- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Recent investigations of neurological developmental disorders have revealed the Rho-family modulators such as Syde and its interactors as the candidate genes. Although the mammalian
Syde proteins are reported to possess GTPase-accelerating activity for RhoA-family proteins, diverse species-specific substrate selectivities and binding partners have been described,
presumably based on their evolutionary variance in the molecular organization. A comprehensive in silico analysis of Syde family proteins was performed to elucidate their molecular functions
and neurodevelopmental networks. Predicted structural modeling of the RhoGAP domain may account for the molecular constraints to substrate specificity among Rho-family proteins. Deducing
conserved binding motifs can extend the Syde interaction network and highlight diverse but Syde isoform-specific signaling pathways in neuronal homeostasis, differentiation, and synaptic
plasticity from novel aspects of post-translational modification and proteolysis. SIMILAR CONTENT BEING VIEWED BY OTHERS IN THE DEVELOPING CEREBRAL CORTEX: AXONOGENESIS, SYNAPSE FORMATION,
AND SYNAPTIC PLASTICITY ARE REGULATED BY _SATB2_ TARGET GENES Article 26 August 2022 SIN-3 FUNCTIONS THROUGH MULTI-PROTEIN INTERACTION TO REGULATE APOPTOSIS, AUTOPHAGY, AND LONGEVITY IN
_CAENORHABDITIS ELEGANS_ Article Open access 22 June 2022 GENE NETWORK ANALYSIS IDENTIFIES DYSREGULATED PATHWAYS IN AN AUTISM SPECTRUM DISORDER CAUSED BY MUTATIONS IN TRANSCRIPTION FACTOR 4
Article Open access 10 February 2025 INTRODUCTION Numerous adhesion molecules regulate synapse development by recruiting presynaptic and postsynaptic components to the synaptic clefts.
Several RhoGTPase-activating proteins (RhoGAPs) have been implicated as critical regulators of these processes. Synapse-defective-1 protein (Syde1) is an essential regulator of the
presynaptic developmental organization. It’s role was demonstrated by the loss of synaptic recruitment and neuronal pathfinding in genetic studies on Syd1, Syde1 ortholog mutants in _C.
elegans_ and _Drosophila_1,2. Scheiffele’s group also demonstrated a similar function for Syde1 in vertebrates by identifying abnormal synaptic clustering in knockout mice brains and
dissociated neuronal cultures3. The N-terminal disordered region of mouse Syde1 is important for the synaptogenic activity and the RhoGAP domain regulates dendritogenesis. The RhoGAP
activity of _Drosophila_ Syd1 regulates the clustering of Bruchpilot (Brp) as the active zone component3,4, suggesting that the domain function and regulation differ among the species.
Although mouse and _Drosophila_ Syde1 possess RhoGAP activity3,4, no GAP activity has been detected in worm Syde1, which binds to GTP-bound MIG-2/Rac4,5. It has been suggested that SYDE1 is
conserved phylogenetically among worms, _Drosophila_, and mice. Yet, given the molecular organization, the activation of Syde1 by upstream signaling, and the overlap between the functions of
Syde2 and Syde1 in mammalian development are unclear2,3. Another knockout study of Syde1 in mice showed that it regulates trophoblast migration and invasion during the development of
placentas by remodeling the cytoskeleton; it is also involved in the fetal growth regulation during gestation6. Regulation of the Syde1 expression by the transcriptional factor glial cell
missing 1 (GCM1) during placentation suggests a mammalian-specific developmental signaling cascade that contrasts with the impairment of synaptic vesicle docking commonly observed among
their mutant organisms3,4,7. Mouse Syde1 forms a complex with Munc18-1 and liprin-α2 through the N-terminal disordered domain, the presynaptic organizers necessary for synaptogenesis. The
Syde1 binding domain in liprin-α2 mediates the interaction with CASK3,8. Liprin-α2 organizes type IIa LAR-RPTP complex through selective liprin-α isoforms, implicating that Syde1 is
subjected to complex regulation in the presynaptic compartment by the neuronal adhesion machinery9. Transcriptomic meta-analysis revealed Syde1 as one of the differentially expressed genes
in the cortex of autistic patients10. A clinical study indicated that Syde2 is also a causative gene for intellectual disability. Nonsense mutations in Syde2 that theoretically generate a
C-terminal- truncated protein devoid of the RhoGAP domain might cause its degradation, and dysregulation of axonal guidance11; the neuronal function of mammalian Syde2 is unknown. Pleckstrin
homology domain-containing family G member 3 (PLEKHG3) possesses a guanidine nucleotide exchange activity and was identified as a Syde2 complex by high-throughput immunoaffinity
screening12. Furthermore, PLEKHG3, a brain-enriched RhoGEF, is a potential candidate genes for autism13. Syde-family proteins are characterized by the presence of RhoGAP and C2 motif in
domain organization. The regions outside the domains have diversity in their sequences, defined mainly by disordered domains that are poorly characterized14. The N-terminal region of Syde1
mediates autoinhibition of the RhoGAP activity. However, the mechanism of integration of the Syde network in the cell-surface molecular architecture remains obscure3,12. Promiscuity of
substrate specificity and the complex regulation of RhoGAP family proteins have been implicated in several studies using cell-based expression systems6,12,15. The structure–function
relationship of the SYDE-family proteins was elucidated by in silico modeling of structural domain by combining sequence and phylogeny. Additionally, the regulatory regions of the Syde
proteins were identified based on the conserved candidate short linear motifs (SLiMs) located in the disordered domains of soluble proteins. The SLiMs provide regulatory flexible interface
prerequisites for the dynamic assembly of protein complexes. Interestingly, predicted interactors highlight the mammalian Syde network involved in neuronal homeostasis, with
post-translational modification and RhoGAP selectivity as synaptic scaffold molecules. The hypothesis of Syde function as a neuronal signaling hub will be worth investigating experimentally
and clinically in the future. METHODS Sequence feature analysis. Syde1 and Syde2 (UniProt accession No. Q6ZW31 and Q5VT97, respectively) were downloaded from UniProt and NCBI Gene database.
Each protein was aligned using the MAFFT multiple sequence alignment software16 and visualized with JalView17. Domain and sequence features were predicted by using InterproScan18 and
secondary structure was predicted with PSIPRED19. Regions outside the predicted domains were assessed for disordered domain including short linear motifs (SLiMs) by MobiDB20, ELM21, and
D2P222 (Supplementary Table S1). SLiMs play critical roles in many biological processes and we predicted SLiMs localized within the disordered domains that are commonly mapped among
orthologs. Since SLiMs have propensity of random occurrences23, binding motifs are also predicted from ANCHOR program24 analyzing the sequence of disordered region and energy for molecular
interaction. Phosphorylation sites and domain organization were also confirmed by Phosphosite in D2P2 and ScanProsite25. S-palmitoylation site was searched from SwissPalm and manually
curated from PUBMED database26,27. Presence of nuclear export signal was examined from sequence and structural aspects by NESdb database28. Phylogenetic analysis. Sequences of Syde orthologs
were downloaded from UniProt database to reconstruct the phylogeny of the protein family. Seventeen Syde1 and vertebrate Syde2 sequences were retrieved and multiple alignment was conducted
with MAFFT using Jalview. Phylogenetic analysis was performed with MEGA11 by Maximum Likelihood based on the JTT model + G (Gamma distributed sites) with 500 bootstrap replicates29. Homology
modeling. Sequences of RhoGAP and C2 domain of each SYDE proteins were submitted to the homology detection method HHpred and multiple alignment-based detection was conducted with
Swiss-model search, taking into account target-template secondary structure similarity30,31. Models for each domain was built and their model quality was assessed and estimated with
transform-restrained Rosetta32, I-Tasser33, and QMEAN34, respectively and all the graphic images were generated by UCSF Chimera software. Evaluation of predicted model was assessed by
Procheck and ERRAT35 (Supplementary Table S2). Molecular docking. Modelled Syde1 and Syde2-RhoA docking analysis were performed by SwarmDock docking program, one of the best flexible docking
performing programs36. The ability of the programs for reproducing the RhoGTPase-RhoGAP interaction was checked by 5c2k and 5c2j for face-to-face. Optimization of inter-atom energies and
side-chains of modeled Syde structures were performed by EGAD program37 and adjusted PDB files were subjected to the SwarmDock web server in full blind mode. Known Syde interaction analysis
and Syde interactors prediction. BioGrid38 and IntAct39 databases were used for compiling a list of experimentally analyzed Syde interactors and manually curated from the literatures (Table
1, Supplementary Table S1). The binding proteins were annotated with the domain architecture and biological processes involved in physiological aspects by retrieving from the UniProt,
InterPro18, KEGG40 database. Details in the interaction were inferred from PubMed with selected keywords for neuronal function. For prediction of Syde interactors we searched for putative
domains and linear motifs putatively mediating the interactions detected in commonly cell-based model or in neurodevelopmental processes. We assume that a predicted Syde linear motif may be
potential interaction site for a novel interactor if the known binding protein is categorized to a class of protein or contains the motif which is already known to bind the corresponding
sequences in the Syde proteins (Table 1). RESULTS Syde proteins have emerged as critical Rho-family regulators in neuronal and embryonic development. However, their physiological roles and
molecular mechanisms in vertebrates are not well known. Therefore, a computational analysis of the SYDE-family proteins was performed by comparing the primary structures of _Drosophila_ and
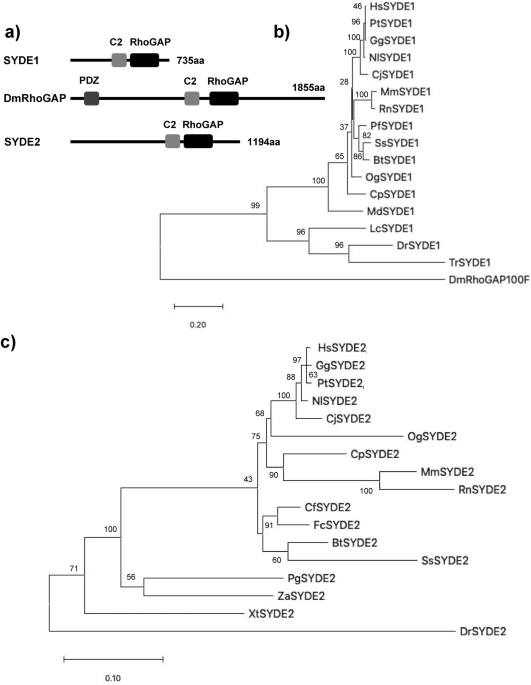
vertebrate orthologs to explore their regulatory and catalytic functions. Homology search using InterproScan identified the C2 and RhoGAP domains that comprised 118 and 207 residues in
SYDE1, and 120 and 216 residues in SYDE2, respectively. These predicted regions are highly conserved between vertebrate Syde1 and Syde2, although each N- or C-terminal sequences is quite
diverse (Figs. 1, 2, 3). In addition to this unique evolutionary variance observed in the disordered Syde domains, crystal structure is not available for any Syde-family protein (Fig. 3a).
Disordered regions are a particular stretch of amino acid patterns or conserved regions with predicted linear motifs that mediate modification by phosphorylation or provide an interface to
specific binding partners for signaling (Fig. 2). Syde1- and 2-binding proteins were examined by BioGrid and IntAct and listed by curating additional relevant literature (Table 1, lower
table). These findings were used to expand for assembling the Syde interaction network (Table 1). The SYDE interactors were selected as the following procedures. Putative interaction motif
scoring e-value lower than 1.0 × 10–2 was selected based on its location within the disordered domain mapped by ELM, D2P2 and Anchor, and phosphorylation site was selected based on the
disordered propensity and predicted site assigned by Phosphosite. Then, the motif conservation through mammalian SYDE1/2 orthologs was also taken account to select the functional interaction
sites. As for evolutional aspects, all the listed motifs were conserved and could be functional in MdSYDE1 (M. domestica), whilst casein kinase II phosphorylation site (MOD_CK2) and Grb2
binding motif (LIG_SH2_Grb2) lacked in LcSYDE1 and FcSYDE2, respectively as indicated in ELM search (Fig. 3b,c). In contrast, ZaSYDE2 was devoid of most of the interaction and
phosphorylation motifs (Figs. 1c and 3c). The putative functional units are described by predicted domains or elements separately as follows. N-TERMINAL REGULATORY REGIONS The intrinsically
disordered region resides in the N-terminus, especially with a more extended form in Syde1 than in Syde2. This portion of mammalian Syde2 proteins shows a relatively low identity between its
paralogs, suggesting functionally divergent roles (Fig. 1). The lower organisms contain a PDZ domain in the middle region with the N-terminal side of C2 domain. However, the corresponding
motif does not reside in the vertebrate Syde proteins (Fig. 1). An alignment of the N-terminus of the vertebrate Syde1 highlights the highly conserved regulatory motifs including the SH2
binding region. But vertebrate Syde2 contains evolutionarily conserved motifs with variable regional sequences outside of the motif. The ATG8 family interacting LIR motif and Crk docking
motifs are located at the N-terminus of Syde1. The 14-3-3zeta and cyclin B/Cdk1-binding site are located within the C-terminal side of the disordered domain (Fig. 3b), and the GSK3
phosphorylation site resides within the linear motif. The Syde1 isoform2 (Q6ZW31-2) lacks 67 residues in the disordered domain but preserves all of the predicted motifs. In contrast, the
N-terminal portion of Syde2 has calcineurin docking LxVP motifs and the motif in the marginal region of the disordered domain was experimentally proven to interact with calcineurin41 (Figs.
2, 3c). Pin1 specifically binds the WW recognition motif represented by phosphorylated serine or threonine residues preceding a proline, and SYDE2 proteins contain a group III/IV motif and
SH3 binding region at the N-terminus. The WW domain for Pin1 recognition and Smek1 binding site for serine/threonine protein phosphatase 4 (PP4)-mediated regulation are conserved among
mammalian Syde2 proteins but not in birds, reptiles, amphibians and fish (Fig. 1, _Z. albicollis_, _P. guttatus_, _X. tropicalis_, and _D. rerio_). Clusters of short-length disordered
domains are located in the middle of Syde2 (Figs. 2, 3). Crk and Grb2 binding motifs reside separately in the disordered regions where highly conserved sequences among vertebrates and
C-terminal variable regions are sequentially organized. THE RHOGAP DOMAINS OF SYDE1/2 AND _DROSOPHILA_ SYD1 The RhoGAP domain of human SYDE1 and SYDE2 proteins show 57.1% sequence homology
with each other. These two RhoGAP domains have similar levels of lower sequence homology to the corresponding domain of _Drosophila_ Syd1 (Q9V7SV) (SYDE1: 46.3%; SYDE2: 45.0%) (Fig. 4a). The
RhoGAP domains of both Syde1 and Syde2 were modeled separately based on tertiary template selection. The HHPred search and potential template candidates by trRosetta and I-Tasser selected
human MgcRacGAP protein (PDB code: 5c2k) for both Syde RhoGAP domains, and ArhGAP2 (N-chimerin PDB code: 3cxl) was selected as the best model for _Drosophila_ Syd1 (Fig. 4b, Supplementary
Fig. S1, Supplementary Table S2). Evaluation of predicted template-based model was performed by Procheck and ERRAT35 (Table 2). Despite the insertion between Syde and template sequences, the
secondary structural elements of the RhoGAP domains in both Syde proteins were superimposed well to MgcRacGAP, which is organized chiefly with helix and turn structures. MgcRacGAP functions
as a GAP activity toward Cdc42 and RhoA and the structures of the docking complexes with MgcRacGAP were experimentally resolved42. The structures of the catalytic core were well
structurally superimposed. Both RhoGAP-Rho interface showed well-matched structures by forming a conserved arginine finger in the GAP domain of Syde proteins bridging the β and γ phosphate
groups of GTP required for its hydrolysis and the cysteine residues located at the substrate interfaces of both proteins are substituted by Ser387 in MgcRacGAP43 (Fig. 4b). Phosphorylation
of serine 387 in MgcRacGAP is known to determine the substrate specificity towards RhoA rather than Rac1 and the replacement of this residue by aspartic acid reduces the RacGAP activity44.
The 3D model quality evaluation of Syde1 and Syde2 revealed closely homologous structures, with QMEAN scores of 0.66 and 0.64, respectively, and _Drosophila_ Syd1 is remotely homologous to
the ArhGAP2 RhoGAP domain with QMEAN score of 0.74. Lower quality regions are located in both Syde1 and Syde2 as insertion between the sixth and seventh α-helices due to partially disordered
loops; otherwise highly positional conservation is shown in the Syde proteins with MgcRacGAP (Supplementary Fig. S1). Both proteins showed similar α-helical distribution patterns, which
could mediate the similar recognition patterns of Rho-family substrates. The SYDE2 isoform (Q5VT97) lacks residues 864–1194, missing RhoGAP and the coiled-coil domains (Fig. 2). Highly
conserved residues among RhoGAP family proteins (Arg436, Lys476, Arg480, Met548, Asn552, and Pro559 in Syde1 numbering) are conserved with MgcRacGAP15,42. Among the vertebrate Syde1 and
Syde2 orthologs, these residues in the Rho-family proteins are highly conserved, although some exceptional cases exist (Syde1 in _D. rerio_ and Syde2 in _O. garnettii_, E7FA87 and H0WYC5,
respectively). Structural analysis with sequence comparison revealed that both Syde1 and Syde2 with MgcRacGAP displayed completely matched orientation of the α-helices that contained the
putative conserved residues interacting with Rho family protein (Fig. 4b, Supplementary Fig. S1). Cdc42-bound MgcRacGAP has different regional structures around the C-terminal α-helices 2
and 3 sequentially flanking the catalytic domain compared to its RhoA-bound model (PDB code: 5c2k and 5c2j). The conserved residues around the arginine finger and α-helix 9 in ArhGAP2 for
Rho-family protein binding are Gly301/Arg304 and Met413/Asn417/Val421/Pro424, respectively, and these residues are conserved with the corresponding residues of DmSyd1 RhoGAP domain15. In the
case of DmSyd1, lower quality of matched region exists in the loop region between α-helix1-2 and 6–7, which is remotely located at the putative interface of the Rho-family protein
(Supplementary Fig. S1). _Drosophila_ Syd1 variable residues in the α-helix 9–10 organizing interface for Rho-family proteins do not match with the corresponding residues in ArhGAP2, but
similar catalytic core EIE sequence within the α-helix2 of ArhGAP2 resides in DmSyd1 as EVE, which is highly conserved among chimerins45. Thus, the substrate interface of DmSyd1 may be
conserved with ArhGAP2 in addition to structural matching. Molecular docking of Syde1 and Syde2 was performed with RhoA and cdc42 using the SwarmDock program to examine the fitting of the
predicted Syde-family proteins to the substrate recognition36. Both predicted Syde1 and Syde2 structures were fitted well to RhoA recognition with a RMSD of 0.760 and 0.657 Å, respectively,
reflecting a highly conserved structural interface with the RhoGAP template (Fig. 5a,b,d). The catalytic arginine finger (Arg436 and Arg854 in Syde1 and 2) was sandwiched by glycine in
P-loop and glutamate in the switch II region of RhoA (Fig. 5e,f). Moreover, a conserved positively charged interface (Lys476 and Lys894; Arg480 and Arg898 in Syde1 and 2) and hydrophilic
regions (Met548 and Met973; Asn552 and Asn977 in Syde1 and Syde2) in the fifth and ninth α-helices, respectively, were close to the RhoA interface (Fig. 5g). RhoB was also selected as SYDE1
binding protein by SwarmDock program with analogy to the predicted SYDE1-RhoA interaction model (Fig. 5c,e,f). No docking model was selected through the molecular docking search for the
interaction of cdc42, RhoC, RhoD, and RhoG with the Syde proteins. THE C2 DOMAIN OF THE SYDE1/2 PROTEINS The InterPro search and comparative sequence analysis indicated that the N- and
C-terminal boundary of the regions of Syde1 C2 domain were less conserved among orthologs than the corresponding regions of Syde2 (Fig. 4c). The human Syde1 and Syde2 proteins show 42.6%
sequence homology in the C2 domain. These C2 domains were modeled separately based on tertiary structure through HHPred search. The best selection was obtained with PLCδ1 protein (PDB code:
1dji) for Syde1 and GIVD cytosolic phospholipase A2 (cPLA2δ, PDB code: 5iz5) for Syde2 (Fig. 4d, Supplementary Fig. S2). The 3D model quality of Syde1 and Syde2 C2 domains was evaluated, and
remotely homologous structures with QMEAN scores of 0.55 and 0.48, were obtained, respectively. The PLCδ1 C2 domain binds Ca2+ in a phosphatidylserine (PS)-dependent manner. The binding
involves Ca2+ binding loops (CBR1-3) located on the same end of the β-sandwich structure with topologically P-family type II46. The CBR3-like region in Syde1 between Asp337 and Arg344
corresponded to Asp708 and Asp714 in PLCδ1 and was oriented toward the outside; other regions including the CBR1 and CBR2 were superimposed well to the PLCδ1 C2 domain. Asp288 and Asp337 in
Syde1 matched the corresponding residues Asp653 and Asp708 as opposed to two Ca2+. However, the other residues in Syde1 were not conserved with those located in the loop region mediating
Ca2+ binding and PS binding in PLCδ147. Both GIVB and GIVD cPLA2 C2 domains have a high affinity for 1-palmitoyl-2-arachidonyl-sn-glycerol-3-phophocholine (PAPC) in the presence of Ca2+. The
basic residues (Lys24/Arg49/Lys52, Lys78 and His44/His82) in the GIVB cPLA2 C2 domain form the interface for binding to PAPC. Although the residues in GIVB are not conserved with GIVD
cPLA2, GIVD possessed a higher affinity for PAPC than GIVB cPLA2 when tested in the in vitro vesicle binding assay48. Moreover, the structural interface organized with these residues of
Syde2 C2 region was superimposed well to GIVD cPLA2 except for the loop region containing Lys24 of GIVD cPLA248. C-TERMINAL REGULATORY REGION IN SYDE1 Syde1 has a C-terminal disordered
domain conserved among mammalian orthologs. The Cys734 palmitoylation site resides at the C-terminus in vertebrate SYDE1 but not in _Drosophila_ and worm Syd126 (Figs. 2, 3b). Reversible
palmitoylation by Golgi-localized DHHC3 allows Syde1 to shuttle between intracellular compartments and plasma membranes. The C-terminal Syde domain contains phosphorylation sites by casein
kinases and the p38 subfamily MAPK and calcineurin interaction sites which are highly conserved among mammalian orthologs. Putative SH3 binding motif is located in the middle of the highly
conserved disordered domain. SYDE1 AND SYDE2 NETWORKS Eleven Syde1 and eight Syde2 interactors were manually curated, and four SYDE1 and two SYDE2 binding proteins were directly retrieved
from literatures (Fig. 6, Table1). Most of interactors belonged to a category of proteins that recognized a conserved linear motif located in the disordered regions. XPO1 is indicated as an
Syde1 complex by high-throughput evidence, and two regions are predicted as the potential nuclear export signatures by XPO1 in Syde1 (Table1). S-palmitoylation of Casein kinase 1γ mediates
phosphorylation and intracellular transport of Lyn at Golgi apparatus49. Golgi-localized DHHC3 specifically palmitoylates Syde1 and casein kinase 1γ might phosphorylate palmitoylated
SYDE126. ELM search selected the SH2 binding motif YINS in SYDE2 that was matched with typical Grb2 binding motif validated by pull-down experiment50,51. MAPK binding D-motif resides at the
C-terminal region in SYDE1, and the sequence has similarity with MKK-type phosphorylation motifs catalyzed by p3852. ELM search hit two 14-3-3 binding motifs in SYDE1, and several
interactors such as Pctaire1 kinase possess similar RLSLP sequence mediating the interaction53. 14-3-3ζs are dimeric with diagonal symmetry wherein two phosphate-binding sites lie in
diagonally opposite position when they form a complex with kinases such as the CaMK and AGC family, including PKA. Cin85 is enriched in the brain and localized in the postsynapse, and
associated with endocytosis regulators such as endophilins and potential SH3 interaction sites were indicated in SYDE1 (Fig. 3b). SYDE1 possesses RxL docking motif for cyclin binding
(DOC_Cyclin) at the N-terminal disordered domain. The sequence was matched to consensus Cdk1/CyclinB complex binding motif identified by Arg/Lys-scanning oriented peptide libraries54. The
LIR (LC3-interacting region) motif consists of core W/F/I-X-X-ɸ (ɸ: L, I, F) sequence required for selective autophagy. Structural studies indicate the following key features: the aromatic
and hydrophobic residues interact with distinct hydrophobic pockets of ubiquitin-like modifiers and an acidic or a phosphorylated Ser/Thr immediately upstream of the core sequence promotes
the interaction55. The LIR motif in SYDE1 satisfied the definition that was categorized as F-LIR based on the aromatic residue. Noncanonical class I SH3 binding motif RQQVSPP resides at the
N-terminus of SYDE2 and brain-specific ArhGAP32 contains the similar motifs that mediate interaction with Crk56,57. GSK3 antagonizes Syd1 and liprin ortholog Syd2 pathway in synaptic vesicle
clustering in _Drosophila_ and sequence alignment suggested that the potential conserved phosphorylation sites lie within the linear motif in both the SYDE1 and SYDE2 as described58 (Table
1). Conserved Crk binding YNPIP sites reside in both SYDE1 and SYDE2, and this Tyr phosphorylated motif shows similarity in the patterns with Crk binding sites in Dab1 and other
substrates59. Calcineurin binding πɸLxVP motif containing polar and polar/hydrophobic residues at position − 2 (π) and − 1 (ɸ), respectively, was selected based on ELM search and its
location in SLiM domain41. SYDE2 contained SFLRPP and RVLSVP sequence matched the πɸLxVP motif by ELM search although the motif selected by ELM in SYDE1 did not fulfill the definition. ELM
search hit Smek4 as the SYDE2 interactor and centrobin fragment containing FRVP showed the similar motif patterns and high affinity to PP4 by ITC and coIP experiments60
(http://slim.icr.ac.uk/motifs/pp4/). As for Pin1 binding motif, multiple S/T-P motifs mediate the interaction in the case of gephyrin and PSD95, and one S/T-P motif sufficiently functions as
Pin1-mediated neuroligin2 binding to gephyrin61,62. SYDE2 contains the typical S/T-P motif similar to the Pin1 binding region in the synaptic components at the N-terminus and other putative
Pin interaction sites (DOC_WW_Pin1_4) were omitted due to low probability in ELM score. Fbw7 binding consensus degron is represented by ɸXɸɸɸTPPXS (ɸ: hydrophobic residue, X: any amino
acid). Phosphorylated Thr and Ser residues interact with WD domain of Fbw7 and these residues were conserved in Fbw7 binding motif in SYDE263. ArhGAP28 has actin-associated RhoAGAP activity
with no Rac1 and Cdc42 selectivity; PLEKHG3 functions as RhoGEF in the brain. Both Syde1 and Syde2 have lower GAP activity toward Cdc42 than RhoA and Rac1, which was consistent with our
prediction model12 (Fig. 5). Most Syde1 interactors such as liprinα2 and Munc18 are categorized as regulators of synaptogenesis including active zone homeostasis. In comparison, Syde2
binding proteins are primarily involved in neuronal differentiation signaling during embryonic brain development. DISCUSSION Rho GTPases are important for cell adhesion, motility,
cytokinesis and contractile responses due to growth factor-induced reorganization of the actin cytoskeleton. Syde1 regulates synaptic exocytosis and its RhoGAP activity is a prerequisite for
dendritogenesis. Although the in vivo function of mammalian Syde2 remains unknown, Syde2 isoform2 lacks most of the RhoGAP domain at the C-terminus, suggesting complex regulation of the
Syde proteins. The homozygous nonsense C.1544C > G mutation (p.(Ser515*) homo) in Syde2 found in an intellectual disability patient generated a C-terminal truncated transcript devoid of
the entire C2 and RhoGAP domain, suggesting a specific role for Syde2 in brain development11. It was reported that the C2 domain of Syde1 in addition to the N-terminal disordered domain has
an autoinhibitory role for RhoGAP activity3; Therefore, the current study may provide the vertebrate-specific Rho-family substrate recognition by the Syde1 RhoGAP domain. Prediction and the
structural quality of RhoGAP domains listed in Supplementary Table S2 satisfied the criteria as the potential template listed by trRosetta and I-Tasser. As for the C2 domain of SYDE1 and
SYDE2, the potential templates were also selected in trRosetta predicting the structure by multiple sequential alignments as well as calculation of inter-residue orientation, but not in
I-Tasser. The predicted model of RhoGAP in Syde1 and Syde2 applied to MgcRacGAP enabled the comparison of the structural interface for its RhoA and Cdc42 (PDB:5c2j), which have different
regional conformations that may constrain substrate recognition. A flexible molecular docking search by SwarmDock selected the interaction of both predicted Syde1 and Syde2 with RhoA but not
cdc42. The result was consistent with the substantial and extremely low GAP activity toward cdc42 in the RhoGAP assay system using FRET sensors3,12. Interestingly, RhoB was also selected as
the SYDE1 substrate although the C-terminal regions of RhoA and RhoB show difference in primary sequences to each other. Neuronal regulation of RhoB activity by RhoGAP remains unknown and
RhoB is known to regulate dendritic morphology and synaptic plasticity64. The functional Syde protein network was expanded and categorized by integrating structure- and interactor-based
prediction with high-throughput evidences: this included inferring the binding partners conserved in lower organisms. Most short linear motifs have a high chance of random occurrence and low
specificity. However, stringent criteria were used for selection of interaction sites65. A predicted binding site and post-translationally modified residues must be conserved among
orthologs and located in a disordered region. Therefore, motif patterns and phosphorylation sites were inferred through several methods20,21,22,25. As for the mammalian Syde proteins, the
RhoGAP activity may be highly regulated by phosphorylation/de-phosphorylation and membrane targeting. Both Syde proteins may regulate synaptic plasticity and dendritic morphology with
cooperative Ca2+-dependent synaptic regulation by Munc18-1 and liprin-α2, Palmitoylation regulates Syde1-specific membrane targeting and casein kinase Iγ might cooperatively regulate
RhoGTPase’s activity at plasma membrane or presynaptic compartment26,27,49. Liprin-α2 and Munc18-1 are known to interact with the Syde1 N-terminal disorder domain and organize active zone
complexes in presynaptic regions3 presumably in a competitive manner with RPTP-liprinα interaction9. Optimal 14-3-3 binding sites satisfy basic residues in positions − 3 to − 5 relative to
the phosphorylated site and kinases in the AGC family, such as PKA/PKG/PKC and CaMK subfamilies, are most commonly implicated in the phosphorylation of the binding sites53,66. Therefore,
Ca2+-dependent scaffold complex formation might be involved in the Syde1 interaction network. Crk and CIN85 containing SH2-SH3 and three SH3 domains, respectively, are known to regulate pre-
and post-synaptic architecture colocalized with PSD95, synaptophysin, and Dock-18067. Syde1 and MgcRacGAP were identified by CIN85 interactors through the SH3-C region recognizing the
PX(P/A)XXR motif, and Syde1 possesses potential SH3 binding sites as the consensus sequence matched with a variety of sequence patterns68,69. The predicted Syde1 interactor cyclin B/Cdk1 is
involved in embryonic neurogenesis with the fate competency in opposition to cyclin D function70. Cyclins have cross-selectivity and Cyclin D recognizes RxL motif, therefore, SYDE1 might
also interact with cyclinD. Casein kinase 2 localizes to the cytosol or membrane in neurons and phosphorylates PACSIN1, thus shutting off Rac1 hydrolysis in the process of dendritic spine
formation71. Neuronal Pin1’s prolyl cis/trans isomerase activity mediates NMDA dissociation from the PSD95 complex and structural arrangement of voltage-gated K+ channels upon neuronal
excitability72. Syde2’s RhoGAP activity might be regulated by Syde2-specific Pin1 binding through the N-terminal disorder region in contrast to the autoinhibitory conformational changes in
SYDE1. The Smek1 and PP4 catalytic subunit PP4c are required for neurogenesis73 and Grb2 is recruited to Ca2+-dependent kinase Pyk2 and ErbB2/3 receptors upon Nrg1 signaling during cerebral
cortical development74. PLEKHG3 is highly expressed in the brain and microdeletion, including the gene, which is known to cause mild mental retardation with spherocytosis13,75. Crk is known
to be recruited to Dab1 upon Reelin signaling76 and may recognize Syde1 and Syde2 proteins through the SH2 domain. Liprin2 converging with PP2A phosphatase are regulated by GSK3β
phosphorylation as downstream signaling cascades of Syd1 during neurogenesis in _Drosophila_58. Predicted phosphorylation by GSK3β may be required for both mammalian Sydes signaling involved
in the exocytosis of synaptic vesicles (Table 1)58. The LxVP binding pocket interacts with calcineurin for comprehensive analysis of its substrates41. The current search additionally
predicted the N-terminal calcineurin binding motif in the Syde2 disordered domain. Linkage of calcineurin with Syde2 implies synaptic activity-dependent AMPA and NMDA receptor function77.
Lastly, Syde-family proteins may be regulated by autophagy and proteasomal degradation during neuronal development. LC3/GABARAP protein noncovalently binds to the canonical ATG8
family-interacting motif (AIM), a core motif consisting of W/F/I-X-X-L/I/V, where any two amino acids flank the aromatic and hydrophobic amino acid during the process of phagophore formation
in the autophagy pathway63. Syde1 might be regulated by ATG8-family-mediated signaling through the AIM motif by coupling with a reversible DHHC3/7-mediated palmitoylation. Interestingly
Syde2 also contains degradation signals with reflecting the relatively lower Syde2 expression than Syde1 in the developing brain (www.brainspan.org). Fbw7 is one of the listed SCF-E3 ligases
involved in neuronal differentiation78,79 and the regulation of astrocyte generation, and proteasome-mediated degradation of Syde2 may be involved in its regulation of RhoGTPase during
neurodevelopment. REFERENCES * Hallam, S. J., Goncharov, A., McEwen, J., Baran, R. & Jin, Y. Syd-1, a presynaptic protein with PDZ, C2 and rhoGAP-like domains specifies axon identity in
_C. elegans_. _Nat. Neurosci._ 5, 1137–1146 (2002). CAS PubMed Google Scholar * Xu, Y. & Quinn, C. C. SYD-1 promotes multiple developmental steps leading to neuronal connectivity.
_Mol. Neurobiol._ 53, 6768–6773 (2016). CAS PubMed Google Scholar * Wentzel, C. _et al._ mSYD1A, a mammalian synapse-defective-1 protein, regulates synaptogenic signaling and vesicle
docking. _Neuron_ 78, 1012–1023 (2013). CAS PubMed PubMed Central Google Scholar * Spinner, M. A., Walla, D. A. & Herman, T. G. _Drosophila_ Syd-1 has RhoGAP activity that is
required for presynaptic clustering of Bruchpilot/ELKS but not Neurexin-1. _Genetics_ 208, 705–716 (2018). CAS PubMed Google Scholar * Xu, Y., Taru, H., Jin, Y. & Quinn, C. C. Syd-1c,
Unc-40 (DCC) and Sax-3 (Robo) function interdependently to promote axon guidance by regulating the MIG-2 GTPase. _PLoS Genet._ 11, e1005185 (2015). PubMed PubMed Central Google Scholar *
Lo, H. F. _et al._ Association of dysfunctional synapse defective 1 (SYDE1) with restricted fetal growth—SYDE1 regulates placental cell migration and invasion. _J. Pathol._ 241, 324–336
(2017). CAS PubMed Google Scholar * Ramesh, N. _et al._ Antagonistic interaction between two neuroligins and coordinate pre- and postsynaptic assembly. _Curr. Biol._ 31, 1–15 (2021).
Google Scholar * Wei, Z. _et al._ Liprin-mediated large signaling complex organization revealed by the liprinα/Cask and liprinα/liprinβ complex structures. _Mol. Cell_ 43, 586–598 (2011).
CAS PubMed Google Scholar * Wakita, M. _et al._ Structural insights into selective interaction between type IIa receptor protein tyrosine phosphatases and liprin-α. _Nat. Commun._ 11,
649–658 (2020). ADS CAS PubMed PubMed Central Google Scholar * Rahman, M. R. _et al._ Comprehensive analysis of RNA-seq gene expression profiling of brain transcriptomes reveals novel
genes, regulators, and pathways in autism spectrum disorder. _Brain Sci._ 10, 747 (2020). CAS PubMed Central Google Scholar * Anazi, S. _et al._ Clinical genomics expands the morbid
genome of intellectual disability and offers a high diagnostic yield. _Mol. Psychiatry_ 22, 615–624 (2016). PubMed Google Scholar * Müller, P. M. _et al._ Systems analysis of RhoGEF and
RhoGAP regulatory proteins reveals spatially organized Rac1 signaling from integrin adhesions. _Nat. Cell Biol._ 22, 498–511 (2020). PubMed Google Scholar * Griswold, A. J. _et al._ A de
novo 1.5Mb microdeletion on chromosome 14q23.2–23.3 in a patient with autism and spherocytosis. _Autism Res._ 4, 221–227 (2011). PubMed PubMed Central Google Scholar * Mosaddeghzadeh, N.
& Ahmadian, M. R. The Rho family GTPases: Mechanisms of regulation and signaling. _Cells_ 10, 1831 (2021). CAS PubMed PubMed Central Google Scholar * Amin, E. _et al._ Deciphering
the molecular and functional basis of RhoGAP family proteins. _J. Biol. Chem._ 291, 20353–20371 (2016). CAS PubMed PubMed Central Google Scholar * Katoh, K., Rozewicki, J. & Yamada,
K. D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. _Brief. Bioinform._ 20, 1160–1166 (2019). CAS PubMed Google Scholar * Waterhouse,
A. M., Procter, J. B., Martin, D. M. A., Clamp, M. & Barton, G. J. Jalview Version2—A multiple sequence alignment editor and analysis workbench. _Bioinformatics_ 25, 1189–1191 (2009).
CAS PubMed PubMed Central Google Scholar * Blum, M. _et al._ The InterPro protein families and domains database: 20 years on. _Nucleic Acids Res._ 49, D344-354 (2020). PubMed Central
Google Scholar * Buchan, D. W. A., Minneci, F., Nugent, T. C. O., Bryson, K. & Jones, D. T. Scalable web services for the PSIPRED protein analysis workbench. _Nucleic Acids Res._ 41,
W349–W357 (2013). PubMed PubMed Central Google Scholar * Potenza, E., Di Domenico, T., Walsh, I. & Tosatto, S. C. E. MobiDB 2.0: An improved database of intrinsically disordered and
mobile proteins. _Nucleic Acid. Res._ 43, D315–D320 (2015). CAS PubMed Google Scholar * Dinkel, H. _et al._ ELM 2016-data update and new functionality of eukaryotic linear motif resource.
_Nucleic Acids Res._ 44, D294–D300 (2016). ADS CAS PubMed Google Scholar * Oates, M. E. _et al._ D2P2: Database of disordered protein predictions. _Nucleic Acids Res._ 41, D508–D516
(2012). PubMed PubMed Central Google Scholar * Tompa, P., Davey, N. E., Gibson, T. J. & Babu, M. M. A million peptide motifs for the molecular biologist. _Mol. Cell_ 55, 161–169
(2014). CAS PubMed Google Scholar * Mészáros, B., Simon, I. & Dosztányl, Z. Prediction of protein binding regions in disordered proteins. _PLoS Comput. Biol._ 5, e1000376 (2009). ADS
PubMed PubMed Central Google Scholar * de Castro, E. _et al._ ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins.
_Nucleic Acids. Res._ 34, W362-365 (2006). PubMed PubMed Central Google Scholar * Oku, S., Takahashi, N., Fukata, Y. & Fukata, M. In silico screening for palmitoyl substrates reveals
a role for DHHC1/3/10(zDHHC1/3/11)-mediated neurochondrin palmitoylation in its targeting to rab5-positive endosomes. _J. Biol. Chem._ 288, 19816–19829 (2013). CAS PubMed PubMed Central
Google Scholar * Blanc, M. _et al._ SwissPalm: Protein palmitoylation database. _F1000 Res._ 4, 26 (2015). Google Scholar * Xu, D., Farmer, A., Collett, G., Grishin, N. V. & Chook, Y.
M. Sequence and structural analyses of nuclear export signals in the NESb database. _Mol. Biol. Cell_ 23, 3677–3693 (2012). CAS PubMed PubMed Central Google Scholar * Tamura, K.,
Stecher, G. & Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. _Mol. Biol. Evol._ 38, 3022–3027 (2021). CAS PubMed PubMed Central Google Scholar * Söding, J.,
Biegert, A. & Lupas, A. N. The HHpred interactive server for protein homology detection and structure prediction. _Nucleic Acids Res._ 33, W244–W248 (2005). PubMed PubMed Central
Google Scholar * Schwede, T., Kopp, J., Guex, N. & Peitsch, M. C. SWISS-MODEL: An automated protein homology-modeling server. _Nucleic Acids Res._ 31, 3381–3385 (2003). CAS PubMed
PubMed Central Google Scholar * Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. _Nat. Protoc._ 5, 725–738
(2010). CAS PubMed PubMed Central Google Scholar * Yang, J. _et al._ Improved protein structure prediction using predicted interresidue orientations. _Proc. Natl. Acad. Sci._ 117,
1496–1503 (2020). CAS PubMed PubMed Central Google Scholar * Benkert, P., Künzli, M. & Schwede, T. QMEAN server for protein model quality estimation. _Nucleic Acids Res._ 37,
W510–W514 (2009). CAS PubMed PubMed Central Google Scholar * Messaoudi, A., Belguith, H. & Hamida, B. Homology modeling and virtual screening approaches to identify potent inhibitors
of VEB-1 β-lactamase. _Theor. Biol. Med. Model._ 10, 22 (2013). CAS PubMed PubMed Central Google Scholar * Torchala, M., Moal, I. H., Chaleil, R. A. G., Fernandez-Recio, J. & Bates,
P. A. SwarmDock: A server for flexible protein-protein docking. _Bioinformatics_ 29, 807–809 (2013). CAS PubMed Google Scholar * Pokala, N. & Handel, T. M. Protein–protein complex
affinities, models for the unfolded state, and negative design of solubility and specificity. _J. Mol. Biol._ 347, 203–227 (2005). CAS PubMed Google Scholar * Chatraryamontri, A. _et al._
The BioGRID interaction database: 2015 update. _Nucleic Acids Res._ 43, D470–D478 (2015). CAS Google Scholar * Orchard, S. _et al._ The MintAct project—IntAct as a common curation
platform for 11 molecular interaction databases. _Nucleic Acids Res._ 42, D358-363 (2014). CAS PubMed Google Scholar * Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y. & Morishima,
K. KEGG: New perspectives on genomes, pathways, diseases and drugs. _Nucleic Acids Res._ 45, D353–D351 (2017). CAS PubMed Google Scholar * Sheftic, S., Page, R. & Peti, W.
Investigating the human calcineurin interaction network using the πϕLxVP SLiM. _Sci. Rep._ 6, 38920 (2016). ADS CAS PubMed PubMed Central Google Scholar * Matsuura, A. & Lee, H. H.
Crystal structure of GTPase-activating domain from human MgcRacGAP. _Biochem. Biophys. Res. Commun._ 435, 367–372 (2013). CAS PubMed Google Scholar * Mishima, M. & Glotzer, M.
Cytokinesis: A logical GAP. _Curr. Biol._ 13, R589–R591 (2003). CAS PubMed Google Scholar * Minoshima, Y. _et al._ Phosphorylation by AuroraB converts MgcRacGAP to a RhoGAP during
cytokinesis. _Dev. Cell_ 4, 549 (2003). CAS PubMed Google Scholar * Wang, H. _et al._ Phospholipase Cγ/diacylglycerol-dependent activation of β2-chimaerin restricts EGF-induced Rac
signaling. _EMBO J._ 25, 2062–2074 (2006). CAS PubMed PubMed Central Google Scholar * Corbalan-Garcia, S. & Gomez-Fernández, J. C. Signaling through C2 domain: More than one lipid
target. _Biochim. Biophys. Acta_ 1838, 1536–1547 (2014). CAS PubMed Google Scholar * Lomansey, J. W., Cheng, H. F., Roffler, S. R. & King, K. Activation of phospholipase Cδ1 through
C2 domain by a Ca2+-enzyme-phosphatidylserine ternary complex. _J. Biol. Chem._ 274, 21995–22001 (1999). Google Scholar * Ghomashchi, F. _et al._ Interfacial kinetic and binding properties
of mammalian group IVB phospholipaseA2 (cPLA2β) and comparison with the other cPLA2 isoforms. _J. Biol. Chem._ 285, 36100–36111 (2010). CAS PubMed PubMed Central Google Scholar *
Kinoshita-Kikuta, E. _et al._ Protein N-myristoylation-dependent phosphorylation of serine 13 of tyrosine kinase Lyn by casein kinase 1γ at the Golgi during intracellular protein traffic.
_Sci. Rep._ 10, 16273 (2020). CAS PubMed PubMed Central Google Scholar * Tinti, M. _et al._ The SH2 domain interaction landscape. _Cell Rep._ 3, 1293–1305 (2013). CAS PubMed PubMed
Central Google Scholar * Liu, B. A. _et al._ SH2 domains recognize contextual peptide sequence information to determine selectivity. _Mol. Cell. Proteom._ 9(11), 2391–2404 (2010). CAS
Google Scholar * Garai, A. _et al._ Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. _Sci. Signal._ 5, ra74.
https://doi.org/10.1126/scisignal.2003004 (2013). Article CAS Google Scholar * Johnson, C. _et al._ Bioinformatics and experimental survey of 14-3-3-binding sites. _Biochem. J._ 427,
69–78 (2010). CAS PubMed Google Scholar * Suzuki, K. _et al._ Identification of non-Ser/Thr-Pro consensus motifs for Cdk1 and their roles in mitotic regulation of C2H2 zinc finger
proteins and Ect2. _Sci. Rep._ 5, 7929 (2015). CAS PubMed PubMed Central Google Scholar * Rogov, V., Dötsch, V., Johansen, T. & Kirkin, V. Interaction between autophagy receptors and
ubiquitin-like proteins. _Mol. Cell_ 53, 167–178 (2014). CAS PubMed Google Scholar * Teyra, J. _et al._ Comprehensive analysis of the human SH3 domain family reveals a wide variety of
non-canonical specificities. _Structure_ 25, 1598–1610 (2017). CAS PubMed Google Scholar * Moon, S. Y., Zang, H. & Zheng, Y. Characterization of a brain-specific Rho GTPase-activating
protein, p200RhoGAP. _J. Biol. Chem._ 278, 4151–4159 (2003). CAS PubMed Google Scholar * Li, L., Tian, X., Zhu, M., Bulgari, D. & Böhme, M. A. Drosophila Syd-1, Liprin-α, and protein
phosphatase 2A B′ subunit wrd function in a linear pathway to prevent ectopic accumulation of synaptic materials in distal axons. _J. Neurosci._ 34, 8474–8487 (2014). PubMed PubMed Central
Google Scholar * Ballif, B. A. _et al._ Activation of a Dab1/CrkL/C3G/Rap1 pathway in Reelin-stimulated neurons. _Curr. Biol._ 14, 606–610 (2004). CAS PubMed Google Scholar * Ueki, Y.
_et al._ A consensus binding motif for the PP2 protein phosphatase. _Mol. Cell_ 76, 953–964 (2019). CAS PubMed PubMed Central Google Scholar * Antonelli, R. _et al._ Pin1-dependent
signaling negatively affects GABAergic transmission by modulating neuroligin2/gephyrin interaction. _Nat. Commun._ 5, 5066 (2014). ADS CAS PubMed Google Scholar * Zita, M. M. _et al._
Post-phosphorylation propyl isomerization of gephyrin represents a mechanism to modulated glycine receptors function. _EMBO J._ 26, 1761–1771 (2007). PubMed Google Scholar * Hao, B.,
Oehlmann, S., Sowa, M. E., Harper, J. W. & Pavletich, N. P. Structure of a Fbw7-Skp1-Cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. _Mol.
Cell_ 26, 131–143 (2007). CAS PubMed Google Scholar * McNair, K. _et al._ A role for RhoB in synaptic plasticity and the regulation of neuronal morphology. _J. Neurosci._ 30, 3508–3517
(2010). CAS PubMed PubMed Central Google Scholar * Gibson, T. J., Dinkel, H., Roey, K. V. & Diella, F. Experimental detection of short regulatory motifs in eukaryotic proteins: tips
for good practice as well as for bad. _Cell Commun. Signal._ 13, 42 (2015). PubMed PubMed Central Google Scholar * Toyo-oka, K. _et al._ 14-3-3ε and ζ regulate neurogenesis and
differentiation of neuronal progenitor cells in the developing brain. _J. Neurosci._ 34, 12168–12181 (2014). CAS PubMed PubMed Central Google Scholar * Shimokawa, N. _et al._ CIN85
regulates dopamine receptor endocytosis and governs behavior in mice. _EMBO J._ 29, 2421–2432 (2010). CAS PubMed PubMed Central Google Scholar * Rouka, E. _et al._ Differential
recognition preferences of the three src homology 3 (SH3) domains from the adaptor CD2-associated protein (CD2AP) and direct association with Ras and Rab interactors 3 (RIN3). _J. Biol.
Chem._ 290, 25275–25292 (2015). CAS PubMed PubMed Central Google Scholar * Havrylov, S., Rzhepetskyy, Y., Malinowska, A., Drobot, L. & Redowicz, M. J. Proteins recruited by SH3
domains of Ruk/CIN85 adaptor identified by LC-MS/MS. _Proteome Sci._ 7, 21 (2009). PubMed PubMed Central Google Scholar * Hagey, D. W. _et al._ Cyclin-B1/2 and -D1 act in opposition to
coordinate cortical progenitor self-renewal and lineage commitment. _Nat. Commun._ 11, 2898 (2020). ADS CAS PubMed PubMed Central Google Scholar * Schael, S. _et al._ Casein kinase 2
phosphorylation of protein kinase C and casein kinase 2 substrate in neurons (PASCIN)1 protein regulates neuronal spine formation. _J. Biol. Chem._ 288, 9393–9312 (2013). Google Scholar *
Antonelli, R. _et al._ Pin1 modulates the synaptic content of NMDA receptors via proylisomerization of PSD95. _J. Neurosci._ 36, 5437–5447 (2016). CAS PubMed PubMed Central Google Scholar
* Lyu, J. _et al._ Protein phosphatase 4 and smek complex negatively regulate Par3 and promote Neuronal Differentiation of neural stem/progenitor cells. _Cell Rep._ 5, 593–600 (2013). CAS
PubMed Google Scholar * Mei, L. & Xiong, W. C. Neuregulin 1 in neural development, synaptic plasticity and schizophrenia. _Nat. Rev. Neurosci._ 9, 437–452 (2008). CAS PubMed PubMed
Central Google Scholar * Lybaek, H., Oyen, N., Fauske, L. & Houge, G. A 2.1 Mb deleteon adjacent but distal to 14q21q23 paracentric inversion in a family with spherocytosis and severe
learning difficulties. _Clin. Genet._ 74, 553–339 (2008). CAS PubMed Google Scholar * Sekine, K. _et al._ Reelin controls neuronal positioning by promoting cell-matrix adhesion via
inside-out activation of integrin α5β1. _Neuron_ 76, 353–369 (2012). CAS PubMed PubMed Central Google Scholar * Sanderson, J. L., Gorski, J. A. & Dell’Acqua, M. L. NMDA
receptor-dependent LTD requires transient synaptic incorporation of Ca2+-permeable AMPARs mediated by AKAP150-anchored PKA and calcineurin. _Neuron_ 89, 1000–1015 (2016). CAS PubMed PubMed
Central Google Scholar * Matsumoto, A. _et al._ Fbxw7-dependent degradation of notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. _J.
Biol. Chem._ 286, 13753–13764 (2011). Google Scholar * Hoeck, J. D. _et al._ Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. _Nat. Neurosci._
13, 1365–1372 (2010). CAS PubMed Google Scholar * Kirli, K. _et al._ A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. _Elife_ 4, e11466
(2015). PubMed PubMed Central Google Scholar * Owald, D. _et al._ Cooperation of Syd-1 with neurexin synchronizes pre- with postsynaptic assembly. _Nat. Neurosci._ 15, 1219–1226 (2012).
CAS PubMed Google Scholar * Owald, D. _et al._ A Syd-1 homologue regulates pre- and postsynaptic maturation in _Drosophila_. _J. Cell Biol._ 188, 565–579 (2010). CAS PubMed PubMed
Central Google Scholar Download references AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Genetics, Institute for Developmental Research, Aichi Developmental Disability
Center, 713-8 Kamiya-cho, Kasugai, Aichi, 480-0392, Japan Zen Kouchi * Laboratory of Bioinformatics, School of Life Sciences, Tokyo University of Pharmacy and Life Sciences, 1432-1
Horinouchi, Hachioji, 192-0392, Japan Masaki Kojima Authors * Zen Kouchi View author publications You can also search for this author inPubMed Google Scholar * Masaki Kojima View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Z.K. designed the study, constructed the sequence and interaction analysis, wrote the manuscripts. Z.K.
and M.K. performed molecular modeling and analyzed the structural data. CORRESPONDING AUTHOR Correspondence to Zen Kouchi. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits
use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the
Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless
indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory
regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kouchi, Z., Kojima, M. Function of SYDE C2-RhoGAP family as signaling hubs for
neuronal development deduced by computational analysis. _Sci Rep_ 12, 4325 (2022). https://doi.org/10.1038/s41598-022-08147-7 Download citation * Received: 19 September 2021 * Accepted: 02
March 2022 * Published: 12 March 2022 * DOI: https://doi.org/10.1038/s41598-022-08147-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative