- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Propagation of the chromatin landscape across cell divisions is central to epigenetic cell memory. Mechanistic analysis of the interplay between DNA replication, the cell cycle, and
the epigenome has provided insights into replication-coupled chromatin assembly and post-replicative chromatin maintenance. These breakthroughs are critical for defining how proliferation
impacts the epigenome during cell identity changes in development and disease. Here we review these findings in the broader context of epigenetic inheritance across mitotic cell division.
Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54
other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and
online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes
which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY
OTHERS PARENTAL NUCLEOSOME SEGREGATION AND THE INHERITANCE OF CELLULAR IDENTITY Article 26 January 2021 SYMMETRIC INHERITANCE OF PARENTAL HISTONES GOVERNS EPIGENOME MAINTENANCE AND EMBRYONIC
STEM CELL IDENTITY Article Open access 04 September 2023 MITOTIC CHROMATIN MARKING GOVERNS THE SEGREGATION OF DNA DAMAGE Article Open access 16 January 2025 REFERENCES * Bannister, A. J.
& Kouzarides, T. Regulation of chromatin by histone modifications. _Cell Res._ 21, 381–395 (2011). CAS PubMed PubMed Central Google Scholar * Hauer, M. H. & Gasser, S. M.
Chromatin and nucleosome dynamics in DNA damage and repair. _Genes Dev._ 31, 2204–2221 (2017). CAS PubMed PubMed Central Google Scholar * Allshire, R. C. & Madhani, H. D. Ten
principles of heterochromatin formation and function. _Nat. Rev. Mol. Cell Biol._ 19, 229–244 (2018). CAS PubMed Google Scholar * Boyle, A. P. et al. High-resolution mapping and
characterization of open chromatin across the genome. _Cell_ 132, 311–322 (2008). CAS PubMed PubMed Central Google Scholar * Mikkelsen, T. S. et al. Genome-wide maps of chromatin state
in pluripotent and lineage-committed cells. _Nature_ 448, 553–560 (2007). CAS PubMed PubMed Central Google Scholar * Kurimoto, K. & Saitou, M. Epigenome regulation during germ cell
specification and development from pluripotent stem cells. _Curr. Opin. Genet. Dev._ 52, 57–64 (2018). CAS PubMed Google Scholar * Heard, E. & Martienssen, R. A. Transgenerational
epigenetic inheritance: myths and mechanisms. _Cell_ 157, 95–109 (2014). CAS PubMed PubMed Central Google Scholar * Flavahan, W. A., Gaskell, E. & Bernstein, B. E. Epigenetic
plasticity and the hallmarks of cancer. _Science_ 357, eaal2380 (2017). PubMed PubMed Central Google Scholar * Kornberg, R. D. Chromatin structure: a repeating unit of histones and DNA.
_Science_ 184, 868–871 (1974). CAS PubMed Google Scholar * Annunziato, A. T. The fork in the road: histone partitioning during DNA replication. _Genes (Basel)_ 6, 353–371 (2015). CAS
Google Scholar * Siddiqui, K., On, K. F. & Diffley, J. F. X. Regulating DNA replication in eukarya. _Cold Spring Harb. Perspect. Biol._ 5, a012930 (2013). PubMed PubMed Central Google
Scholar * Marchal, C., Sima, J. & Gilbert, D. M. Control of DNA replication timing in the 3D genome. _Nat. Rev. Mol. Cell Biol._ 20, 721–737 (2019). CAS PubMed Google Scholar *
Kschonsak, M. & Haering, C. H. Shaping mitotic chromosomes: from classical concepts to molecular mechanisms. _BioEssays_ 37, 755–766 (2015). CAS PubMed PubMed Central Google Scholar
* Palozola, K. C., Lerner, J. & Zaret, K. S. A changing paradigm of transcriptional memory propagation through mitosis. _Nat. Rev. Mol. Cell Biol._ 20, 55–64 (2019). CAS PubMed PubMed
Central Google Scholar * Hammond, C. M., Strømme, C. B., Huang, H., Patel, D. J. & Groth, A. Histone chaperone networks shaping chromatin function. _Nat. Rev. Mol. Cell Biol._ 18,
141–158 (2017). CAS PubMed PubMed Central Google Scholar * Grover, P., Asa, J. S. & Campos, E. I. H3-H4 histone chaperone pathways. _Annu. Rev. Genet._ 52, 109–130 (2018). CAS
PubMed Google Scholar * Buschbeck, M. & Hake, S. B. Variants of core histones and their roles in cell fate decisions, development and cancer. _Nat. Rev. Mol. Cell Biol._ 18, 299–314
(2017). CAS PubMed Google Scholar * Smolle, M. & Workman, J. L. Transcription-associated histone modifications and cryptic transcription. _Biochim. Biophys. Acta_ 1829, 84–97 (2013).
CAS PubMed Google Scholar * Greenberg, M. V. C. & Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. _Nat. Rev. Mol. Cell Biol._ 20, 590–607
(2019). CAS PubMed Google Scholar * Alabert, C. & Groth, A. Chromatin replication and epigenome maintenance. _Nat. Rev. Mol. Cell Biol._ 13, 153–167 (2012). CAS PubMed Google
Scholar * Luger, K., Dechassa, M. L. & Tremethick, D. J. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? _Nat. Rev. Mol. Cell Biol._ 13,
436–447 (2012). CAS PubMed PubMed Central Google Scholar * Stillman, B. Histone modifications: insights into their influence on gene expression. _Cell_ 175, 6–9 (2018). CAS PubMed
Google Scholar * Alabert, C. et al. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. _Nat. Cell Biol._ 16,
281–293 (2014). CAS PubMed PubMed Central Google Scholar * Sirbu, B. M. et al. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins
on nascent DNA (iPOND) coupled with mass spectrometry. _J. Biol. Chem._ 288, 31458–31467 (2013). CAS PubMed PubMed Central Google Scholar * Li, H. & O’Donnell, M. E. The eukaryotic
CMG helicase at the replication fork: emerging architecture reveals an unexpected mechanism. _BioEssays_ 40, 1700208 (2018). Google Scholar * Burgers, P. M. J. & Kunkel, T. A.
Eukaryotic DNA replication fork. _Annu. Rev. Biochem._ 86, 417–438 (2017). CAS PubMed PubMed Central Google Scholar * Alabert, C. et al. Two distinct modes for propagation of histone
PTMs across the cell cycle. _Genes Dev._ 29, 585–590 (2015). CAS PubMed PubMed Central Google Scholar * Loyola, A., Bonaldi, T., Roche, D., Imhof, A. & Almouzni, G. PTMs on H3
variants before chromatin assembly potentiate their final epigenetic state. _Mol. Cell_ 24, 309–316 (2006). CAS PubMed Google Scholar * Scharf, A. N. D. et al. Monomethylation of lysine
20 on histone H4 facilitates chromatin maturation. _Mol. Cell. Biol._ 29, 57–67 (2009). CAS PubMed Google Scholar * Burgess, R. J. & Zhang, Z. Histone chaperones in nucleosome
assembly and human disease. _Nat. Struct. Mol. Biol._ 20, 14–22 (2013). CAS PubMed PubMed Central Google Scholar * Nagarajan, P. et al. Histone acetyl transferase 1 is essential for
mammalian development, genome stability, and the processing of newly synthesized histones H3 and H4. _PLoS Genet._ 9, e1003518 (2013). CAS PubMed PubMed Central Google Scholar * Xu, M.,
Wang, W., Chen, S. & Zhu, B. A model for mitotic inheritance of histone lysine methylation. _EMBO Rep._ 13, 60–67 (2011). PubMed PubMed Central Google Scholar * Zee, B. M., Levin, R.
S., DiMaggio, P. A. & Garcia, B. A. Global turnover of histone post-translational modifications and variants in human cells. _Epigenetics Chromatin_ 3, 22 (2010). CAS PubMed PubMed
Central Google Scholar * Probst, A. V., Dunleavy, E. & Almouzni, G. Epigenetic inheritance during the cell cycle. _Nat. Rev. Mol. Cell Biol._ 10, 192–206 (2009). CAS PubMed Google
Scholar * Tagami, H., Ray-Gallet, D., Almouzni, G. & Nakatani, Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. _Cell_
116, 51–61 (2004). CAS PubMed Google Scholar * Xu, M. et al. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. _Science_ 328, 94–98 (2010). CAS
PubMed Google Scholar * Saredi, G. et al. H4K20me0 marks post-replicative chromatin and recruits the TONSL–MMS22L DNA repair complex. _Nature_ 534, 714–718 (2016). CAS PubMed PubMed
Central Google Scholar * Nakamura, K. et al. H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. _Nat. Cell Biol._ 21, 311–318 (2019). CAS PubMed
PubMed Central Google Scholar * Pellegrino, S., Michelena, J., Teloni, F., Imhof, R. & Altmeyer, M. Replication-coupled dilution of H4K20me2 guides 53BP1 to pre-replicative chromatin.
_Cell Rep._ 19, 1819–1831 (2017). CAS PubMed PubMed Central Google Scholar * Stillman, B. Chromatin assembly during SV40 DNA replication in vitro. _Cell_ 45, 555–565 (1986). CAS PubMed
Google Scholar * Smith, S. & Stillman, B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. _Cell_ 58,
15–25 (1989). CAS PubMed Google Scholar * Shibahara, K. & Stillman, B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. _Cell_ 96,
575–585 (1999). CAS PubMed Google Scholar * Petryk, N. et al. MCM2 promotes symmetric inheritance of modified histones during DNA replication. _Science_ 361, 1389–1392 (2018). CAS PubMed
Google Scholar * Yu, C. et al. A mechanism for preventing asymmetric histone segregation onto replicating DNA strands. _Science_ 361, 1386–1389 (2018). CAS PubMed PubMed Central Google
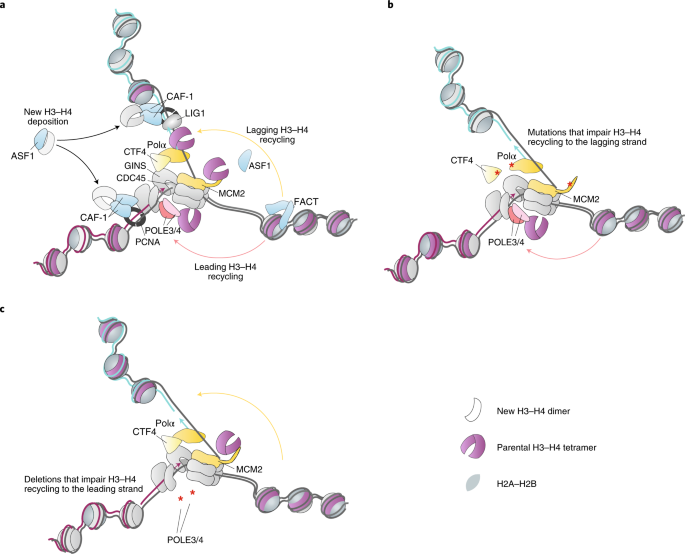
Scholar * Gan, H. et al. The Mcm2-Ctf4-Polα axis facilitates parental histone H3-H4 transfer to lagging strands. _Mol. Cell_ 72, 140–151.e3 (2018). CAS PubMed PubMed Central Google
Scholar * Pospelov, V., Russev, G., Vassilev, L. & Tsanev, R. Nucleosome segregation in chromatin replicated in the presence of cycloheximide. _J. Mol. Biol._ 156, 79–91 (1982). CAS
PubMed Google Scholar * Jackson, V. & Chalkley, R. Histone segregation on replicating chromatin. _Biochemistry_ 24, 6930–6938 (1985). CAS PubMed Google Scholar * Cusick, M. E.,
DePamphilis, M. L. & Wassarman, P. M. Dispersive segregation of nucleosomes during replication of simian virus 40 chromosomes. _J. Mol. Biol._ 178, 249–271 (1984). CAS PubMed Google
Scholar * Jackson, V., Granner, D. K. & Chalkley, R. Deposition of histones onto replicating chromosomes. _Proc. Natl Acad. Sci. USA_ 72, 4440–4444 (1975). CAS PubMed PubMed Central
Google Scholar * Russev, G. & Hancock, R. Assembly of new histones into nucleosomes and their distribution in replicating chromatin. _Proc. Natl Acad. Sci. USA_ 79, 3143–3147 (1982).
CAS PubMed PubMed Central Google Scholar * Crémisi, C., Chestier, A. & Yaniv, M. Assembly of SV40 and polyoma minichromosomes during replication. _Cold Spring Harb. Symp. Quant.
Biol._ 42, 409–416 (1978). PubMed Google Scholar * Ishimi, Y., Komamura-Kohno, Y., Arai, K. & Masai, H. Biochemical activities associated with mouse Mcm2 protein. _J. Biol. Chem._ 276,
42744–42752 (2001). CAS PubMed Google Scholar * Groth, A. et al. Regulation of replication fork progression through histone supply and demand. _Science_ 318, 1928–1931 (2007). CAS
PubMed Google Scholar * Huang, H. et al. A unique binding mode enables MCM2 to chaperone histones H3-H4 at replication forks. _Nat. Struct. Mol. Biol._ 22, 618–626 (2015). CAS PubMed
PubMed Central Google Scholar * Richet, N. et al. Structural insight into how the human helicase subunit MCM2 may act as a histone chaperone together with ASF1 at the replication fork.
_Nucleic Acids Res._ 43, 1905–1917 (2015). CAS PubMed PubMed Central Google Scholar * Foltman, M. et al. Eukaryotic replisome components cooperate to process histones during chromosome
replication. _Cell Rep._ 3, 892–904 (2013). CAS PubMed Google Scholar * Jasencakova, Z. et al. Replication stress interferes with histone recycling and predeposition marking of new
histones. _Mol. Cell_ 37, 736–743 (2010). CAS PubMed Google Scholar * Wang, H., Wang, M., Yang, N. & Xu, R.-M. Structure of the quaternary complex of histone H3-H4 heterodimer with
chaperone ASF1 and the replicative helicase subunit MCM2. _Protein Cell_ 6, 693–697 (2015). CAS PubMed PubMed Central Google Scholar * Zasadzińska, E. et al. Inheritance of CENP-A
nucleosomes during DNA replication requires HJURP. _Dev. Cell_ 47, 348–362.e7 (2018). PubMed PubMed Central Google Scholar * Douglas, M. E., Ali, F. A., Costa, A. & Diffley, J. F. X.
The mechanism of eukaryotic CMG helicase activation. _Nature_ 555, 265–268 (2018). CAS PubMed PubMed Central Google Scholar * Georgescu, R. et al. Structure of eukaryotic CMG helicase at
a replication fork and implications to replisome architecture and origin initiation. _Proc. Natl Acad. Sci. USA_ 114, E697–E706 (2017). CAS PubMed PubMed Central Google Scholar *
Saxton, D. S. & Rine, J. Epigenetic memory independent of symmetric histone inheritance. _eLife_ 8, e51421 (2019). PubMed PubMed Central Google Scholar * Simon, A. C. et al. A Ctf4
trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome. _Nature_ 510, 293–297 (2014). CAS PubMed PubMed Central Google Scholar * He, H. et al. Coordinated
regulation of heterochromatin inheritance by Dpb3-Dpb4 complex. _Proc. Natl Acad. Sci. USA_ 114, 12524–12529 (2017). CAS PubMed PubMed Central Google Scholar * Bellelli, R. et al.
POLE3-POLE4 is a histone H3-H4 chaperone that maintains chromatin integrity during DNA replication. _Mol. Cell_ 72, 112–126.e5 (2018). CAS PubMed PubMed Central Google Scholar * Iida, T.
& Araki, H. Noncompetitive counteractions of DNA polymerase and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in _Saccharomyces cerevisiae_. _Mol. Cell. Biol._ 24,
217–227 (2004). CAS PubMed PubMed Central Google Scholar * Bellelli, R. et al. Polε instability drives replication stress, abnormal development, and tumorigenesis. _Mol. Cell_ 70,
707–721.e7 (2018). CAS PubMed PubMed Central Google Scholar * Mejlvang, J. et al. New histone supply regulates replication fork speed and PCNA unloading. _J. Cell Biol._ 204, 29–43
(2014). CAS PubMed PubMed Central Google Scholar * Liu, S. et al. RPA binds histone H3-H4 and functions in DNA replication-coupled nucleosome assembly. _Science_ 355, 415–420 (2017). CAS
PubMed Google Scholar * Evrin, C., Maman, J. D., Diamante, A., Pellegrini, L. & Labib, K. Histone H2A-H2B binding by Pol α in the eukaryotic replisome contributes to the maintenance
of repressive chromatin. _EMBO J._ 37, e99021 (2018). PubMed PubMed Central Google Scholar * Clément, C. et al. High-resolution visualization of H3 variants during replication reveals
their controlled recycling. _Nat. Commun._ 9, 3181 (2018). PubMed PubMed Central Google Scholar * Gurova, K., Chang, H.-W., Valieva, M. E., Sandlesh, P. & Studitsky, V. M. Structure
and function of the histone chaperone FACT - resolving FACTual issues. _Biochim. Biophys. Acta. Gene Regul. Mech._ 1861, 892–904 (2018). CAS Google Scholar * Kurat, C. F., Yeeles, J. T.
P., Patel, H., Early, A. & Diffley, J. F. X. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. _Mol. Cell_ 65, 117–130 (2017).
CAS PubMed PubMed Central Google Scholar * Abe, T. et al. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. _J. Biol.
Chem._ 286, 30504–30512 (2011). CAS PubMed PubMed Central Google Scholar * Tsunaka, Y., Fujiwara, Y., Oyama, T., Hirose, S. & Morikawa, K. Integrated molecular mechanism directing
nucleosome reorganization by human FACT. _Genes Dev._ 30, 673–686 (2016). CAS PubMed PubMed Central Google Scholar * Wang, T. et al. The histone chaperone FACT modulates nucleosome
structure by tethering its components. _Life Sci. Alliance_ 1, e201800107 (2018). PubMed PubMed Central Google Scholar * Mayanagi, K. et al. Structural visualization of key steps in
nucleosome reorganization by human FACT. _Sci. Rep._ 9, 10183 (2019). PubMed PubMed Central Google Scholar * Liu, Y. et al. FACT caught in the act of manipulating the nucleosome. _Nature_
577, 426–431 (2020). CAS PubMed Google Scholar * Chereji, R. V. & Clark, D. J. Major determinants of nucleosome positioning. _Biophys. J._ 114, 2279–2289 (2018). CAS PubMed PubMed
Central Google Scholar * Meyer, C. A. & Liu, X. S. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. _Nat. Rev. Genet._ 15, 709–721 (2014).
CAS PubMed PubMed Central Google Scholar * Ramachandran, S. & Henikoff, S. Transcriptional regulators compete with nucleosomes post-replication. _Cell_ 165, 580–592 (2016). CAS
PubMed PubMed Central Google Scholar * Vasseur, P. et al. Dynamics of nucleosome positioning maturation following genomic replication. _Cell Rep._ 16, 2651–2665 (2016). CAS PubMed
PubMed Central Google Scholar * Fennessy, R. T. & Owen-Hughes, T. Establishment of a promoter-based chromatin architecture on recently replicated DNA can accommodate variable
inter-nucleosome spacing. _Nucleic Acids Res._ 44, 7189–7203 (2016). CAS PubMed PubMed Central Google Scholar * Gutiérrez, M. P., MacAlpine, H. K. & MacAlpine, D. M. Nascent
chromatin occupancy profiling reveals locus- and factor-specific chromatin maturation dynamics behind the DNA replication fork. _Genome Res._ 29, 1123–1133 (2019). PubMed PubMed Central
Google Scholar * Stewart-Morgan, K. R., Reverón-Gómez, N. & Groth, A. Transcription restart establishes chromatin accessibility after DNA replication. _Mol. Cell_ 75, 284–297.e6 (2019).
CAS PubMed Google Scholar * Smith, D. J. & Whitehouse, I. Intrinsic coupling of lagging-strand synthesis to chromatin assembly. _Nature_ 483, 434–438 (2012). CAS PubMed PubMed
Central Google Scholar * Yadav, T. & Whitehouse, I. Replication-coupled nucleosome assembly and positioning by ATP-dependent chromatin-remodeling enzymes. _Cell Rep._ 15, 715–723
(2016). CAS PubMed PubMed Central Google Scholar * Kaplan, N. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. _Nature_ 458, 362–366 (2009). CAS PubMed Google
Scholar * Anderson, J. D. & Widom, J. Poly(dA-dT) promoter elements increase the equilibrium accessibility of nucleosomal DNA target sites. _Mol. Cell. Biol._ 21, 3830–3839 (2001). CAS
PubMed PubMed Central Google Scholar * Ramachandran, S., Ahmad, K. & Henikoff, S. Capitalizing on disaster: establishing chromatin specificity behind the replication fork.
_BioEssays_ 39, 1600150 (2017). Google Scholar * Petruk, S. et al. Delayed accumulation of H3K27me3 on nascent DNA is essential for recruitment of transcription factors at early stages of
stem cell differentiation. _Mol. Cell_ 66, 247–257.e5 (2017). CAS PubMed PubMed Central Google Scholar * Petruk, S. et al. Structure of nascent chromatin is essential for hematopoietic
lineage Specification. _Cell Rep._ 19, 295–306 (2017). CAS PubMed PubMed Central Google Scholar * Iwafuchi-Doi, M. & Zaret, K. S. Pioneer transcription factors in cell reprogramming.
_Genes Dev._ 28, 2679–2692 (2014). PubMed PubMed Central Google Scholar * Owens, N. et al. CTCF confers local nucleosome resiliency after DNA replication and during mitosis. _eLife_ 8,
e47898 (2019). PubMed PubMed Central Google Scholar * Reverón-Gómez, N. et al. Accurate recycling of parental histones reproduces the histone modification landscape during DNA
replication. _Mol. Cell_ 72, 239–249.e5 (2018). PubMed PubMed Central Google Scholar * Madamba, E. V., Berthet, E. B. & Francis, N. J. Inheritance of histones H3 and H4 during DNA
Replication In Vitro. _Cell Rep._ 21, 1361–1374 (2017). CAS PubMed Google Scholar * Schlissel, G. & Rine, J. The nucleosome core particle remembers its position through DNA
replication and RNA transcription. _Proc. Natl Acad. Sci. USA_ 116, 20605–20611 (2019). CAS PubMed PubMed Central Google Scholar * Escobar, T. M. et al. Active and repressed chromatin
domains exhibit distinct nucleosome segregation during DNA replication. _Cell_ 179, 953–963.e11 (2019). CAS PubMed PubMed Central Google Scholar * Hansen, K. H. et al. A model for
transmission of the H3K27me3 epigenetic mark. _Nat. Cell Biol._ 10, 1291–1300 (2008). CAS PubMed Google Scholar * Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living
cells. _Cell_ 149, 1447–1460 (2012). CAS PubMed PubMed Central Google Scholar * Coleman, R. T. & Struhl, G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a
_Drosophila HOX_ gene. _Science_ 356, eaai8236 (2017). PubMed PubMed Central Google Scholar * Laprell, F., Finkl, K. & Müller, J. Propagation of Polycomb-repressed chromatin requires
sequence-specific recruitment to DNA. _Science_ 356, eaai8266 (2017). Google Scholar * Gaydos, L. J., Wang, W. & Strome, S. Gene repression. H3K27me and PRC2 transmit a memory of
repression across generations and during development. _Science_ 345, 1515–1518 (2014). CAS PubMed PubMed Central Google Scholar * Audergon, P. N. C. B. et al. Epigenetics. Restricted
epigenetic inheritance of H3K9 methylation. _Science_ 348, 132–135 (2015). CAS PubMed PubMed Central Google Scholar * Ragunathan, K., Jih, G. & Moazed, D. Epigenetics. Epigenetic
inheritance uncoupled from sequence-specific recruitment. _Science_ 348, 1258699 (2015). PubMed Google Scholar * Aygün, O., Mehta, S. & Grewal, S. I. S. HDAC-mediated suppression of
histone turnover promotes epigenetic stability of heterochromatin. _Nat. Struct. Mol. Biol._ 20, 547–554 (2013). PubMed PubMed Central Google Scholar * Zentner, G. E. & Henikoff, S.
Regulation of nucleosome dynamics by histone modifications. _Nat. Struct. Mol. Biol._ 20, 259–266 (2013). CAS PubMed Google Scholar * Dodd, I. B., Micheelsen, M. A., Sneppen, K. &
Thon, G. Theoretical analysis of epigenetic cell memory by nucleosome modification. _Cell_ 129, 813–822 (2007). CAS PubMed Google Scholar * Pesavento, J. J., Yang, H., Kelleher, N. L.
& Mizzen, C. A. Certain and progressive methylation of histone H4 at lysine 20 during the cell cycle. _Mol. Cell. Biol._ 28, 468–486 (2008). CAS PubMed Google Scholar * Sweet, S. M.
M., Li, M., Thomas, P. M., Durbin, K. R. & Kelleher, N. L. Kinetics of re-establishing H3K79 methylation marks in global human chromatin. _J. Biol. Chem._ 285, 32778–32786 (2010). CAS
PubMed PubMed Central Google Scholar * Alabert, C. et al. Domain model explains propagation dynamics and stability of histone H3K27 and H3K36 methylation landscapes. _Cell Rep._ 30,
1223–1234.e8 (2020). PubMed Google Scholar * Bonnet, J. et al. Quantification of proteins and histone marks in _Drosophila_ embryos reveals stoichiometric relationships impacting chromatin
regulation. _Dev. Cell_ 51, 632–644.e6 (2019). CAS PubMed Google Scholar * Reinberg, D. & Vales, L. D. Chromatin domains rich in inheritance. _Science_ 361, 33–34 (2018). CAS PubMed
Google Scholar * Laugesen, A., Højfeldt, J. W. & Helin, K. Molecular mechanisms directing PRC2 recruitment and H3K27 methylation. _Mol. Cell_ 74, 8–18 (2019). CAS PubMed PubMed
Central Google Scholar * Oksuz, O. et al. Capturing the onset of PRC2-mediated repressive domain formation. _Mol. Cell_ 70, 1149–1162.e5 (2018). CAS PubMed PubMed Central Google Scholar
* Poepsel, S., Kasinath, V. & Nogales, E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. _Nat. Struct. Mol. Biol._ 25, 154–162 (2018).
CAS PubMed PubMed Central Google Scholar * Højfeldt, J. W. et al. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. _Nat. Struct. Mol. Biol._ 25, 225–232
(2018). PubMed PubMed Central Google Scholar * Wang, X. & Moazed, D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. _Science_ 356, 88–91
(2017). CAS PubMed PubMed Central Google Scholar * Yu, R., Wang, X. & Moazed, D. Epigenetic inheritance mediated by coupling of RNAi and histone H3K9 methylation. _Nature_ 558,
615–619 (2018). CAS PubMed PubMed Central Google Scholar * Howe, F. S., Fischl, H., Murray, S. C. & Mellor, J. Is H3K4me3 instructive for transcription activation? _BioEssays_ 39,
1–12 (2017). CAS PubMed Google Scholar * Hörmanseder, E. et al. H3K4 methylation-dependent memory of somatic cell identity inhibits reprogramming and development of nuclear transfer
embryos. _Cell Stem Cell_ 21, 135–143.e6 (2017). PubMed PubMed Central Google Scholar * Lauberth, S. M. et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and
selective gene activation. _Cell_ 152, 1021–1036 (2013). CAS PubMed PubMed Central Google Scholar * Thomas, L. R. et al. Interaction with WDR5 promotes target gene recognition and
tumorigenesis by MYC. _Mol. Cell_ 58, 440–452 (2015). CAS PubMed PubMed Central Google Scholar * Cano-Rodriguez, D. et al. Writing of H3K4Me3 overcomes epigenetic silencing in a
sustained but context-dependent manner. _Nat. Commun._ 7, 12284 (2016). CAS PubMed PubMed Central Google Scholar * Flury, V. et al. The histone acetyltransferase Mst2 protects active
chromatin from epigenetic silencing by acetylating the ubiquitin ligase Brl1. _Mol. Cell_ 67, 294–307.e9 (2017). CAS PubMed PubMed Central Google Scholar * Bernstein, B. E. et al. A
bivalent chromatin structure marks key developmental genes in embryonic stem cells. _Cell_ 125, 315–326 (2006). CAS PubMed Google Scholar * Liu, L., Michowski, W., Kolodziejczyk, A. &
Sicinski, P. The cell cycle in stem cell proliferation, pluripotency and differentiation. _Nat. Cell Biol._ 21, 1060–1067 (2019). CAS PubMed PubMed Central Google Scholar * Larson, A.
G. & Narlikar, G. J. The role of phase separation in heterochromatin formation, function, and regulation. _Biochemistry_ 57, 2540–2548 (2018). CAS PubMed Google Scholar * Rowley, M.
J. & Corces, V. G. Organizational principles of 3D genome architecture. _Nat. Rev. Genet._ 19, 789–800 (2018). CAS PubMed Google Scholar * Nagano, T. et al. Cell-cycle dynamics of
chromosomal organization at single-cell resolution. _Nature_ 547, 61–67 (2017). CAS PubMed PubMed Central Google Scholar * Wooten, M. et al. Asymmetric histone inheritance via
strand-specific incorporation and biased replication fork movement. _Nat. Struct. Mol. Biol._ 26, 732–743 (2019). CAS PubMed PubMed Central Google Scholar * Ishiuchi, T. et al. Early
embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. _Nat. Struct. Mol. Biol._ 22, 662–671 (2015). CAS PubMed Google Scholar * Cheloufi, S. et al.
The histone chaperone CAF-1 safeguards somatic cell identity. _Nature_ 528, 218–224 (2015). CAS PubMed PubMed Central Google Scholar * Song, Y. et al. CAF-1 is essential for _Drosophila_
development and involved in the maintenance of epigenetic memory. _Dev. Biol._ 311, 213–222 (2007). CAS PubMed Google Scholar * Nakano, S., Stillman, B. & Horvitz, H. R.
Replication-coupled chromatin assembly generates a neuronal bilateral asymmetry in _C. elegans_. _Cell_ 147, 1525–1536 (2011). CAS PubMed PubMed Central Google Scholar * Volk, A. et al.
A CHAF1B-dependent molecular switch in hematopoiesis and leukemia pathogenesis. _Cancer Cell_ 34, 707–723.e7 (2018). CAS PubMed PubMed Central Google Scholar * Cheng, L. et al. Chromatin
assembly factor 1 (CAF-1) facilitates the establishment of facultative heterochromatin during pluripotency exit. _Nucleic Acids Res._ 47, 11114–11131 (2019). PubMed PubMed Central Google
Scholar * Yadav, T., Quivy, J.-P. & Almouzni, G. Chromatin plasticity: a versatile landscape that underlies cell fate and identity. _Science_ 361, 1332–1336 (2018). CAS PubMed Google
Scholar * Yu, C. et al. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. _Mol. Cell_ 56, 551–563 (2014). CAS
PubMed PubMed Central Google Scholar * Xu, C. & Corces, V. G. Nascent DNA methylome mapping reveals inheritance of hemimethylation at CTCF/cohesin sites. _Science_ 359, 1166–1170
(2018). CAS PubMed PubMed Central Google Scholar * Xu, C. & Corces, V. G. Genome-wide mapping of protein-DNA interactions on nascent chromatin. _Methods Mol. Biol._ 1766, 231–238
(2018). CAS PubMed PubMed Central Google Scholar * Charlton, J. et al. Global delay in nascent strand DNA methylation. _Nat. Struct. Mol. Biol._ 25, 327–332 (2018). CAS PubMed PubMed
Central Google Scholar * Smith, D. J., Yadav, T. & Whitehouse, I. Detection and sequencing of Okazaki fragments in _S. cerevisiae_. _Methods Mol. Biol._ 1300, 141–153 (2015). CAS
PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS K.R.S.-M. is supported by a postdoctoral fellowship from the Lundbeck Foundation and a Marie Curie Individual
Fellowship (MSCA-IF-2016 no. 747332). Research in the Groth laboratory is supported by the Lundbeck Foundation (R198-2015-269), the European Research Council (ERC CoG no. 724436),
Independent Research Fund Denmark (7016-00042B; 4092-00404B), the Novo Nordisk Foundation (NNF14CC0001; NNF14OC0012839), the NEYE foundation and the Danish Cancer Society. AUTHOR INFORMATION
Author notes * Nataliya Petryk Present address: Epigenetics and Cell Fate, UMR7216 CNRS, University of Paris, Paris, France AUTHORS AND AFFILIATIONS * The Novo Nordisk Foundation Center for
Protein Research (CPR), University of Copenhagen, Copenhagen, Denmark Kathleen R. Stewart-Morgan & Anja Groth * Biotech Research and Innovation Centre (BRIC), University of Copenhagen,
Copenhagen, Denmark Kathleen R. Stewart-Morgan, Nataliya Petryk & Anja Groth Authors * Kathleen R. Stewart-Morgan View author publications You can also search for this author inPubMed
Google Scholar * Nataliya Petryk View author publications You can also search for this author inPubMed Google Scholar * Anja Groth View author publications You can also search for this
author inPubMed Google Scholar CONTRIBUTIONS K.R.S.-M., N.P. and A.G. conceived, prepared figures for and wrote the manuscript. CORRESPONDING AUTHOR Correspondence to Anja Groth. ETHICS
DECLARATIONS COMPETING INTERESTS A.G. is CSO and co-founder of Ankrin Therapeutics. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional
claims in published maps and institutional affiliations. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Stewart-Morgan, K.R., Petryk, N. & Groth, A.
Chromatin replication and epigenetic cell memory. _Nat Cell Biol_ 22, 361–371 (2020). https://doi.org/10.1038/s41556-020-0487-y Download citation * Received: 12 September 2019 * Accepted:
18 February 2020 * Published: 30 March 2020 * Issue Date: April 2020 * DOI: https://doi.org/10.1038/s41556-020-0487-y SHARE THIS ARTICLE Anyone you share the following link with will be able
to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative