- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT AT-1/SLC33A1 is a key member of the endoplasmic reticulum (ER) acetylation machinery, transporting acetyl-CoA from the cytosol into the ER lumen where acetyl-CoA serves as the
acetyl-group donor for Nε-lysine acetylation. Dysfunctional ER acetylation, as caused by heterozygous or homozygous mutations as well as gene duplication events of _AT-1/SLC33A1_, has been
linked to both developmental and degenerative diseases. Here, we investigate two models of AT-1 dysregulation and altered acetyl-CoA flux: AT-1S113R/+ mice, a model of AT-1
haploinsufficiency, and AT-1 sTg mice, a model of AT-1 overexpression. The animals display distinct metabolic adaptation across intracellular compartments, including reprogramming of lipid
metabolism and mitochondria bioenergetics. Mechanistically, the perturbations to AT-1-dependent acetyl-CoA flux result in global and specific changes in both the proteome and the
acetyl-proteome (protein acetylation). Collectively, our results suggest that AT-1 acts as an important metabolic regulator that maintains acetyl-CoA homeostasis by promoting functional
crosstalk between different intracellular organelles. SIMILAR CONTENT BEING VIEWED BY OTHERS AUTOPHAGY REGULATION BY ACETYLATION—IMPLICATIONS FOR NEURODEGENERATIVE DISEASES Article Open
access 22 January 2021 ENDOPLASMIC RETICULUM ACETYLTRANSFERASES ATASE1 AND ATASE2 DIFFERENTIALLY REGULATE RETICULOPHAGY, MACROAUTOPHAGY AND CELLULAR ACETYL-COA METABOLISM Article Open access
12 April 2021 COX17 ACETYLATION VIA MOF–KANSL COMPLEX PROMOTES MITOCHONDRIAL INTEGRITY AND FUNCTION Article Open access 09 October 2023 INTRODUCTION The acetyl-CoA transporter, AT-1 (also
referred to as SLC33A1), is a key member of the endoplasmic reticulum (ER) acetylation machinery, transporting acetyl-CoA from the cytosol into the ER lumen where acetyl-CoA serves as the
acetyl-group donor for Nε-lysine acetylation1,2. Gene duplications in _AT-1/SLC33A1_ have been identified in patients with autistic-like features, intellectual disability, and dysmorphic
features; heterozygous mutations in _AT-1/SLC33A1_ are associated with a familial form of spastic paraplegia, while homozygous mutations are associated with developmental delay and premature
death3,4,5,6,7. Mouse models of altered AT-1 expression are effective models of human AT-1-linked diseases8,9,10. Both cell-based and mouse-based experiments support the conclusion that
AT-1 activity regulates ER proteostasis by maintaining the balance between quality control and the induction of reticulophagy1,2,8,9,10,11,12,13,14. In this study, we investigate the
outcomes of dysregulated AT-1 activity on intracellular acetyl-CoA homeostasis. Changes in the intracellular acetyl-CoA flux caused by hypoactive or hyperactive AT-1 could conceivably
influence phenotypes beyond the secretory pathway; in particular the metabolic effects of these genetic manipulations have yet to be defined. For this purpose, we examine the hepatic
molecular signatures of AT-1S113R/+ mice8, a model of AT-1 haploinsufficiency, and AT-1 sTg mice10, a model of global AT-1 overexpression. The animals display distinct metabolic
reprogramming across several intracellular compartments and pathways that is achieved through specific changes in both the proteome and the acetyl-proteome (protein acetylation). When taken
together, our results suggest that AT-1 is an essential component of an intracellular communication network that promotes functional crosstalk between different cellular compartments and
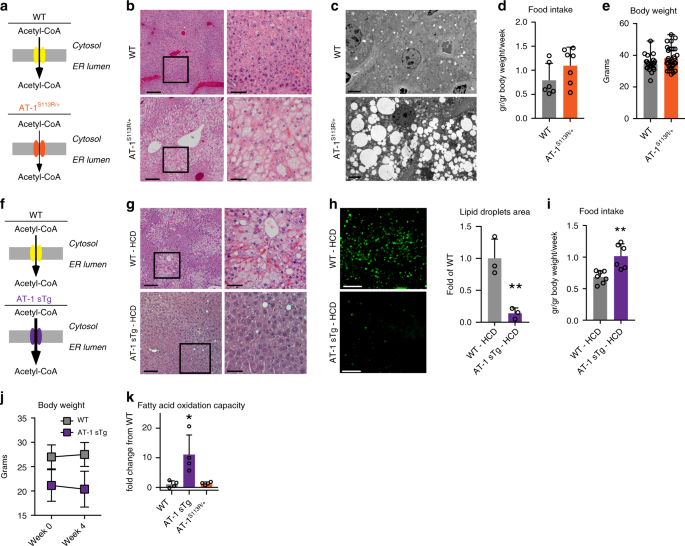
organelles to maintain acetyl-CoA homeostasis. RESULTS ABERRANT AT-1 ACTIVITY ALTERS LIPID METABOLISM AT-1S113R/+ mice are haploinsufficient for AT-1 activity (Fig. 1a), resulting in a ~50%
decrease in acetyl-CoA transport from the cytosol into the ER lumen8. Histological assessment revealed that AT-1S113R/+ mice have increased propensity to liver steatosis, which could be
documented by both hematoxylin and eosin (H&E) (Fig. 1b) and electron microscopy (Fig. 1c). Although observed with both regular (40.2% carbohydrate, 4.7% fat) and breeder (44.7%
carbohydrate, 9% fat) diets, the steatosis was much more common in animals fed the breeder diet. Importantly, the animals displayed no differences in food intake or body weight (Fig. 1d, e).
Biochemical analysis revealed a ~60% increase in free acetyl-CoA available within the cytosol and a marked accumulation of fatty acids and triglycerides in the liver (Table 1). Finally, the
serum lipid profiles were not different between wild-type (WT) and AT-1S113R/+ mice (Table 2), indicating that the steatosis was not associated with dyslipidemia. Contrary to AT-1S113R/+,
AT-1 sTg mice that systemically overexpress AT-1 had increased transport of acetyl-CoA from the cytosol to the ER lumen (Fig. 1f)10. If indeed the propensity of AT-1S113R/+ to develop
steatosis is caused by increased acetyl-CoA availability in the cytosol, then AT-1 sTg mice should be resistant, even when challenged with a lipogenic high-carbohydrate diet (HCD). To test
this, both WT and AT-1 sTg animals were fed a HCD (70% carbohydrates, 5.2% fat) for 4 weeks15,16. As expected, WT mice on the HCD showed diffuse steatosis and lipid accumulation in the liver
sections (Fig. 1g, h); however, AT-1 sTg on the HCD showed normal parenchyma and no histological evidence of lipid accumulation (Fig. 1g, h). We previously reported that AT-1 sTg eat more
than their WT littermates10, and although similar behavior was observed over the 4 weeks of HCD feeding (Fig. 1i), there was no increase in body weight (Fig. 1j), suggesting that the changes
in lipid metabolism were not due to altered food intake. When on a regular diet, AT-1 sTg mice showed reduced levels of cytosolic acetyl-CoA, fatty acids, and triglycerides as compared with
WT littermates (Table 3). On the HCD, WT mice displayed a marked increase in fatty acids and triglycerides, whereas AT-1 sTg remained overall resistant to the lipogenic challenge (Table 3).
Although there was a modest increase in triglycerides, the levels remained well below those of their WT littermates (Table 3). Furthermore, WT mice responded to the HCD by increasing serum
triglyceride levels, while AT-1 sTg mice did not (Table 4). An increase in oxidation of fatty acids is one possible explanation for the hepatic lipid-storage-resistance phenotype of AT-1 sTg
mice. Consistent with this, fatty acid oxidative capacity was greater in primary hepatocytes from AT-1 sTg mice compared with WT controls (Fig. 1k). When taken together, the results
obtained with AT-1S113R/+ and AT-1 sTg mice indicate that changes in AT-1 activity and acetyl-CoA flux from the cytosol to the ER lumen can cause important changes in the availability of
acetyl-CoA within the cytosol, impacting lipid metabolism and propensity to hepatic steatosis. ABERRANT AT-1 ACTIVITY AFFECTS THE PROTEOME To dissect the mechanism(s) underlying the above
metabolic adaptation, we used quantitative mass spectrometry coupled with liquid chromatography and investigated possible proteomic changes within the liver of AT-1 sTg and AT-1S113R/+
animals17,18. We found 2056 and 373 expression levels of proteins that were significantly (_P_ < 0.05, Fisher’s method) altered in AT-1 sTg and AT-1S113R/+ mice, respectively (Fig. 2a,
see also Supplementary Data 1). The cumulative difference in the distribution indicated that there are more downregulated proteins in AT-1S113R/+ (mean FC = −0.122) than in AT-1 sTg (mean FC
= −0.060) (Fig. 2b). Collectively, 61 proteins were affected in both models of AT-1 dysregulation; of them, 22 are associated with the secretory pathway, 8 with the mitochondria, 7 with the
nucleus, and 24 with the cytosol (Fig. 2c, d). Comparing the proteomic response in the two mouse models, opposing changes in abundance were detected for 14 of these proteins, while the
remaining 47 changed in the same direction in both models (Fig. 2d). To assess the functional consequences of these protein expression changes, we conducted Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis using the Over-Representation Analysis method. Enriched pathways passed a Benjamini–Hochberg test with a FDR < 0.05. Twenty-nine pathways were enriched in
the AT-1 sTg, and 14 pathways were enriched in AT-1S113R/+, of which 6 were overlapping (Fig. 2e). These KEGG pathways were then categorized into broad key pathways (Fig. 2f). In addition to
secretory pathway and mitochondrial processes anticipated based on prior work in the neuron-specific mouse model of AT-1 overexpression9, here, other pathways were also identified. Pathways
responsive to changes in AT-1 included lipid metabolism, glucose metabolism, and amino acid metabolism. KEGG pathways enriched in both models of AT-1 dysregulation included carbon
metabolism, fatty acid degradation, fatty acid metabolism, butanoate metabolism, PPAR signaling, and BCAA degradation. Despite both models perturbing the activity of AT-1, we observed unique
protein expression consequences in each model. Specifically, AT-1S113R/+ displayed overall more downregulated proteins, and distinct enrichment in many mitochondrial pathways and lipid
metabolism pathways. In contrast, AT-1 sTg displayed a wider distribution of protein expression and a wider array of enriched pathways: transcription, translation, secretory pathway, lipid
metabolism, glucose metabolism, mitochondria adaptation, and amino acid metabolism, indicating that the overexpression of AT-1 causes highly global protein expression changes. These data
show that AT-1 responsive proteins and pathways extend to metabolic pathways not previously known to be affected by changes in the cytosol-to-ER acetyl-CoA flux. ABERRANT AT-1 ACTIVITY
AFFECTS THE ACETYL-PROTEOME Acetyl-CoA-dependent processes not only include fatty acid metabolism and the citric acid cycle but also Nε-lysine acetylation, which can occur in the nucleus and
cytoplasm19, ER2,14, mitochondria20, and the peroxisome21. To assess the ability of AT-1 and acetyl-CoA availability to modulate protein function through lysine acetylation, we used a mass
spectrometry method that elucidates the stoichiometry of site-specific lysine acetylation within proteomes22. We quantified the steady-state acetylation stoichiometry on lysine sites at the
proteome level, which ranged from < 1 to 99%. In the AT-1 sTg and AT-1S113R/+ liver, 3341 and 3003 lysine sites were identified, respectively. When compared with WT, 375 were
significantly changing in the AT-1 sTg, and 415 were significantly changing in the AT-1S113R/+ (Fig. 3a; see also Supplementary Data 2). The distribution of stoichiometry changes (Fig. 3b)
as well as the average number of lysine sites per protein identified (Fig. 3c) were significantly different between the two models (see also later). These differences can possibly be
explained by changes in the availability of cytosolic acetyl-CoA as a result of less (in the AT-1S113R/+ mice) or more (in the AT-1 sTg mice) AT-1-mediated transport into the ER lumen. A
protein with multiple lysine sites can display varying stoichiometry on each site, leading to multiple acetylated protein forms capable of influencing protein stability and activity.
Comparison of the AT-1 models revealed a 12% (88 sites) overlap in significantly changing acetylation sites, and 21% (119 proteins) overlap in modified proteins (Fig. 3d). To assess the
functional implications of these posttranslational modifications, we conducted the KEGG pathway analysis of the acetyl-proteome. We found a compelling 57% overlap in enriched pathways in
both models of AT-1 dysregulation, which include lipid metabolism, amino acid metabolism, mitochondria, and secretory pathway and proteostasis (Fig. 3e). The 88 acetylation sites that were
detected in both models are localized as follows: 26 in the secretory pathway, 18 in the mitochondria, 33 in the cytosol, and 11 in the nucleus. Strikingly, 85 of the acetylation sites had
increased acetylation stoichiometry in one mouse model and decreased acetylation stoichiometry in the other model, whereas only 3 acetylation sites had stoichiometry changes in the same
direction in both models compared with WT (Fig. 3f). With the exception of eight acetylated sites, all of the lysine sites in the AT-1S113R/+ showed hyperacetylation, whereas the lysine site
in the AT-1 sTg showed hypoacetylation. Lastly, we evaluated the subcellular location of all acetylated proteins significantly different from WT; the general trends again indicate
hypoacetylation in AT-1 sTg and hyperacetylation in AT-1S113R/+ (Fig. 3g). When broken down by compartment, this distribution was again maintained across the secretory pathway, mitochondria,
nucleus and cytosol (Fig. 3h). This differential response likely reflects changes in substrate availability. Indeed, the increased transport of cytosolic acetyl-CoA into the ER lumen in
AT-1 sTg mice reduced acetyl-CoA availability in the cytosol, whereas the reduced transport to the ER in AT-1S113R/+ mice had the opposite effect. These results strongly suggest that these
coordinated posttranslational acetyl modifications are intimately linked to the levels of cytosolic acetyl-CoA, and that AT-1 activity is a major driver of protein acetylation in these mouse
models. AT-1 CONTROLS CELL METABOLISM AND MITOCHONDRIA BIOENERGETICS Comparison of the proteomic and acetyl-stoichiometric data indicate that the metabolic reprogramming induced by changes
in AT-1 activity and acetyl-CoA availability is quite complex with proteins being regulated either at the level of the proteome or acetylome, or at both levels (Fig. 4a). Comparison of the
two data sets demonstrated that there are 105 and 35 acetylation sites in the AT-1 sTg and AT-1S113R/+ models, respectively, that are significantly changed at both the protein and
posttranslational (acetylation) level as compared with WT. To further understand how these different modes of regulation can integrate into specific metabolic responses, we focused on the
two metabolic clusters which are immediately relevant to the liver phenotype (see Fig. 1) and were heavily highlighted in the pathway analysis—lipid and mitochondrial metabolism. Proteins
that were identified in the enriched KEGG pathways from either model at the level of the proteome and acetyl-proteome were included in the cluster analysis. Factors regulated at the level of
the proteome and/or acetylome were represented within each cluster (Fig. 4b, c; see also Supplementary Tables 1, 2). Again, these results reveal the far-reaching metabolic impact of changes
in AT-1 activity and consequent acetyl-CoA availability. An important and unexpected finding of this study is the fact that the mitochondria dynamically adapt to changes in acetyl-CoA
transport from the cytosol to the ER lumen imparted by the activity of the ER membrane acetyl-CoA transporter, AT-1. Also unexpected is that fact that the mitochondria adaptation can involve
changes in stoichiometry of lysine acetylation that was expected to be buffered from acetyl-CoA levels in the cytosol. In line with this finding, we investigated the functional significance
of the mitochondria adaptive response. Direct visualization of the mitochondria by structure-illumination microscopy (SIM) in primary cultured hepatocytes revealed significant expansion of
the mitochondria network in AT-1 sTg mice (Fig. 4d), with significant increases detected in both mitochondrial area and volume (Fig. 4e). Mitochondrial remodeling was associated with a
significant upregulation of the mRNA levels of the transcriptional co‐activator _peroxisome proliferator‐activated receptor‐γ_ (_PPAR‐γ_) _co‐activator 1α_ (_PGC‐1α_) (Fig. 4f), which is an
important regulator of mitochondrial biogenesis and mitochondrial function23,24. These results are in line with the expansion of PPAR signaling observed at the level of the proteome (see
Fig. 2; also discussed later). Next, we used U-13C glutamine isotope tracing to measure metabolites in primary hepatocytes and determine the functional significance of the TCA/citric acid
cycle adaptation identified in the proteome and acetyl stoichiometry data. The results revealed increased levels of 13C-labeled alpha-ketoglutarate and fumarate, increased total levels of
alpha-ketoglutarate, and a trend (_P_ = 0.055, Student’s _t_ test) toward increased malate levels in AT-1S113R/+ mice (Fig. 4g; see also Supplementary Data 3 and Supplementary Table 3). No
significant differences were observed in TCA/citric acid metabolites in AT-1 sTg animals. Together, data shown here suggest that AT-1 sTg and AT-1S113R/+ mice are adapting in a different
capacity to AT-1 activity and acetyl-CoA availability. In AT-1 sTg mice, we detected proteomic enrichment in PPAR signaling and acetylated protein enrichment in the TCA cycle. These changes
were reflected functionally in an overall expansion in the mitochondria, but without a change in overall levels of TCA metabolites. In contrast, in AT-1S113R/+ mice, we detected an increase
in engagement of the TCA/citric acid cycle, which was reflected in the proteomic pathway enrichment of PPAR signaling, TCA cycle, and oxidative phosphorylation and acetyl proteoform
enrichment in PPAR signaling and the TCA cycle. The increased fatty acid oxidation capacity observed in AT-1 sTg mice (see Fig. 1k) is also reflected in the expansion of the mitochondria
network (Fig. 4d) and the adaptive response of the carnitine-acylcarnitine translocase machinery (Slc25a20, Cpt1a and Cpt2) and the hydroxyacyl-CoA dehydrogenase/3-ketoacyl-CoA
thiolase/enoyl-CoA hydratase trifunctional enzyme (Hadh, Hadha, and Hadhb). In fact, we found Hadhb, Cpt2, and Cpt1a to be modified at the proteome level, Hadh and Hadha at the acetylome
level, and Slc25a20 both at the proteome and acetylome level (Fig. 4b, c; see also Supplementary Tables 1, 2). Together, these adaptive changes at the proteome and acetylome level integrate
to promote the oxidation of fatty acids within the mitochondria. DISCUSSION Cell-based and mouse-based studies support the conclusion that AT-1 activity regulates ER proteostasis by
maintaining the balance between quality control and the induction of reticulophagy1,2,8,9,10,11,12,13,14. However, changes in intracellular acetyl-CoA flux caused by hypoactive or
hyperactive AT-1 could conceivably influence cellular events beyond those already established; in particular the metabolic effects of these genetic manipulations have not been defined. In
this study, we used two models of AT-1 dysregulation and altered acetyl-CoA flux: AT-1S113R/+ mice, a model of AT-1 haploinsufficiency, and AT-1 sTg mice, a model of AT-1 overexpression. We
discovered that AT-1 activity has pleiotropic effects that go beyond quality control and the induction of reticulophagy. Specifically, the animals display distinct metabolic adaptation
across intracellular compartments, including reprogramming of lipid metabolism and mitochondria bioenergetics. Phenotypically, this adaptive response in the AT-1S113R/+ model leads to
spontaneous steatosis and increased engagement of the TCA cycle. In contrast, in the AT-1 sTg model, it leads to resistance to diet-induced steatosis and expansion of the mitochondria
network. Mechanistically, this functional adaptation is achieved by global reprograming of several biological pathways caused by specific changes in both the proteome and the acetyl-proteome
(protein acetylation). Importantly, the proteome and acetyl-proteome of AT-1 mouse models support these cellular phenotypes—also showing dramatic changes across lipid metabolism and
mitochondria-related pathways. Therefore, AT-1 has emerged as a key regulator in intracellular acetyl-CoA homeostasis, which has far reaching consequences within the cell’s metabolism. To
maintain homeostasis, the cell must ensure crosstalk between different organelles and compartments. It seems likely that key cellular metabolites reflect the immediate activity of metabolic
enzymes as well as the functional metabolic state of intracellular organelles. In this way, signaling molecules and/or availability respond to extracellular and/or intracellular changes and
allow for implementation of the appropriate adaptive response. Our results indicate that changes in acetyl-CoA flux from the cytosol to the ER lumen, as caused by reduced or increased AT-1
activity, cause significant metabolic adaptation. These results suggest that the cytosol-to-ER flux of acetyl-CoA is a branch of the more general nutrient-signaling-pathway2,25,26,27 that
enables rapid modulation and reprogramming of different intracellular activities upon fluctuation of metabolites/nutrients. Mechanistically, AT-1 responsive proteins are involved in the
secretory pathway, mitochondria, lipid, glucose, and amino acid metabolism. The AT-1 response includes changes in acetyl proteoforms in a manner that appears to be highly coordinated and
occurs within different cellular organelles and compartments. These data suggest that acetyl-CoA not only serves as a cytosolic sensor but that the status of cytosol-to-ER acetyl-CoA flux is
an essential component of an intracellular communication network in place to maintain acetyl-CoA homeostasis. Overall, the results support the conclusion that the flux of acetyl-CoA from
the cytosol to the ER lumen can act as a metabolic regulator, and directly impacts crosstalk between different intracellular organelles and compartments. METHODS ANIMALS All animal
experiments were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals and received ethical approval by the Institutional Animal Care and Use Committee of
the University of Wisconsin‐Madison. Generation of AT-1S113R/+ animals was achieved by crossing mice carrying the _Slc33a1-_S113R mutation to WT animals8. AT-1S113R/+ mice were studied
between 5 and 7 months of age. Generation of AT-1 sTg mouse was achieved by crossing Rosa26:tTA mice with pTRE AT-1 mice to generate ROSA26:tTA;pTRE-AT-1 (AT-1 sTg mice)10. AT-1 sTg mice
were studied at ~3 months of age. Age-matched wild-type (WT) littermates were used as controls. Males were used for the experiments in this study. DIETS The following diets were used
throughout our study: regular chow, teklad 8604; breeder chow, teklad 2019; high-carbohydrate diet (HCD), teklad 98090. HEPATOCYTE ISOLATION Primary mouse hepatocytes were prepared by
anesthetizing the animals by intraperitoneal injection of 0.4 mg/g freshly prepared 2, 2, 2-tribromoethanol (Avertin). The abdominal cavity was opened and a cannula was inserted into the
inferior vena cava. The flow of perfusion buffer (HBSS, 59 mM HEPES, 0.6 mM EGTA; pH 7.5) began, and the portal vein was immediately cut. The liver was then perfused with collagenase buffer
(HBSS, 0.25 mg/ml of collagenase type IV (Sigma-Aldrich C5138), 7 mM CaCl2, 53 mM HEPES, pH 7.5). Next, the liver was excised, and cells were harvested for tissue culture experiments by
cutting into the liver capsule. Hepatocytes were passed through a 70 -μm cell strainer and centrifuged (100 × _g_, 5 min), re-suspended in washing media (M199, 1× glutamax), centrifuged
again (100 × _g_, 5 min), re-suspended in plating media (10% FBS, 1× glutamax, 1× penicillin/streptomycin/glutamine, 1× BSA) and plated. For immunocytochemistry, hepatocytes were plated at
100,000 cells per collagen-coated coverslip (GC-12, Neuvitro), and transfected with CellLight Mitochondria-GFP BacMam 2.0 (ThermoFisher Scientific) overnight. Transfected hepatocytes were
fixed with paraformaldehyde (4%, 15710, Electron Microscopy Sciences) for 10 min, followed by permeabilization with 0.1% Triton-X100 (Roche Applied Science) for 5 min and incubation in
blocking buffer (10% BSA, 5% goat serum in PBS) for one hour. Nuclei were stained with DAPI (62248, ThermoFisher Scientific) during the blocking step. Cells were washed three times in PBS
and prolong diamond antifade mountant (P36965, ThermoFisher Scientific) was used to mount the coverslips. Images were acquired using Structured Illumination Microscopy (Nikon SIM), and
analyzed in Imaris imaging software (Bitplane, Oxford Instruments) using the Surfaces module. LIPID DETERMINATIONS Whole liver samples were homogenized in PBS and the following assays were
performed: Coenzyme A was measured using the CoA assay kit (K367, BioVision); free fatty acid was measured using the free fatty acid assay kit (ab65341, Abcam); triglycerides were measured
using the triglyceride assay kit (ab65336, Abcam); the total cholesterol was measured using the cholesterol assay kit (ab65359, Abcam). Cytosolic acetyl-CoA was isolated from the total
liver, and was measured using the acetyl-CoA assay kit (ab87546, Abcam). To isolate the cytosol, the liver was homogenized (in 10 mM HEPES, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 1 mM DTT,
protease and phosphatase inhibitors), then centrifuged (600 × _g_, 10 min). The supernatant was collected and centrifuged again (89,300 × _g_, 90 min) for analysis. For lipid serum analytes,
blood was collected transcardially from mice with an insulin syringe. Blood was clotted on ice for 15 min, then centrifuged at 200×_g_ for 10 min at 4 °C and the supernatant was collected.
The lipid serum profile was performed by the University of Wisconsin-Madison Clinical laboratory. REAL-TIME PCR Real-time PCR was performed using the Roche 480 lightcycler and Sybr Green
Real Time PCR Master Mix (Life Technologies). Gene expression levels were normalized against GAPDH levels and expressed as fold of change from control. The cycling parameters for PGC1-α Pan
(all PGC1-α isoforms) were as follows: 95 °C, 15 s; 60 °C, 60 s, for a maximum for 40 cycles. Primer sets were as follows: _PGC-1α (pan)_: 5′-TGATGTGAATGACTTGGATACAGACA-3′ (sense),
5′-GCTCATTGTTGTACTGGTTGGATATG-3′ (antisense); GAPDH: 5′-AGGTCGGTGTGAACGGATTTG-3′(sense), 5′-TGTAGACCATGTAGTTGAGGTCA-3′ (antisense). HISTOLOGY Transmission electron microscopy was performed
at the Electron Microscopy facility of the William S. Middleton Memorial Veterans Hospital (Madison, WI). Tissue sections were observed using the transmission electron microscopy (H-600;
Hitachi) at 75 kV. For LipidTox staining, liver sections were collected and immediately placed in the Optimal Cutting Temperature medium (ThermoFisher Scientific, San Jose, CA, USA), and
stored at −80 °C until processed. Sections were sliced on a microtome cryostat (Microm HM 505 N), and processed using HCS LipidTOX™ Neutral Lipid Stains protocol (ThermoFisher Scientific,
San Jose, CA, USA). Processed slides were imaged on a macroconfocal microscope (Leica TCS-LSI). For H&E staining, the liver sections were collected and immediately placed in 10% neutral
buffered formalin overnight. Sections were then embedded in paraffin using standard techniques. Sections were sliced on a microtome; they underwent deparaffinization and rehydration, and
processed for H&E staining. Processed slides were imaged on a Zeiss Axiovert 200 inverted fluorescent microscope. FATTY ACID OXIDATION Mitochondria fuel oxidation was determined using a
Seahorse XF mitochondria Fuel Flex Test kit (Agilent Technologies, Wilmington, DE, USA). Primary hepatocytes from mice were plated at 1.0 × 104 cells/well on a 96-well collagen-coated
microplate overnight. One hour before the assay, the media was changed to 180 µL/well of XF base media (Seahorse XF DMEM, 1 mM pyruvate, 2 mM glutamine, 10 mM glucose; pH 7.4) and placed at
37 °C non-CO2 incubator for 1 h. The following three fuel pathway inhibitors were added during the assay: UK5099 (to inhibit glucose oxidation), Etomoxir (to inhibit long chain fatty acid
oxidation), and BPTES (to inhibit glutamine oxidation). Fatty acid oxidation capacity was tested by injecting UK5099 (2.0 μM final) and BPTES (3.0 μM final) after the third measurement of
baseline oxygen consumption. Six rate measurements proceeding this injection, Etomoxir (4.0 μM final) was injected. The equation for FAO capacity was [1-((baseline OCR-UK5099/BPTES
OCR)/(baseline OCR – all inhibitors OCR))] and was expressed as a function of cellular oxygen consumption rate. SUBCELLULAR FRACTIONATION In order to improve our ability to capture changes
at the subcellular compartment level, the liver was processed through a crude fractionation protocol prior to quantitative proteomics and stoichiometry of acetylation. The liver was
homogenized in Buffer A (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2; pH 7.4) and passed through a 70-μm cell strainer before centrifugation (900 × _g_, 5 min). The pellet was washed in PBS and
centrifuged (900×_g_, 5 min), twice and retained. For the cytosolic and mitochondrial fraction, the original supernatant was centrifuged again (21,000 × _g_, 2 min); supernatant was retained
and re-centrifuged and the final supernatant contained the cytosolic fraction. The pellet containing the mitochondria was resuspended in Buffer B (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2;
320 mM sucrose, pH 7.4) and centrifuged (3500 × _g_, 5 min), twice, and the final pellet was retained. QUANTITATIVE PROTEOMICS Quadruplicate liver samples of the cytosol, mitochondria, and
nucleus were homogenized, then lysed in lysis buffer (8 M urea, 50 mM Tris, pH = 8, 5 mM CaCl2, 20 mM NaCl, 1 EDTA-free Roche protease inhibitor tablet, and 1 Roche PhosSTOP phosphatase
inhibitor tablet) with a probe sonicator for three pulses at 60 W, 20 kHz for 15 s, each followed by a 30 s pause for cooling at 4 °C. Crude lysates were then centrifuged at 14,000 × _g_ for
5 min, after which the supernatant was collected and protein concentrations were measured by Pierce BCA Protein Assay (Pierce, Rockford, IL, USA) according to the manufacture’s protocol.
Lysate containing 400 μg proteins was reduced in 5 mM dithiothreitol (DTT) at room temperature for 1 h, followed by alkylation in 15 mM iodoacetamide (IAA) for 30 min in the dark. Alkylation
was quenched by adding DTT to 5 mM. The resulting solution was then diluted with Tris buffer (pH = 8) to 0.9 M urea, and proteins were digested with trypsin (Promega, Madison, WI) at 1:50
enzyme to protein ratio at 37 °C for 18 h. Digestion was quenched by adding trifluoroacetic acid (TFA) to a final concentration of 0.3% and desalted with C18 SepPak cartridges (Waters,
Milford, MA, USA). Peptides were dried under vacuum and reconstituted in 0.5 M TEAB before labeling. For each genotype, samples were assigned to two batches of 4-plex dimethylated leucine
(DiLeu) tags each in biological duplicate (two samples and two littermate controls per batch). In all, 4 mg of each DiLeu tags were suspended in anhydrous DMF and combined with
4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methyl-morpholinium tetrafluoroborate (DMTMM) and N-methylmorpholine (NMM) at 0.6 × molar ratios to tags. The mixture was vortexed at room temperature
for 1 h. Following centrifugation, the supernatant was immediately mixed with 400 μg tryptic peptides from one condition. Peptides were labeled at a 10:1 label to peptide mass ratio and
vortexed at room temperature for 2 h. The reaction was quenched by adding 5% NH2OH to the final concentration of 0.25%, after which samples were dried under vacuum. Each batch of labeled
peptides was combined, respectively, as 4-plex mixtures. The mixtures were purified by strong cation exchange liquid chromatography (SCX LC) with a PolySULFOETHYL A column (200 mm × 2.1 mm,
5 μm, 300 Å, PolyLC, Columbia, MD). Elutes containing labeled peptides were collected by a FC-4 fraction collector (Rainin Dynamax) and dried under vacuum. The cleaned samples were then
fractionated with a Kinetex C18 column (5 μm, 100 Å, Phenomenex, Torrance, CA, USA), and a binary mobile phase at pH = 10 (mobile phase A is 10 mM aqueous ammonium formate and mobile phase B
is 10 mM ammonium formate in 90% ACN). Gradient was set as following: 0–3 min 1% B phase; B phase linearly increased to 35% from 3 to 50 min, then quickly increased to 60, 70, and 100% in 4
min, 4 min, and 2 min, respectively, after which the column was washed by 100% B phase for 15 min. Eluents in the linear gradient were collected by 2-min intervals and combined into ten
fractions. Each fraction was dried under vacuum. Peptides in each fraction were reconstituted in 0.1% formic acid (FA) and subjected to reversed phase LC-MS/MS analysis with an Orbitrap
Fusion Lumos Tribrid mass spectrometer (ThermoFisher Scientific, San Jose, CA, USA) interfaced with a Dionex Ultimate 3000 UPLC system (ThermoFisher Scientific, San Jose, CA, USA). Peptides
were loaded onto a 75-μm inner diameter microcapillary column custom-packed with 15 cm of Bridged Ethylene Hybrid C18 particles (1.7 μm, 130 Å, Waters). Labeled peptide were separated with a
90 min gradient from 3 to 30% ACN with 0.1% FA, followed by 10 min to 75% ACN and then 10 min to 95% ACN. After that, the column was equilibrated at 3% ACN for 15 min to prepare for the
next injection. The mass spectrometer was operated in a top 20 data-dependent acquisition mode. Survey scans of peptide precursors from _m/z_ 350 to 1500 were performed at a resolving power
of 60 K and an AGC target of 2 × 105 with a maximum injection time of 100 ms. The top 20 intense precursor ions were selected and subjected to the HCD fragmentation at a normalized collision
energy of 30% followed by tandem MS acquisition at a resolving power of 15 K and an AGC target of 5 × 104, with a maximum injection time of 100 ms and a lower mass limit of _m/z_ 110.
Precursors were subjected to a dynamic exclusion of 45 s with a 10 ppm mass tolerance. Raw files were processed with Proteome Discoverer 2.1 engine (ThermoFisher Scientific, San Jose, CA,
USA) with Byonic search engine (Protein Metrics Inc, San Carlos, CA, USA). Spectra were searched against the Uniprot _Mus musculus_ reviewed database with trypsin as the enzyme and maximum
two missed cleavages. The parent mass error tolerance was set to be 50 ppm, and fragment mass tolerance was 0.02 Da. Fixed modifications included DiLeu labels on peptide N-termini and lysine
residues (+145.12801 Da) and carbamidomethylation on cysteine residues (+57.02146 Da). Dynamic modifications included oxidation of methionine residues (+15.99492 Da). Identifications were
filtered to 1% peptide and protein FDR. Quantitation was performed in Proteome Discoverer with a reporter ion integration tolerance of 20 ppm for the most confident centroid. Only the PSMs
that contained all reporter ion channels were considered, and protein quantitative ratios were determined using a minimum of one unique quantified peptide. Reporter ion ratio values for
protein groups were exported to Microsoft Excel, and all fractions were combined for downstream analysis (see statistics section for processing). Proteins that had _P_ < 0.05 (Fisher’s
method) were filtered as significant changes. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set
identifier PXD013736. STOICHIOMETRY OF ACETYLATION Analysis was conducted using 200 μg of protein from the liver mitochondrial and cytosolic subcellular fractions, which were denatured in
urea buffer (8 M urea (deionized), 500 mM ammonium bicarbonate pH = 8.0, 5 mM DTT)28. Samples were incubated for 20 min at 60 °C at 1500 rpm using the Eppendorf ThermoMixer® C. Cysteines
were alkylated with 50 mM iodoacetamide and incubated for 20 min Chemical acetylation using two rounds of ~20 µmol heavy isotopic D6-acetic anhydride (Cambridge Isotope Laboratories). The pH
of the samples was spot checked and raised back to ~8.5 after each chemical labeling step. Samples were diluted using 100 mM ammonium bicarbonate pH = 8.0 to 2 M urea and digested with
1:100 trypsin at 37 °C for 4 h. Samples were then diluted to 1 M urea prior to a second digestion by gluC (1:100). Chemically acetylated peptides were resuspended into ~2 mL of HPRP buffer A
(100 mM Ammonium Formate pH = 10) and injected onto a preequilibrated Phenomenex Gemini® NX-C18 column (5 µm, 110 Å, 150 × 2.0 mm) with 2% Buffer B (10% Buffer A, 90% acetonitrile).
Peptides were separated with a Shimadzu LC-20AT HPLC system using a 2%–40% Buffer B linear gradient over 60 min at 0.4 mL/min flow rate, collecting 24 fractions throughout the length of the
gradient. Fractions were dried down using a speedvac and pooled by concatenation into six final fractions. The samples were analyzed using data-independent acquisition (DIA) analysis by a
Thermo Q-Exactive Orbitrap coupled to a Dionex Ultimate 3000 RSLC nano UPLC with a Waters Atlantic reverse phase column (100 μm × 150 mm). For data-independent acquisition (DIA), the MS
survey scan was performed in profile mode with a resolution of 70,000, AGC of 1e6, maximum fill time of 100 ms in the scan range between 400 and 1000 m/z. The survey scan was followed 30 DIA
scans in profile mode with a resolution of 35,000, AGC 1e6, 20 m/z window, and NCE of 30. The source voltage was set at 2000 V and capillary temperature at 250 °C. To deconvolute and
analyze the DIA spectra, a spectral library containing all light and heavy acetyl-lysine feature pairs was generated. Spectral library samples were processed identically to the experimental
samples, except they were treated with C12-acetic anhydride (Sigma) and analyzed using data-dependent acquisition (DDA) mass spectrometry analysis. Using the openly available MaxQuant
(v1.6.1) software package, we performed a database search to find peptides present in the DDA samples analyzed. Carbamidomethylation (C) was set as a fixed modification, and oxidation (M)
and acetyl (K) were set as variable modifications. Trypsin and GluC were set as the digestion enzymes, with the max number of missed cleavages set to five. DDA runs from both the
mitochondrial and cytosolic fractions were run to make one combined library. Heavy acetyl fragment ion pairs were generated in silico, such that the spectral library would contain both the
light (endogenous) acetylation peaks and the heavy (chemical) acetylation peaks. The experimental samples were processed using Spectronaut (v10) using the generated spectral library. The
subcellular fraction experimental samples were processed separately. The data were processed using an in-house R script, which can be accessed through the GitHub link:
https://doi.org/10.5281/zenodo.3238525, such that stoichiometry was calculated from the ratio of endogenous (light) fragment ion peak area over the total (endogenous and chemical) fragment
ion peak area. We performed an isotopic envelope correction of the heavy-labeled peak to remove any contribution from naturally occurring isotopes from the light labeled peak. All fractions
were combined for downstream analysis; proteins that were _P_ < 0.05 compared with WT were filtered as significant changes (see Statistics section for processing. The raw data, processed
data, spectral library, and the analysis logs describing the settings for the Spectronaut analyses have been deposited to the ProteomeXchange Consortium via the MassIVE partner repository
with the data set identifier PXD014013. QUANTITATIVE POST-ACQUISITION DATA SET ANALYSIS Pathway analysis was performed using the online tool WebGestalt (www.webgestalt.org), using _Mus
musculus_ as the organism, Over-Representation Analysis as the method, and the KEGG pathway database. Uniprot identifiers of proteins passing a _p_-value threshold of 0.05 were used as the
input and genome_protein-coding as the reference gene set. Additional parameters were 5 as the minimum and 500 as the maximum number of genes for a category, Benjamini–Hochberg for multiple
test adjustment to an FDR of 0.05. To determine subcellular localization, proteins were categorized according to their primary Uniprot annotation. Secretory pathway localization includes the
endoplasmic reticulum, Golgi apparatus, proteasome, lysosome, extracellular, and secreted proteins. The nucleus includes chromosome and nucleus. Cytoplasm includes cytoskeleton, cytosol,
ribosomes, and all other organelles. The heatmap was generated in Perseus (v. 1.5.8.5)29 using Log2 transformed fold change from the control. Hierarchical clustering was performed using
Euclidean as the distance metric and complete as the linkage criterion. Cluster analysis was determined using KEGG pathways, which arose during pathway analysis. For the lipid metabolism
cluster, all proteins which were found in the following KEGG pathways in either of the AT-1 models and in either the proteome or acetyl stoichiometry were included: fatty acid degradation,
peroxisome, butanoate metabolism, proponoate metabolism, biosynthesis of unsaturated fatty acid, and fat digestion. For the mitochondria-related cluster, all proteins that were found in the
following KEGG pathways were included: PPAR signaling pathway, oxidative phosphorylation, TCA cycle, Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease. These proteins were
the input for a STRING analysis. Proteins with no interactions were hidden, with the minimum required interaction score set at high confidence (0.7). Active interaction sources included all
sources except textmining. METABOLOMICS Hepatocytes were plated at 900,000 cells per 35 mm collagen-coated plate. All cells received a media change of pre-media (10% FBS, 1%
penicillin/streptomycin/glutamine in the Dulbecco’s modified Eagle medium (DMEM)) 2 h before 13C media was added. After the 2 h pre-incubation, cells were either immediately snap-frozen in
liquid nitrogen or given 13C media (10% FBS, 1 mM sodium pyruvate, 25 mM 13C5-labeled glutamine (99%, Cambridge Isotope Laboratories) in glutamine-free DMEM) for 30 min and then snap-frozen
with liquid nitrogen. Plates of cultured hepatocytes and media were kept frozen at −80 °C until time of extraction. Media (500 µL) was lyophilized by vacuum centrifugation. Metabolites were
extracted directly from plated hepatocytes or from lyophilized media on ice with 4 °C 7:2:1 HPLC-grade methanol:water:chloroform. After addition of extraction solvent, cells were immediately
scraped from the plate using a cell scraper; media samples were vortexed and probe sonicated for 10 s. Resulting extracts were transferred to microcentrifuge tubes and incubated at 4 °C for
5 min, then centrifuged at 10000 × _g_ for 10 min at 4 °C to pellet precipitated protein. Extracts were aliquoted into glass autosampler vials and dried by vacuum centrifugation (150 µL for
gas chromatography analysis). For GC-MS analysis, dried extract was derivatized for 90 min with 20 mg/mL methoxyamine hydrochloride in pyridine at 20 °C (25 µL) and then with MSTFA for 30
min at 37 °C (25 uL). Samples were analyzed by GC-Orbitrap; 1 µL of sample, split 1:10, was injected onto a TraceGOLD TG-5SilMS GC column (cat. no. 26096–1420, Thermo Scientific).
Temperature was held at 50 °C for 1 min, then ramped to 320 °C at a rate of 11 °C/min, then held at 320 °C for 4.40 min. Molecules were analyzed with positive electron-impact (EI)-Orbitrap
full scan of 50–650 m/z range. For data analysis, selected m/z and retention times were used to quantify metabolites and their isotopic distributes; peak areas were quantified using Thermo’s
Tracefinder application. We corrected for naturally occurring 13C isotopes using tool written in MATLAB30 and performed batch-correction using the ComBat function in R31,32. Metabolomics
raw MS files are available on the public repository MassIVE with the accession code ID MSV000083885 (ftp://massive.ucsd.edu/MSV000083885). Additional information can be found in
Supplementary Table 3 and Supplementary Data 3. STATISTICS Data analysis was performed in Graphpad Prism v 7.02 (GraphPad Software, Inc) and R v3.5.1. Unless otherwise specified, data are
expressed as mean ± standard deviation. For the proteomics, fold changes were computed within each DiLeu batch experiment, an F-test was used to test for equivalent variance among groups,
and a Student’s _t_ test was performed assuming equal or unequal variance according to the results of the F-test. A final fold change was calculated by averaging the two experiments
together, and the _p-_values of the two separate DiLeu experiments were combined using Fisher’s method as implemented in the R package _metap_ (R version 3.5.1). For the stoichiometry of
acetylation, a one-way ANOVA test was performed. Comparison of the proteins and acetyl proteoforms distribution was performed using a Kolmogorov–Smirnov test. For all other analyses,
comparison of the means was performed using a Student’s _t_ test. The following statistical significance was used: *_P_ < 0.05; **_P_ < 0.005; #_P_ < 0.0005. REPORTING SUMMARY
Further information on research design is available in the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY The mass spectrometry proteomics data that support the
findings of this study have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the accession code PXD013736. The acetyl-proteomics data that support the
findings of this study have been deposited to the ProteomeXchange Consortium via the MassIVE partner repository with the accession code PXD014013. The metabolomics data that support the
findings of this study have been deposited in the MassIVE repository with the accession code MSV000083885 (ftp://massive.ucsd.edu/MSV000083885).The R script that was used to process the
acetyl-proteomics data have been deposited on Github with the identifier (search terms: AT1 Acetylation Stoich) (https://doi.org/10.5281/zenodo.3238525). The authors declare that all other
data supporting the findings of this study are available within the paper and its supplementary tables. CODE AVAILABILITY The in house R script that was used to process the acetyl-proteomics
data have been deposited on Github with the identifier (search terms: AT1 Acetylation Stoich) (https://doi.org/10.5281/zenodo.3238525). The README file found on Github describes how the
input data for the scripts can be accessed through ProteomeXchange accession code PXD014013. REFERENCES * Jonas, M. C., Pehar, M. & Puglielli, L. AT-1 is the ER membrane acetyl-CoA
transporter and is essential for cell viability. _J. Cell Sci._ 123, 3378–3388 (2010). Article CAS Google Scholar * Farrugia, M. A. & Puglielli, L. Nε-lysine acetylation in the
endoplasmic reticulum–a novel cellular mechanism that regulates proteostasis and autophagy. _J. Cell Sci._ 131, jcs221747 (2018). Article Google Scholar * Huppke, P. et al. Mutations in
SLC33A1 cause a lethal autosomal-recessive disorder with congenital cataracts, hearing loss, and low serum copper and ceruloplasmin. _Am. J. Hum. Genet._ 90, 61–68 (2012). Article CAS
Google Scholar * Chiplunkar, S. et al. Huppke-Brendel syndrome in a seven months old boy with a novel 2-bp deletion in SLC33A1. _Metab. Brain Dis._ 31, 1195–1198 (2016). Article Google
Scholar * Lin, P. et al. A missense mutation in SLC33A1, which encodes the acetyl-CoA transporter, causes autosomal-dominant spastic paraplegia (SPG42). _Am. J. Hum. Genet._ 83, 752–759
(2008). Article CAS Google Scholar * Sanders, S. J. et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with
autism. _Neuron_ 70, 863–885 (2011). Article CAS Google Scholar * Prasad, A. et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. _G3
(Bethesda)_ 2, 1665–1685 (2012). Article CAS Google Scholar * Peng, Y. et al. Deficient import of acetyl-CoA into the ER lumen causes neurodegeneration and propensity to infections,
inflammation, and cancer. _J. Neurosci._ 34, 6772–6789 (2014). Article CAS Google Scholar * Hullinger, R. et al. Increased expression of AT-1/SLC33A1 causes an autistic-like phenotype in
mice by affecting dendritic branching and spine formation. _J. Exp. Med._ 213, 1267–1284 (2016). Article CAS Google Scholar * Peng, Y. et al. Increased transport of acetyl-CoA into the
endoplasmic reticulum causes a progeria-like phenotype. _Aging Cell_ 17, e12820 (2018). Article Google Scholar * Pehar, M., Jonas, M. C., Hare, T. M. & Puglielli, L. SLC33A1/AT-1
protein regulates the induction of autophagy downstream of IRE1/XBP1 pathway. _J. Biol. Chem._ 287, 29921–29930 (2012). Article CAS Google Scholar * Peng, Y. & Puglielli, L. N-lysine
acetylation in the lumen of the endoplasmic reticulum: a way to regulate autophagy and maintain protein homeostasis in the secretory pathway. _Autophagy_ 12, 1051–1052 (2016). Article CAS
Google Scholar * Peng, Y. et al. Improved proteostasis in the secretory pathway rescues Alzheimer's disease in the mouse. _Brain_ 139, 937–952 (2016). Article Google Scholar * Pehar,
M., Lehnus, M., Karst, A. & Puglielli, L. Proteomic assessment shows that many endoplasmic reticulum (ER)-resident proteins are targeted by N(epsilon)-lysine acetylation in the lumen of
the organelle and predicts broad biological impact. _J. Biol. Chem._ 287, 22436–22440 (2012). Article CAS Google Scholar * Li, X., Lian, F., Liu, C., Hu, K. Q. & Wang, X. D.
Isocaloric pair-fed high-carbohydrate diet induced more hepatic steatosis and inflammation than high-fat diet mediated by miR-34a/SIRT1 axis in mice. _Sci. Rep._ 5, 16774 (2015). Article
ADS CAS Google Scholar * Ishimoto, T. et al. High-fat and high-sucrose (western) diet induces steatohepatitis that is dependent on fructokinase. _Hepatology_ 58, 1632–1643 (2013). Article
CAS Google Scholar * Frost, D. C., Greer, T. & Li, L. High-resolution enabled 12-plex DiLeu isobaric tags for quantitative proteomics. _Anal. Chem._ 87, 1646–1654 (2015). Article
CAS Google Scholar * Xiang, F., Ye, H., Chen, R., Fu, Q. & Li, L. N,N-dimethyl leucines as novel isobaric tandem mass tags for quantitative proteomics and peptidomics. _Anal. Chem._
82, 2817–2825 (2010). Article CAS Google Scholar * Kouzarides, T. Acetylation: a regulatory modification to rival phosphorylation? _EMBO J._ 19, 1176–1179 (2000). Article CAS Google
Scholar * Anderson, K. A. & Hirschey, M. D. Mitochondrial protein acetylation regulates metabolism. _Essays Biochem._ 52, 23–35 (2012). Article CAS Google Scholar * Pougovkina, O. et
al. Mitochondrial protein acetylation is driven by acetyl-CoA from fatty acid oxidation. _Hum. Mol. Genet._ 23, 3513–3522 (2014). Article CAS Google Scholar * Baeza, J. et al.
Quantifying dynamic protein acetylation using quantitative stoichiometry. Preprint at https://www.biorxiv.org/content/10.1101/472530v1.full (2018). * Villena, J. A. New insights into PGC-1
coactivators: redefining their role in the regulation of mitochondrial function and beyond. _FEBS J._ 282, 647–672 (2015). Article CAS Google Scholar * Scarpulla, R. C., Vega, R. B. &
Kelly, D. P. Transcriptional integration of mitochondrial biogenesis. _Trends Endocrinol. Metab._ 23, 459–466 (2012). Article CAS Google Scholar * Nieborak, A. & Schneider, R.
Metabolic intermediates - cellular messengers talking to chromatin modifiers. _Mol. Metab._ 14, 39–52 (2018). Article CAS Google Scholar * Kaelin, W. G. Jr. & McKnight, S. L.
Influence of metabolism on epigenetics and disease. _Cell_ 153, 56–69 (2013). Article CAS Google Scholar * Pehar, M. et al. Altered longevity-assurance activity ofp53: p44 in the mouse
causes memory loss, neurodegeneration and premature death. _Aging Cell_ 9, 174–190 (2010). Article CAS Google Scholar * Baeza, J. et al. Stoichiometry of site-specific lysine acetylation
in an entire proteome. _J. Biol. Chem._ 289, 21326–21338 (2014). Article Google Scholar * Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics
data. _Nat. Methods_ 13, 731–740 (2016). Article CAS Google Scholar * MATLAB Release R2015b. (2015). * Leek, J. T. et al. _sva: Surrogate Variable Anaysis_. R package version 3.28.0.
(2018). * R: A language and environment for statistical computing (R Foundation for Statistical Computing, 2017). Download references ACKNOWLEDGEMENTS We thank Dr. John Svaren for critical
reading of an early version of this paper. We thank Karl Miller for technical help. This work was supported by the NIH (NS094154 and AG053937 to L.P.; AG057408 to L.P. and R.M.A.; GM65386 to
J.M.D.; R01DK071801, R01AG052324, and P41GM108538 to L.L.; P41 GM108538 to J.J.C.); a core grant to the Waisman Center from NICHD-U54 HD090256. I.A.D. was supported by T32 AG000213 and T32
GM007507. SIM imaging was performed at the Biochemistry Optical Core of the University of Wisconsin‐Madison (Madison, WI). The Orbitrap instruments were purchased through the support of an
NIH shared instrument grant (NIH-NCRR S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. This work was also supported
using resources and facilities of the William S. Middleton Memorial Veterans Hospital (Madison, WI, USA). AUTHOR INFORMATION Author notes * Qing Yu Present address: Harvard Medical School,
Boston, MA, USA * Porsha R. Howell Present address: Harvard T.H. Chan School of Public Health, Boston, MA, USA * Maggie S. Burhans Present address: Fred Hutchinson Cancer Research Center,
Seattle, WA, USA * These authors contributed equally: Alexis J. Lawton, Yajing Peng, Qing Yu. AUTHORS AND AFFILIATIONS * Department of Medicine, University of Wisconsin-Madison, Madison, WI,
USA Inca A. Dieterich, Yajing Peng, Timothy W. Rhoads, Porsha R. Howell, Maggie S. Burhans, Rozalyn M. Anderson & Luigi Puglielli * Waisman Center, University of Wisconsin-Madison,
Madison, WI, USA Inca A. Dieterich, Yajing Peng & Luigi Puglielli * Neuroscience Training Program, University of Wisconsin-Madison, Madison, WI, USA Inca A. Dieterich * Department of
Biomolecular Chemistry and the Wisconsin Institute for Discovery, University of Wisconsin-Madison, Madison, WI, USA Alexis J. Lawton, Eric A. Armstrong & John M. Denu * School of
Pharmacy and Department of Chemistry, University of Wisconsin-Madison, Madison, WI, USA Qing Yu, Yusi Cui & Lingjun Li * Department of Chemistry, Biomolecular Chemistry and Genome Center
of Wisconsin, University of Wisconsin-Madison, Madison, WI, USA Katherine A. Overmyer & Joshua J. Coon * Morgridge Institute for Research, Madison, WI, USA Katherine A. Overmyer &
Joshua J. Coon * Geriatric Research Education Clinical Center, Veterans Affairs Medical Center, Madison, Wisconsin, WI, USA Rozalyn M. Anderson & Luigi Puglielli * Department of
Neuroscience, University of Wisconsin‐Madison, Madison, WI, USA Luigi Puglielli Authors * Inca A. Dieterich View author publications You can also search for this author inPubMed Google
Scholar * Alexis J. Lawton View author publications You can also search for this author inPubMed Google Scholar * Yajing Peng View author publications You can also search for this author
inPubMed Google Scholar * Qing Yu View author publications You can also search for this author inPubMed Google Scholar * Timothy W. Rhoads View author publications You can also search for
this author inPubMed Google Scholar * Katherine A. Overmyer View author publications You can also search for this author inPubMed Google Scholar * Yusi Cui View author publications You can
also search for this author inPubMed Google Scholar * Eric A. Armstrong View author publications You can also search for this author inPubMed Google Scholar * Porsha R. Howell View author
publications You can also search for this author inPubMed Google Scholar * Maggie S. Burhans View author publications You can also search for this author inPubMed Google Scholar * Lingjun Li
View author publications You can also search for this author inPubMed Google Scholar * John M. Denu View author publications You can also search for this author inPubMed Google Scholar *
Joshua J. Coon View author publications You can also search for this author inPubMed Google Scholar * Rozalyn M. Anderson View author publications You can also search for this author
inPubMed Google Scholar * Luigi Puglielli View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS I.A.D., A.J.L., Y.P., Q.Y., K.A.O., Y.C., E.A.A.
and P.R.H. performed the experiments and analyzed the data. T.W.R. analyzed the data. M.S.B., L.L., J.M.D., J.J.C., R.M.A. and L.P. provided critical advice for the experiments. L.P.
designed the overall study. L.P. and I.A.D. wrote the paper with input from all authors. CORRESPONDING AUTHOR Correspondence to Luigi Puglielli. ETHICS DECLARATIONS COMPETING INTERESTS
J.M.D. is a co-founder of Galilei BioScience Inc, devoted to the development of small-molecule effectors for SIRT6. Remaining authors have no conflict of interests to disclose. ADDITIONAL
INFORMATION PEER REVIEW INFORMATION: _Nature Communications_ would like to thank Suzanne Jackowski and other, anonymous, reviewers for their contributions to the peer review of this work.
Peer review reports available. PUBLISHER’S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE SUPPLEMENTARY DATASETS REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the
article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use
is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Dieterich, I.A., Lawton, A.J., Peng, Y. _et al._ Acetyl-CoA flux regulates the
proteome and acetyl-proteome to maintain intracellular metabolic crosstalk. _Nat Commun_ 10, 3929 (2019). https://doi.org/10.1038/s41467-019-11945-9 Download citation * Received: 01 March
2019 * Accepted: 08 August 2019 * Published: 02 September 2019 * DOI: https://doi.org/10.1038/s41467-019-11945-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to
read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative






