studio (v.1.3.1093). RESULTS DISTRIBUTION OF AAA SNPS IN THE GENOME AND THEIR GENE
REGULATION The literature search identified 86 SNPs related to AAA, 48 of which originated from the recorded GWAS databases, and 38 were from AAA gene association studies in which SNPs
frequency in AAA patients was at least 3% higher than in the control group. The variant-gene mapping procedure performed with SNPs snpXplorer AnnotateMe tool showed that most of the
identified SNPs were annotated based on their chromosomal position (n = 49), followed by their eQTL GTEx tissue expression (n = 28) and direct gene coding region (n = 9) (Fig. 2A,B),
implying the possible effect of the SNPs of interest in AAA. The specificity of each SNPs was observed since the majority of the SNPs (_N_ = 57 variants) are mapped and annotated for one
gene (Fig. 2C). We observed a random distribution of SNPs across different chromosomes. Chromosome 3 and 9 predominantly harbored most of the AAA SNPs (Fig. 2D). No SNPs were found on
chromosome 17, X, and Y, suggesting that AAA SNPs identified in this study are not sex-linked (Fig. 2D). On average, each chromosome contains 4 SNPs, with chromosomes 3 and 9 in contain the
most SNPs (Fig. 2E). In the chromosome 3, we found 8 SNPs that affect a single gene (_TGFBR2)_ and 1 SNP that affects 9 different genes (_ITIH4, DNAH1, SFMBT1, GNL3, GLYCTK, NT5DC2, STIMATE,
GLYCTK-AS1, MUSTN1_). In the chromosome 9, we found 3 SNPs that affect a single gene (TGFBR1). The complete SNP chromosome mapping in a 1:250,000 scale of pixel to base pair can be seen in
Supplementary Fig. 1. Single SNP can affect multiple genes depending on their genomic location. Most of the SNPs associated with AAA reside in the intronic, regulatory, upstream, downstream,
or intergenic region of their respective gene (Fig. 2F). In particular, SNPs with high CADD pathogenicity scores (> 10) result in non-synonymous, noncoding change, and stop gained
mutations. A high CADD pathogenicity score is indicative of the deleterious effect of the SNP, as compared to other possible mutations within the human genome29. SNPS AND THE ASSOCIATED
GENES ARE LINKED WITH HIGH PATHOGENICITY IN AAA We identified 15 SNPs affecting 20 genes with a CADD (Combined Annotation Dependent Depletion) pathogenicity score above 10 (Table 1). CADD
scores correlate with pathogenicity, disease severity, regulatory effects, and complex trait associations. A score greater than 10 indicates that the nucleotide substitution is predicted to
be the 10% most deleterious substitution within the human genome, a score of 20 or greater indicates the 1% most deleterious, a score of 30 or greater indicates the 0.1% most deleterious and
so on. We found 1 SNP with a CADD score above 30 (rs5516, _KLK1_), 1 SNP with a CADD score above 20 (rs1801133, _MTHFR_), and 13 SNPs with a CADD score above 10. We confirmed the
association between SNPs with high CADD pathogenicity scores with their frequency on the retrieved 38 AAA-gene association studies (Table 2). We identified four SNPs and associated genes
consisting of the SNP rs5516 (_KLK1_), rs1800795 (_IL-6_), rs2230806 (_ABCA1_), and rs243865 (_MMP-2_) have both higher CADD scores and frequency in AAA patients. However, we observed that a
high CADD score does not necessarily correlate with frequency in AAA. BIOLOGICAL TRAITS ASSOCIATED WITH AAA SNPS Using SnpXplorer AnnotateMe platform, gene enrichment analysis from the
GWAS-catalog associations of the AAA SNPs showed strong correlations with various lipid metabolism pathways such as LDL-cholesterol measurement (52%), total cholesterol measurement (34%),
triglyceride measurement (26%), HDL-cholesterol measurement (23%), and CRP measurement (14%) (Fig. 3A). Only 49% of all tested SNPs were found to be directly labeled with the AAA trait, this
may suggest that the remaining 51% have an indirect association with AAA occurrence through various lipid metabolism pathways and genes. As for associations with other cardiovascular
diseases, AAA and CAD share 25% of the same SNPs, while AAA and MI only share 10%. In addition to SNPs, we present the GWAS-catalog associations of the genes associated with AAA SNPs.
AAA-SNPs associated genes have correlations with various diseases such as coronary artery disease (21%), myocardial infarction (12%), and type II diabetes (10%). AAA SNPs-associated genes
were also found to be associated with genes relating to total cholesterol measurement (17%), LDL-cholesterol measurement (15%), CRP measurement (12%), HDL-cholesterol measurement (10%), and
triglyceride measurement (9%) (Fig. 3B). GENE ONTOLOGY ANALYSIS OF AAA SNPS Gene-set enrichment analysis was done with the genes associated with AAA SNPs, using Gene Ontology (GO) as the
gene-set source. The gene-set enrichment was then visualized using REVIGO25. Annotated GO terms were selected using a 2-step process as described by Tesi et al. (2021) in the snpXplorer web
server24. The GO term clustering and annotation in Fig. 4 depict the most prominent and significant biological processes and associated genes in AAA based on the snpXplorer annotation
algorithm. The AAA-associated genes were found to be prominently involved in cell population proliferation, followed by regulation of cell population proliferation (_p_ = 3.19 × 10−8),
muscle cell proliferation (_p_ = 3.08 × 10−6), regulation of smooth muscle cell proliferation (_p_ = 4.97 × 10−6), regulation of plasma lipoprotein particle levels (_p_ = 1.58 × 10−5),
regulation of protein kinase activity (_p_ = 2.15 × 10−5), regulation of phosphorus metabolic process (_p_ = 2.77 × 10−5), and blood circulation (_p_ = 4.63 × 10−5). Images processed using
SNPsXplorer showing that these annotated terms were found to be most significant according to the semantic similarity (Supplementary Fig. 2) and a dynamic cut tree algorithm for term-based
clustering and p values (Supplementary Fig. 3). The most significant gene ontology terms associated with the SNPs associated genes are summarized in Table 3. SIGNIFICANT SIGNALING NETWORKS
ASSOCIATED WITH AAA SNPS To assess the interaction between SNPs associated genes related to AAA, gene–gene interactomes were constructed and plotted using GeneMANIA plugins in Cytoscape
(Fig. 5). The most significant Genes related to top 25 GO annotated functions (Table 4) were included to further grouped and visualized on the network module to see their network association
according to their shared pathways, co-localization, co-expression, and physical interaction (Fig. 5A). As a hallmark of pathological remodeling in AAA, we observed the strongest
interaction between _TGFB1_ with the _TGFB1_ receptor family and notably strong co-localization with _MMP2_ and _MMP9_, suggesting the role of _TGFB1_ pathway as one of the reminiscent
factors that could link the presence of SNPs to the development of AAA. This analysis also includes several additional genes predicted to convey a strong interaction with some of our gene
candidates. 14 genes were associated with lipid metabolism pathways; regulation of plasma lipoprotein level, regulation of lipid localization, regulation of lipid transport, and regulation
of cholesterol transport (Fig. 5B). 10 genes were associated with extracellular matrix (ECM) organization (Fig. 5C). 11 genes were associated with smooth muscle cell proliferation pathways
(Fig. 5D). 10 genes were associated with reactive oxygen species metabolism (Fig. 5E). IL-6, TGFB1, and TGFB1 receptor family shared three modules, the most out of all genes. IL-6 is
involved in lipid metabolism, ECM organization, and smooth muscle cell proliferation pathways. While TGFB1 and the TGFB1 receptor family are involved in ECM organization, smooth muscle cell
proliferation, and ROS metabolism pathways. CLINICAL CHARACTERISTICS OF AAA PATIENTS Out of the AAA candidate gene association studies in the literature, we found 15 studies with complete
clinical data, totaling a sample size of 10.956 (5676 control & 5280 case). The available clinical data were age (mean), men (n), aortic diameter (mm, mean), smoking (current &
past), hypertension, diabetes, CAD (coronary artery disease), PAD (Peripheral Artery Disease), and dyslipidemia. Data variabilities between each variables are different depending on the
availability. A detailed summary of each study can be found in Supplementary Table 1. The non-disease control and AAA case groups shared a similar average age (69.1 and 70.9 years). Several
clinical characteristics, such as a history of smoking (past or current, _p_ = 0.037), hypertension (_p_ = 0.013), and dyslipidemia (_p_ = 0.042), were positively associated with AAA.
Conversely, there were no significant differences in the presence of diabetes, CAD, and PAD between the two populations. This association is summarized in Fig. 6. DISCUSSION As a complex
multifactorial disease, there is little evidence pinpointing the specific genetic predisposition that could contribute to the development of AAA. Previous studies have identified several
SNPs associated with AAA, but little is known about the underlying mechanisms and biological significance of the identified SNPs to AAA pathobiology. In this integrative in silico analysis,
we used snpXplorer AnnotateMe platform to merge the SNPs candidate from the GWAS catalog and previously published AAA clinical cohorts. We further identify several SNPs with possible
pathological signaling pathways associated with AAA as well as their correlative risk factors in these cohorts. In the present study, we calculated the frequency of the top 20 SNPs in AAA
compared to non-disease control and measured the CADD pathogenicity score in order to observe their association with the risk of developing AAA. Here, we identified SNP rs5516, a stop-gained
mutation for _KLK1_ (Kallikrein 1), with the highest pathogenicity score and a significantly high frequency in AAA (17.8%). Indeed, previous SNPs studies performed in both Australian and
Asian cohorts have shown the association of rs5516 with AAA30,43. Our analysis identified other interesting polymorphism profiles associated with significant genes that correlated with AAA.
We identified a high frequency of rs1800629 for _TNF-α_, and rs1800795 for _IL-6_, inflammatory cytokines well-known to mediate the inflammatory response and smooth muscle cell proliferation
during AAA development44,45,46. Indeed, Jablonska et al_._ reported that the mutation detected in both of the alleles increased the risk of AAA formation among heterozygous carriers31.
However, our result show that CADD score did not necessarily correlate with the frequency of AAA in the observed studies. The SNP rs1801133, located in the coding region of _MTHFR_
(Methylenetetrahydrofolate Reductase), was one of the highest-scoring SNPs, yet it’s CT/CC allele was only 3% more expressed in Greek AAA patients than in controls34. However, in this study,
the analysis of the SNPs in the UK cohort revealed that the frequency of SNP rs1801133 was significantly more frequent. This is most likely due to the population, socio-economical,
environmental and genome differences in these two cohorts living in two different geographic areas. Indeed, the meta-analysis performed for SNPs on _MTHFR_ showed a discrepancy regarding the
protective or pathogenic role of either SNP rs1801133 or _MTHFR_47,48,49. Therefore, this result contextualizes the plasticity of population-specific SNPs and its phenotypic effect to the
development of AAA. Several other SNPs with high frequency in AAA such as rs2230806 (_ABCA1_), rs3775290 (_TLR3_), and rs10757278 (_CDKN2B_) were also strongly associated with significant
AAA pathways50,51,52. The loss of _CDKN2B_ promotes p53-dependent smooth muscle cell apoptosis and aneurysm formation. In parallel, our analysis revealed high pathogenicity of regulatory
_CDKN2B_ SNPs (CADD score: 10.42), where this SNP was observed in Chinese and European cohorts39,53 and mark the possible genetic-environment interplay in AAA development. However, we were
unable to validate its shared associated network in AAA-related pathways along with other gene candidates listed in this study. Therefore, a deeper understanding of the role of _CDKN2B_ in
AAA is warranted. The profile of SNPs found in the DNA between genes can act as an essential biological marker with a strong association with the disease. Interestingly, our descriptive
analysis conducted from the gene association clinical studies described significant traits related to the well-described risk factors of AAA such as CAD and dyslipidemia pointing a
possibility that specific SNPs profile might factor on the progression of these risk factors, and eventually fuel the expansion of AAA. For example, our analysis showed rs429358 polymorphism
in _APOE_, a canonical transporter of cholesterol particles54, had a significant pathogenicity score in AAA patients. However, this SNP was found to be unassociated with AAA in an
Australian cohort55. This discrepancy could rely on the characteristics of the cohorts and the type of analysis performed in each study. Genotyping of specific _APOE_ alleles was performed
in the Australian AAA cohort in 640 samples compared to GWAS population study from 7600 AAA cases using DNA sequencing cross referenced with DNA variants library from European-descent
veterans across USA23. The sensitivity of the meta-analysis, difference in geographic cohorts studied and the power of the sample size were likely more amenable to capture associations of
AAA with rs429358 _APOE_ SNPs polymorphism. This further emphasizes the need to perform large-scale worldwide multi-centered studies including populations from disparate groups to uncover
the full spectrum of drivers of AAA genetic vulnerability. Notably, a previous multi-ethnic cohort study has described that the polymorphism of APOE had a significant association with
dyslipidemia in Asian ethnic groups56. The _APOE_ gene is polymorphic and co-exists as APOE-ε2, -ε3 and -ε4 alleles57. APOE-ε4 has been shown to associate with increased in LDL-cholesterol
levels and higher cardiovascular risk58. The highest frequency of APOE-ε4 carrier was observed in the Malay population and was associated with high LDL-C levels. These observations suggest
that the inherent geographical genetic background of certain ethnic groups could present a diverse portfolio of SNPs associated with AAA, which warrants further studies. The main strength of
this study is the stringency of our method to include the most recent GWAS catalogs in AAA and combine the data with other available cohorts in the literature to comprehensively identify
SNPs with a strong association with AAA. We have summarized and visualized the genomic distribution and frequency of each SNP using the most recent bioinformatic software (snpXplorer)24 to
identify their associated molecular pathways and variant consequences. We have integrated this result into pathway enrichment analysis to focus on the biological interaction of the
associated genes in the AAA pathogenesis. We acknowledge several limitations in our study. Considering the limited genomic studies performed in AAA, the number of samples included in our
analysis is lower than other similar in silico SNPs analysis in other diseases. This limitation hindered us from performing a meta-analysis on each SNPs candidate to validate its consistency
between each geographical origin and to confirm the region-specific susceptibility of AAA59. Indeed, the small number of cohorts is also a major limitation in performing and assessing the
predictive potential of these SNPs candidates in AAA. Moreover, the technological advances in the development of sequencing such as whole-exome sequencing could facilitate the identification
of SNPs in the human genomic data. However, this would require significant financial resources and might not be applicable in each clinical setting, thus limiting the potential to use SNPs
profile as a predictive marker to identify AAA. In conclusion, we have identified a significant profile of polymorphism associated with important risk factors such as dyslipidemia and the
main pathobiological pathways of AAA development. Further investigation in large population studies will be necessary to confirm this finding and to finally reveal the specific genetic
heritage of individuals carrying a risk of developing AAA. DATA AVAILABILITY GWAS datasets analyzed for this study are available in GWAS catalog with the trait label “abdominal aortic
aneurysm” (EFO_0004214). The datasets were derived from the following public domain resources: https://www.ebi.ac.uk/gwas/. Non-GWAS studies are openly available at locations cited in the
reference section. REFERENCES * Sakalihasan, N. _et al._ Abdominal aortic aneurysms. _Nat. Rev. Dis. Primers_ 4, 34 (2018). Article Google Scholar * Dale, M. A., Ruhlman, M. K. &
Baxter, B. T. Inflammatory cell phenotypes in AAAs. _Arterioscler. Thromb. Vasc. Biol._ 35, 1746–1755 (2015). Article CAS Google Scholar * Bogunovic, N. _et al._ Impaired smooth muscle
cell contractility as a novel concept of abdominal aortic aneurysm pathophysiology. _Sci. Rep._ 9, 6837 (2019). Article ADS Google Scholar * Didangelos, A. _et al._ Extracellular matrix
composition and remodeling in human abdominal aortic aneurysms: A proteomics approach. _Mol. Cell. Proteom._ 10, 8 (2011). Article Google Scholar * Golledge, J. & Norman, P. E.
Atherosclerosis and abdominal aortic aneurysm. _Arterioscler. Thromb. Vasc. Biol._ 30, 1075–1077 (2010). Article CAS Google Scholar * Harrison, S. C. _et al._ Genetic association of
lipids and lipid drug targets with abdominal aortic aneurysm: A meta-analysis. _JAMA Cardiol._ 3, 26–33 (2018). Article Google Scholar * Hernesniemi, J. A., Vänni, V. & Hakala, T. The
prevalence of abdominal aortic aneurysm is consistently high among patients with coronary artery disease. _J. Vasc. Surg._ 62, 232-240.e3 (2015). Article Google Scholar * Forsdahl, S. H.,
Singh, K., Solberg, S. & Jacobsen, B. K. Risk factors for abdominal aortic aneurysms. _Circulation_ 119, 2202–2208 (2009). Article Google Scholar * Robert, F. & Pelletier, J.
Exploring the impact of single-nucleotide polymorphisms on translation. _Front. Genet._ 9, 507 (2018). Article CAS Google Scholar * Buroker, E. SNPs, transcriptional factor binding sites
and disease. _Biomed. Genet. Genom._ 2, 1–9 (2017). Google Scholar * Shen, L. X., Basilion, J. P. & Stanton, V. P. Single-nucleotide polymorphisms can cause different structural folds
of mRNA. _Proc. Natl. Acad. Sci. U. S. A._ 96, 7871–7876 (1999). Article ADS CAS Google Scholar * Sampson, J. N. _et al._ Analysis of heritability and shared heritability based on
genome-wide association studies for thirteen cancer types. _J. Natl. Cancer Inst._ 107, 279 (2015). Article Google Scholar * Yang, J., Zeng, J., Goddard, M. E., Wray, N. R. & Visscher,
P. M. Concepts, estimation and interpretation of SNP-based heritability. _Nat. Genet._ 49, 1304–1310 (2017). Article CAS Google Scholar * van de Luijtgaarden, K. M. _et al._ Risk of
abdominal aortic aneurysm (AAA) among male and female relatives of AAA patients. _Vasc. Med._ 22, 112–118 (2017). Article Google Scholar * Webster, M. W. _et al._ Ultrasound screening of
first-degree relatives of patients with an abdominal aortic aneurysm. _J. Vasc. Surg._ 13, 9–14 (1991). Article CAS Google Scholar * Rossaak, J. I. _et al._ Familial abdominal aortic
aneurysms in the Otago region of New Zealand. _Cardiovasc. Surg._ 9, 241–248 (2001). Article CAS Google Scholar * Joergensen, T. M. M. _et al._ Editor’s choice—High heritability of
liability to abdominal aortic aneurysms: A population based twin study. _Eur. J. Vasc. Endovasc. Surg._ 52, 41–46 (2016). Article CAS Google Scholar * van de Luijtgaarden, K. M. _et al._
First genetic analysis of aneurysm genes in familial and sporadic abdominal aortic aneurysm. _Hum. Genet._ 134, 881–893 (2015). Article Google Scholar * Srinivasan, S., Clements, J. A.
& Batra, J. Single nucleotide polymorphisms in clinics: Fantasy or reality for cancer?. _Crit. Rev. Clin. Lab. Sci._ 53, 29–39 (2016). Article CAS Google Scholar * Tey, H. J. &
Ng, C. H. Computational analysis of functional SNPs in Alzheimer’s disease-associated endocytosis genes. _PeerJ_ 7, e7667 (2019). Article Google Scholar * Kaur, S. _et al._ Role of single
nucleotide polymorphisms (SNPs) in common migraine. _Egypt. J. Neurol. Psychiatry Neurosurg._ 55, 47 (2019). Article Google Scholar * Christiansen, M. K. _et al._ Coronary artery
disease-associated genetic variants and biomarkers of inflammation. _PLoS ONE_ 12, e0180365 (2017). Article Google Scholar * Klarin, D. _et al._ Genetic architecture of abdominal aortic
aneurysm in the million veteran program. _Circulation_ 142, 1633–1646 (2020). Article Google Scholar * Tesi, N., van der Lee, S., Hulsman, M., Holstege, H. & Reinders, M. J. T.
snpXplorer: A web application to explore human SNP-associations and annotate SNP-sets. _Nucleic Acids Res._ 49, W603–W612 (2021). Article CAS Google Scholar * Supek, F., Bošnjak, M.,
Škunca, N. & Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. _PLoS ONE_ 6, e21800 (2011). Article ADS CAS Google Scholar * Shannon, P. _et al._
Cytoscape: A software environment for integrated models of biomolecular interaction networks. _Genome Res._ 13, 2498–2504 (2003). Article CAS Google Scholar * Franz, M. _et al._ GeneMANIA
update 2018. _Nucleic Acids Res._ 46, W60–W64 (2018). Article CAS Google Scholar * R: The R Project for Statistical Computing. https://www.r-project.org/. * CADD: Predicting the
deleteriousness of variants throughout the human genome | Nucleic Acids Research | Oxford Academic. https://academic.oup.com/nar/article/47/D1/D886/5146191. * Biros, E. _et al._ A single
nucleotide polymorphism in exon 3 of the kallikrein 1 gene is associated with large but not small abdominal aortic aneurysm. _Atherosclerosis_ 217, 452–457 (2011). Article CAS Google
Scholar * Jabłońska, A. _et al._ Polymorphisms in the IL-6 and TNF-α gene are associated with an increased risk of abdominal aortic aneurysm. _Int. J. Cardiol._ 329, 192–197 (2021). Article
Google Scholar * Zhao, L. _et al._ Correlation between ABCA1 gene polymorphism and aopA-I and HDL-C in abdominal aortic aneurysm. _Med. Sci. Monit._ 22, 172–176 (2016). Article CAS
Google Scholar * Saracini, C. _et al._ Polymorphisms of genes involved in extracellular matrix remodeling and abdominal aortic aneurysm. _J. Vasc. Surg._ 55, 171-179.e2 (2012). Article
Google Scholar * Saratzis, A. _et al._ Association between seven single nucleotide polymorphisms involved in inflammation and proteolysis and abdominal aortic aneurysm. _J. Vasc. Surg._ 61,
1120-1128.e1 (2015). Article Google Scholar * Zuo, S. _et al._ Potential interactions between genetic polymorphisms of the transforming growth factor-β pathway and environmental factors
in abdominal aortic aneurysms. _Eur. J. Vasc. Endovasc. Surg._ 50, 71–77 (2015). Article CAS Google Scholar * Rašiová, M. _et al._ An association between rs7635818 polymorphism located on
chromosome 3p12.3 and the presence of abdominal aortic aneurysm. _Physiol. Res._ 70, 193–201 (2021). Article Google Scholar * Saratzis, A. _et al._ C-reactive protein polymorphism
rs3091244 is associated with abdominal aortic aneurysm. _J. Vasc. Surg._ 60, 1332–1339 (2014). Article Google Scholar * Jabłońska, A. _et al._ TLR2 2029C/T and TLR3 1377C/T and -7C/A
polymorphisms are associated with the occurrence of abdominal aortic aneurysm. _J. Immunol._ 204, 2900–2909 (2020). Article Google Scholar * Wei, Y. _et al._ Association of polymorphisms
on chromosome 9p21.3 region with increased susceptibility of abdominal aortic aneurysm in a Chinese Han population. _J. Vasc. Surg._ 59, 879–885 (2014). Article Google Scholar * Galora, S.
_et al._ Association of rs1466535 LRP1 but not rs3019885 SLC30A8 and rs6674171 TDRD10 gene polymorphisms with abdominal aortic aneurysm in Italian patients. _J. Vasc. Surg._ 61, 787–792
(2015). Article Google Scholar * Crkvenac Gregorek, A., Gornik, K. C., Polancec, D. S. & Dabelic, S. Association of 1166A>C AT1R, -1562C>T MMP-9, ACE I/D, and CCR5Δ32
polymorphisms with abdominal aortic aneurysm in croatian patients. _Genet. Test Mol. Biomark._ 20, 616–623 (2016). Article CAS Google Scholar * Zuo, D. _et al._ Combination of miR-125b
and miR-27a enhances sensitivity and specificity of AFP-based diagnosis of hepatocellular carcinoma. _Tumor Biol._ 37, 6539–6549 (2016). Article CAS Google Scholar * Zhang, Y. _et al._
Association of the KLK1 rs5516 G allele and the ACE D allele with aortic aneurysm and atherosclerotic stenosis. _Medicine_ 95, e5120 (2016). Article CAS Google Scholar * Xiong, J., Wu,
Z., Chen, C. & Guo, W. Chronic obstructive pulmonary disease effect on the prevalence and postoperative outcome of abdominal aortic aneurysms: A meta-analysis. _Sci. Rep._ 6, 25003
(2016). Article ADS CAS Google Scholar * Tripsianis, G. _et al._ Coexpression of IL-6 and TNF-α: Prognostic significance on breast cancer outcome. _Neo_ 61, 205–212 (2014). Article CAS
Google Scholar * Knobloch, J. _et al._ TNFα-induced airway smooth muscle cell proliferation depends on endothelin receptor signaling, GM-CSF and IL-6. _Biochem. Pharmacol._ 116, 188–199
(2016). Article CAS Google Scholar * Jones, B. _et al._ Collagen fibril abnormalities in human and mice abdominal aortic aneurysm. _Acta Biomater._ 110, 129–140 (2020). Article CAS
Google Scholar * Strauss, E., Waliszewski, K., Gabriel, M., Zapalski, S. & Pawlak, A. L. Increased risk of the abdominal aortic aneurysm in carriers of the MTHFR 677T allele. _J. Appl.
Genet._ 44, 85–93 (2003). Google Scholar * Liu, J. _et al._ Hyperhomocysteinaemia is an independent risk factor of abdominal aortic aneurysm in a Chinese Han population. _Sci. Rep._ 6,
17966 (2016). Article ADS CAS Google Scholar * Jabłońska, A. _et al._ Analysis of host Toll-like receptor 3 and RIG-I-like receptor gene expression in patients with abdominal aortic
aneurysm. _J. Vasc. Surg._ 68, 39S-46S (2018). Article Google Scholar * Nishihara, M. _et al._ The role of IL-6 in pathogenesis of abdominal aortic aneurysm in mice. _PLoS ONE_ 12,
e0185923 (2017). Article Google Scholar * Leeper, N. J. _et al._ Loss of CDKN2B promotes p53-dependent smooth muscle cell apoptosis and aneurysm formation. _Arterioscler. Thromb. Vasc.
Biol._ 33, e1–e10 (2013). Article CAS Google Scholar * Bown, M. J. _et al._ Association between the coronary artery disease risk locus on chromosome 9p21.3 and abdominal aortic aneurysm.
_Circul. Cardiovasc. Genet._ 1, 39–42 (2008). Article Google Scholar * Khalil, Y. A., Rabès, J.-P., Boileau, C. & Varret, M. APOE gene variants in primary dyslipidemia.
_Atherosclerosis_ 328, 11–22 (2021). Article CAS Google Scholar * Golledge, J. _et al._ Apolipoprotein E genotype is associated with serum C-reactive protein but not abdominal aortic
aneurysm. _Atherosclerosis_ 209, 487–491 (2010). Article CAS Google Scholar * Tan, C. E. _et al._ APOE polymorphism and lipid profile in three ethnic groups in the Singapore population.
_Atherosclerosis_ 170, 253–260 (2003). Article CAS Google Scholar * Singh, P. P., Singh, M. & Mastana, S. S. APOE distribution in world populations with new data from India and the
UK. _Ann. Hum. Biol._ 33, 279–308 (2006). Article CAS Google Scholar * El-Lebedy, D., Raslan, H. M. & Mohammed, A. M. Apolipoprotein E gene polymorphism and risk of type 2 diabetes
and cardiovascular disease. _Cardiovasc. Diabetol._ 15, 12 (2016). Article Google Scholar * Fedorova, L. _et al._ Analysis of common SNPs across continents reveals major genomic
differences between human populations. _Genes (Basel)_ 13, 1472 (2022). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS B.R lab is supported by the National Institute of
Health (R01 HL146627 and R01HL149927). M.S is funded by the AHA postdoctoral award (907602). L.M. is supported by the German Center for Cardiovascular Research (DZHK), the German Research
Council (DFG) sponsored TRR267, the National Institute of Health (NIH; 1R011HL150359-01), the Bavarian State Ministry of Health and Care through the research project DigiMed Bayern, as well
as the Swedish Heart-Lung-Foundation (20210450) and Swedish Research Council (Vetenkapsrådet, 2019-01577). P.T. is supported by the California Tobacco Related Disease Research Program of the
University of California (T29IR0636) the VA Office of Research and Development (BX-003362-01). KG is supported by the National Institute of Health (R01 HL15674). AUTHOR INFORMATION Author
notes * These authors contributed equally: Chrysania Lim and Muhammad Yogi Pratama. AUTHORS AND AFFILIATIONS * Division of Vascular and Endovascular Surgery, Department of Surgery, New York
University Langone Medical Center, New York, USA Chrysania Lim, Muhammad Yogi Pratama, Cristobal Rivera, Michele Silvestro, Thomas Maldonado & Bhama Ramkhelawon * Department of
Biomedicine, Indonesia International Institute for Life-Sciences (i3L), Jakarta, Indonesia Chrysania Lim & Muhammad Yogi Pratama * Department of Cell Biology, New York University Langone
Medical Center, New York, USA Muhammad Yogi Pratama, Cristobal Rivera, Michele Silvestro & Bhama Ramkhelawon * VA Palo Alto Health Care System, Palo Alto, CA, USA Philip S. Tsao *
Department of Medicine, Stanford University School of Medicine, Stanford, CA, USA Philip S. Tsao * Department of Vascular and Endovascular Surgery, Technical University Munich, Munich,
Germany Lars Maegdefessel * German Center for Cardiovascular Research (DZHK), Partner Site Munich Heart Alliance, Berlin, Germany Lars Maegdefessel * Department of Medicine, Karolinska
Institute, Stockholm, Sweden Lars Maegdefessel * Department of Surgery, University of Michigan, Ann Arbor, MI, USA Katherine A. Gallagher Authors * Chrysania Lim View author publications You
can also search for this author inPubMed Google Scholar * Muhammad Yogi Pratama View author publications You can also search for this author inPubMed Google Scholar * Cristobal Rivera View
author publications You can also search for this author inPubMed Google Scholar * Michele Silvestro View author publications You can also search for this author inPubMed Google Scholar *
Philip S. Tsao View author publications You can also search for this author inPubMed Google Scholar * Lars Maegdefessel View author publications You can also search for this author inPubMed
Google Scholar * Katherine A. Gallagher View author publications You can also search for this author inPubMed Google Scholar * Thomas Maldonado View author publications You can also search
for this author inPubMed Google Scholar * Bhama Ramkhelawon View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS B.R. conceptually developed and
supervised the project. C.L. collected, performed formal analysis, wrote the original draft. M.Y.P. curated, visualized and validated the data, wrote the original draft. C.R. curated and
visualized the data. P.T. performed formal analyses. T.M., K.G., P.T., L.M, M.S, C.R. provided feedback and discussion. CORRESPONDING AUTHOR Correspondence to Bhama Ramkhelawon. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims
in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the
article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your
intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence,
visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Lim, C., Pratama, M.Y., Rivera, C. _et al._ Linking single nucleotide
polymorphisms to signaling blueprints in abdominal aortic aneurysms. _Sci Rep_ 12, 20990 (2022). https://doi.org/10.1038/s41598-022-25144-y Download citation * Received: 12 September 2022 *
Accepted: 25 November 2022 * Published: 05 December 2022 * DOI: https://doi.org/10.1038/s41598-022-25144-y SHARE THIS ARTICLE Anyone you share the following link with will be able to read
this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative