- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT FAM210B (family with sequence similarity 210 member B) is a novel protein that has been linked to tumor development. However, its role and underlying mechanisms in lung
adenocarcinoma (LUAD) progression remain largely unexplored. In this study, FAM210B was observed to be down-regulated in LUAD cells. Analyses of public datasets revealed that decreased
expression of FAM210B predicts poor survival. Accordingly, in vitro and in vivo studies have confirmed the inhibitory role of FAM210B on the growth and tumor metastasis of LUAD cells.
RNA-seq analysis further indicated that FAM210B plays a role in regulating innate immune-related signaling pathways in LUAD cells, particularly involving the production of type I interferon
(IFN-α/β). Specifically, FAM210B activates STAT1/IRF9/IFIT3 axis by upregulating IFN-α/β expression, leading to the inhibition of proliferation and migration of LUAD cells. Furthermore,
TOM70 (Translocase of outer mitochondrial membrane 70, also named as TOMM70) has been identified as a functional interacting partner of FAM210B in its modulation on the expression of
IFN-α/β, as well as the proliferative and metastatic phenotypes of LUAD cells. In conclusion, our study indicates that FAM210B is an important suppressor of cellular viability and mobility
during lung cancer progression. SIMILAR CONTENT BEING VIEWED BY OTHERS RNA M6A READER YTHDF2 FACILITATES LUNG ADENOCARCINOMA CELL PROLIFERATION AND METASTASIS BY TARGETING THE
AXIN1/WNT/Β-CATENIN SIGNALING Article Open access 13 May 2021 RNA BINDING PROTEIN ALYREF REGULATES FERROPTOSIS TO FACILITATE LUAD GROWTH AND METASTASIS VIA PROMOTING SLC7A11 MRNA STABILITY
Article Open access 08 January 2025 DDX17 PROMOTES THE GROWTH AND METASTASIS OF LUNG ADENOCARCINOMA Article Open access 22 October 2022 INTRODUCTION Lung cancer is a highly malignant form of
cancer with high morbidity and fatality rates, making it a major global health concern [1, 2]. There are two main subtypes of lung cancer: non-small cell lung cancer (NSCLC) and small cell
lung cancer (SCLC). NSCLC accounts for 85% of lung cancers, mainly comprised of lung adenocarcinoma (LUAD, 40%) and lung squamous cell carcinoma [3]. Despite advancements being made in tumor
therapeutic strategies in the recent years, the five-year survival rate for individuals suffering from lung cancer remains poor [3]. Further understanding of LUAD pathogenesis is critical
for the development of new therapy for this devastating illness. Type I interferons (IFN) are major immune response regulators mainly consist of several IFN-α subtypes and one IFN-β subtype,
with otherwise the less-reported IFN-ɛ, −κ, −τ, and −ω subtypes. These proteins exert their functions through binding to a heterodimer receptor of IFNAR1/IFNAR2. The ligand-receptor complex
recruits and phosphorylates the signal transducers and activators of transcription 1 (STAT1) and STAT2, which then interact with interferon regulatory factor 9 (IRF9), and is translocated
to the nucleus to activate the transcription of interferon-stimulated genes (ISGs) [4, 5]. Both the paracrine and autocrine signaling of IFN have been reported to play important roles in
tumor development, respectively, by regulating the anti-tumor immune response or the cellular processes and functions of the tumor cells [6,7,8,9,10,11]. Taking part in the
Chromosome-centric Human Proteome Project (C-HPP), our team has been recently dedicated to studying the role of newly identified genes on Chromosome 20 in tumor development, as demonstrated
by our previous research on ZSWIM1’s involvement in the pathogenesis of LUAD [12] and the role of C20orf24 in promoting colorectal cancer progression [13]. _FAM210B_ (family with sequence
similarity 210 member B, C20orf108) is another newly identified gene located on Chromosome 20 and is widely expressed in human tissues [14]. Based on its significant regulatory effect on
LUAD cell proliferation in a preliminary screening assay, we have selected this gene for further studies aiming to elucidate its functions in relation to LUAD cancer development. Up to date,
little has been known about its physiological function, and even less about the underlying molecular mechanisms. In the context of tumor development, downregulation of FAM210B has been
linked to decreased survival rates for tumor patients and increased growth and metastasis of tumor cells [15, 16]. In ovarian cancer, the loss of FAM210B was found to reduce the expression
of pyruvate dehydrogenase kinase 4 (PDK4) and glycolysis, promoting epithelial-to-mesenchymal transition (EMT), migration, and invasion [15]. In hepatocellular carcinoma (HCC), knockdown of
FAM210B protein was associated with increased activation of MAPK signaling and AKT signaling pathways [16]. Moreover, altered expression of FAM210B in peripheral blood has been identified as
part of a 16-gene signature for predicting immune-related adverse events of severe diarrhea during anti-CTLA-4 immunotherapy [17]. FAM210B was found to be associated with mammary tumor
incidence in rats [18]. These previous studies suggested a possible association of this gene with cancer. Further in-depth exploration in the field appears meaningful. In this study, we
demonstrate the significant inhibitory role of FAM210B in LUAD growth and metastasis and explore the related molecular mechanism. By using RNA-seq assay and interactome analysis, we show
that FAM210B activates the innate immune pathway in LUAD cells by interacting with TOM70 (Translocase of outer mitochondrial membrane 70, also named TOMM70), leading to the production of
type I interferon and ultimately inhibiting the growth and metastasis of LUAD cells. Our data provide novel insights into the pathogenesis of LUAD and identify FAM210B as a potential therapy
target for this disease. MATERIALS AND METHODS CELL LINES AND CULTURE Human bronchial epithelial (HBE) cells and human non-small cell lung adenocarcinoma cell lines H1299 and A549 were
obtained from Cell Resource Center, Institute of Life Science Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM (Gibco BRL, USA) and supported with 10% FBS (Procell Life
Science & Technology Co., Ltd, Wuhan, China) in a humidified chamber at 37 °C with 5% CO2 as described previously [12]. OVERALL SURVIVAL ANALYSIS Kaplan-Meier (KM) Plotter
(https://kmplot.com/analysis/) was utilized to analyze overall survival from the TCGA-LUAD dataset (_n_ = 504) and two GEO datasets, namely GSE31210 (_n_ = 226) and GSE30219 (_n_ = 85). The
TCGA-LUAD dataset comprises a total of 513 LUAD cases; however, 9 cases were excluded from the overall survival analysis due to missing status information, resulting in a final analysis of
504 patients. The GSE30219 dataset includes a total of 293 lung tumor patients, of which 85 cases were classified as adenocarcinoma (ADC) based on histological classification. Since our
study primarily focuses on the adenocarcinoma subtype, we selected these 85 cases for survival curve analysis. Hazard ratios (HRs) with 95% confidence intervals and log-rank _P_ values were
calculated to assess the prognostic significance of FAM210B expression. PLASMID CONSTRUCTION The purchased pEGFP-N1-FAM210B plasmid (iGEbio Co., Ltd, Guangzhou, China) was used as template
to amplify the FAM210B coding sequence (CDS) of 591 bp. The pCMV-FLAG-based plasmid FLAG-FAM210B was then constructed using seamless cloning technology (Vazyme Biotech Co., Ltd, Nanjing,
China). ESTABLISHMENT OF STABLE CELLS Cells stably expressing FLAG-FAM210B were established using methods similar to those used in previous studies [12]. Briefly, recombinant FLAG-FAM210B
cDNA was constructed and then inserted into the lentiviral vector PLVX-IRES-EGFP-PuroR. This recombinant plasmid, in conjunction with packaging plasmids psPAX2 and pMD2.G, was employed to
generate lentiviruses in HEK-293T cells carrying PLVX-EGFP-FLAG-FAM210B. After 48 h, the culture medium carrying the lentiviruses was collected and used to infect H1299 cells, A549 cells,
and luciferase-expressing A549 cells, resulting in the generation of stable cells (OE-FAM210B and OE-V, or OE-FAM210B-Luc or OE-V-Luc). REAL-TIME QUANTITATIVE REVERSE TRANSCRIPTION PCR
(QRT-PCR) ASSAYS Total RNA extraction and cDNA quantification were performed using standard methods. The gene-specific primer sequences used in this study are provided in Table S1. SMALL
INTERFERING RNA TRANSFECTION The small interfering RNA (siRNA) sequences designed in this study are provided in Table S2. Cells were transfected with siRNA using Lipofectamine@2000
(Invitrogen, CA, USA) according to the manufacturer’s instructions. COLONY FORMATION ASSAY 1 × 103 cells/well were seeded into 6-well plates and cultured for 10–15 days. The resulting
colonies were fixed with 4% paraformaldehyde for 30 min, stained with crystal violet for 15 min, and counted by ImageJ software. TRANSWELL ASSAY The Transwell assay was performed following
the protocol described previously [12]. Migration assays were conducted using Transwell chambers with polycarbonate membranes (24 wells, 8 μm pore size), while invasion assays utilized
Matrigel-coated chambers (24 wells, 8 μm pore size). H1299 or A549 cells (8–10 × 104/well) were seeded into the upper chamber containing serum-free DMEM. After an incubation period of 8–10
h, cells that migrated to the basal side of the membrane were fixed and stained for subsequent microscopic evaluation using an Olympus microscope (Tokyo, Japan). WOUND HEALING ASSAY A549
cells of OE-Vector and OE-FAM210B or H1299 cells transfected with GFP-Vector or GFP-FAM210B were cultured in 6 cm petri dishes until reaching confluency. A linear wound was created by
scratching, and the petri dishes were then placed on the stage of an inverted microscope equipped with phase-contrast optics to capture sequential images of the cells at 5-min intervals over
a period of 24 h to observe the process of wound healing. The recording process was controlled using Cyto-MINI software (Guangzhou Shipu Photoelectric Technology Co. LTD, Guangzhou, China)
as described previously [12]. MTT ASSAY 1 × 103 cells/well were seeded into 96-well plates and cultured for the indicated durations. Then, 200 μl of MTT working solution (0.5 mg/ml) was
added to each well and incubated for 4 h. After that, 150 μl of dimethyl sulfoxide was added and incubated for 10 min. The absorbance at 570 nm was measured to assess cell proliferation at
each time point. WESTERN BLOTTING ASSAY The Western blotting assay was conducted following previously described methods [12, 19, 20]. Antibodies used in this study are as following: β-actin
(#66009-1-Ig), LaminA/C (#10298-1-AP), GAPDH (#10494-1-AP), STAT1 (#10144-2-AP), IRF9 (#14167-1-AP), TOM70 (#14528-1-AP), and Vimentin (#10366-1-AP) from Proteintech (Wuhan, China);
E-cadherin (#3195) from Cell Signaling Technology (Massachusetts, USA). Other antibodies used in this study were p-STAT1 (#AF3300, Affinity Biosciences, OH, USA), anti-p-IRF3 (#AF3438,
Affinity Biosciences, OH, USA), IFN-α (#DF6086, Affinity Biosciences, OH, USA), IRF3 (#a19717, ABclonal, China), IFN-β (#R381675, Zenbio, USA), FLAG (#M185-3, MBL, Tokyo, Japan), and FAM210B
(#NBP2-14523, NOVUS, CO, USA). NUCLEAR-CYTOPLASMIC SEPARATION ASSAYS H1299 cells transfected with either GFP-FAM210B or GFP-Vector were utilized to conduct nuclear-cytoplasmic separation
assays. The Nuclear-Cytosol Extraction Kit (#P1200, Beijing Pulilai Gene Technology Co., Ltd, China) was employed to separate nuclear and cytoplasmic fractions according to the
manufacturer’s instructions. Following this separation, western blotting was performed to analyze protein expression levels in the different fractions. The blots were probed with specific
antibodies against IRF3 (#a19717, ABclonal, China) and phosphorylated IRF3 (#AF3438, Affinity Biosciences, OH, USA) to assess the localization and expression of these proteins. RNA-SEQ ASSAY
H1299 cells were transfected with GFP-FAM210B or FAM210B siRNA for RNA-seq analysis. Total RNA was extracted using TRIzol reagent (Invitrogen, USA). Equal amounts of total RNA from three
independent experiments were combined to create a library for RNA-seq, following the standard protocol provided by BGI (BGI-Shenzhen, Shenzhen, China). The library was prepared using the
BGISEQ-500RS High-throughput sequencing kit (PE100, V3.0, MGI Tech Co., Ltd), as previously described [21, 22]. Differential gene expression analysis was conducted using the edgeR package,
considering a _P_ value threshold of less than 0.01. To perform Gene Ontology (GO) analysis on these differentially expressed genes (DEGs), the Database for Annotation, Visualization, and
Integrated Discovery (DAVID) platform was utilized. CO-IMMUNOPRECIPITATION (CO-IP) ASSAY The cell samples were lysed in EBC buffer supplemented with a proteasome inhibitor cocktail (CWbio
Co., Ltd, Taizhou, China), as well as NaF, Na3VO4, and PMSF [12]. 1 mg of cell lysate was incubated with 2 μg of primary antibody overnight at 4 °C. Protein A/G PLUS-Agarose (Santa Cruz
Biotechnology, Texas, USA) was then added to the mixture and incubated for 4–6 h. The immune complexes were subsequently subjected to five washes with EBC buffer and separated using western
blotting. IMMUNOFLUORESCENCE (IF) ASSAY H1299 cells were transfected either with pGFP-FAM210B and FLAG-TOM70 (for the colocalization of FAM210B and TOM70 proteins), or only with pGFP-FAM210B
(for the localization of p-IRF3). After fixation, the cells were incubated with anti-TOM70 (#14528-1-AP, 1:200) or anti-p-IRF3 (#AF3438, Affinity Biosciences, OH, USA) overnight at 4 °C.
The cells were then incubated with Alexa Fluor 594-conjugated secondary antibody (ZSGBBIO Co., Ltd, Beijing, China), and the nuclei were stained with DAPI. To observe the colocalization of
GFP-FAM210B and mitochondria in live cells, H1299 cells were transfected with pGFP-FAM210B for 48 h and then stained with MitoTracker Red (100 nM, Thermo Fisher Scientific) for 20 min. These
samples were observed and imaged using a confocal microscope (LSM 900, Carl Zeiss). MASS SPECTROMETRY ANALYSIS H1299 cells stably expressing FAM210B (OE-FLAG-FAM210B) were lysed with lysis
buffer. The lysates were then subjected to standard co-IP with anti-FLAG antibody, while non-immune IgG was used as the control group. The immunoprecipitated complexes were separated by
SDS-PAGE and visualized through silver staining. The proteins in the gel were subsequently extracted using an enzymatic hydrolysis method similar to that described previously [12, 23]. The
resulting peptide samples were analyzed on an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, MA, USA). The raw data obtained from the mass spectrometer were automatically
analyzed by the Sequest HT engine of the Proteome Discoverer (PD, Thermo Fisher Scientific) v2.1.1.21 against UniProtKB/Swiss-Prot _Homo sapiens_ protein database with the default settings.
ETHICS STATEMENT All animal-related procedures in this study were approved by the Experimental Animal Ethics Committee of Jinan University (Approval No: IACUC-20220219-04). IN VIVO ANIMAL
EXPERIMENTS The mice were purchased from Beijing Weitong Lihua Laboratory Animal Co., LTD (Beijing, China), and were raised under SPF standard conditions. For the xenograft experiment, A549
cells or H1299 cells stably overexpressing FLAG-FAM210B (OE-FAM210B) and their respective control cells (1.5–2.0 × 106 cells/100 μL/mouse, _n_ = 6, based on random allocation) were injected
into the subcutaneous area of the armpit of BALB/c nude mice following previously described methods [24]. The tumor growth rate was monitored every 2 days, and the volume of tumor was
calculated using the formula: _V_ = (length x width2)/2. At 30–40 days of subcutaneous inoculation, the mice were euthanized by cervical dislocation, and the tumors were collected for
western blot and histological analysis. No blinding was done. For the in vivo metastasis experiment, each female NCG mouse (NOD-Prkdcem26Cd52Il2rgem26Cd22, aged 4–6 weeks, Beijing Weitong
Lihua Laboratory Animal Co., LTD, Beijing, China) received an injection of luciferase-expressing OE-FAM210B-Luc or OE-V-Luc A549 cells (2 × 106 cells/100 μL PBS/mouse, _n_ = 6, based on
random allocation) via the tail vein. After 40 days post-injection, bioluminescent imaging was performed using a Xenogen IVIS Lumina II system (PerkinElmer, MA, USA) to visualize the
presence of lung metastases. Additionally, lung tissue sections were subjected to Hematoxylin and Eosin (H&E) staining to detect tumor metastasis in the lungs. STATISTICAL ANALYSIS The
mean ± SEM values from three independent experiments were presented in this study. Data visualization and statistical analysis were performed using GraphPad Prism 6 software. Statistical
significance was determined by performing Student’s _t_-test, or two-way ANOVA, with _P_ < 0.05 considered as significant. RESULTS FAM210B’S EXPRESSION IS POSITIVELY ASSOCIATED WITH THE
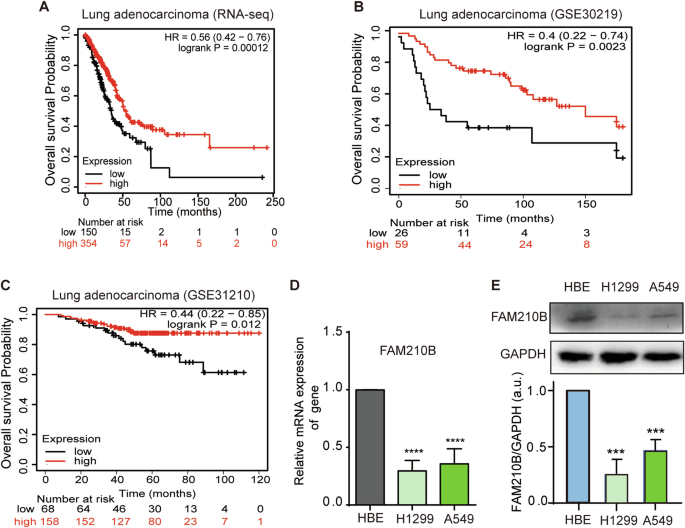
PROGNOSIS OF LUAD PATIENTS We investigated the correlation between FAM210B expression and overall survival in patients with LUAD. Kaplan-Meier analysis showed a statistically significant
link between high FAM210B expression and improved overall survival among LUAD patients (Fig. 1A–C), suggesting a positive correlation between FAM210B expression and the prognosis of LUAD.
Furthermore, FAM210B mRNA and protein levels in A549 and H1299 LUAD cells were significantly lower than that of normal HBE cells (Fig. 1D–E), hinting FAM210B might be involved in the
progression of LUAD. However, we did not observe differential expression of the _FAM210B_ gene in normal and tumor tissues of LUAD patients when analyzing TCGA data (Fig. S1A). In evaluating
the expression of FAM210B across different stages of LUAD, specifically the terminal respiratory unit (TRU), proximal-inflammatory (PI), and proximal-proliferative (PP) subtypes, as
referenced in Collisson et al.‘s study [25], the results shown in Fig. S1B indicated that tumor samples in the TRU stage (which harbored the majority of EGFR-mutated tumors) exhibited a
slight increase in expression compared to the PP and PI stages. However, the implications of this increase require further investigation. FAM210B SUPPRESSED LUAD CELL PROLIFERATION IN VITRO
AND LUAD TUMOR GROWTH IN VIVO We proceeded to investigate the impact of FAM210B on LUAD cell proliferation. Given that FAM210B expression is higher in A549 cells than in H1299 cells, we
knocked down FAM210B in A549 cells. Figure 2A shows the successful knockdown of FAM210B in A549 cells. The MTT assay revealed that silencing FAM210B significantly enhanced A549 cell
proliferation (Fig. 2B). Similarly, the colony formation assay demonstrated a notable increase in the number of A549 cell colonies following FAM210B knockdown (Fig. 2C). Conversely, stable
overexpression of FLAG-FAM210B in H1299 cells led to decreased cellular viability and colony forming capacity (Fig. 2D). Similar results were also obtained in A549 cells (Fig. 2E), with a
more pronounced decrease in cell viability on day 5 compared to H1299 cells, possibly due to the stronger suppressive effect of FAM210B overexpression on A549 cell viability. In vivo
experiments showed that stable overexpression of FAM210B in both H1299 and A549 cells inhibited tumor growth in xenograft mouse models, resulting in a reduction in tumor volume (Fig. 2F–I,
Fig. S2A–D). Immunohistochemistry analysis revealed a downregulation of both the proliferation marker Ki67 and the endothelial marker CD31 (indicating vascular density) from tumor tissues of
A549 and H1299 when overexpressing FAM210B (Fig. 2J, K, Fig. S2E), suggesting that FAM210B suppressed xenograft tumor growth and angiogenesis. FAM210B SUPPRESSED THE MIGRATION AND INVASION
OF LUAD CELLS IN VITRO AND THE TUMOR METASTASIS OF LUAD TUMOR IN VIVO Next, we tested the impact of FAM210B on LUAD metastasis ability. Both transient and stable overexpression of FAM210B in
H1299 and A549 cells led to a significant decrease in cell migration and invasion (Fig. 3A, B). Conversely, knockdown of FAM210B enhanced the migratory and invasive capabilities of H1299
cells (Fig. 3C). Consistently, FAM210B overexpression led to increased expression of E-cadherin (an epithelial marker) and decreased expression of Vimentin (mesenchymal markers), whereas
FAM210B silencing had the opposite effect on E-cadherin and Vimentin (Fig. 3D, E). These findings were further supported by wound healing assays, demonstrating delayed wound closure in A549
and H1299 cells overexpressing FAM210B (Fig. 3F). Metastasis suppression by FAM210B was then validated in the tail vein injection model, as evidenced by the reduced fluorescence intensity
observed in OE-FAM210B group compared to control group (Fig. 3G). The A549-injected mice had shrunk lungs covered by pulmonary metastatic nodules, while those of mice injected with A549
cells overexpressing FAM210B displayed normal form and much smoother surfaces (Fig. 3H, left panel). Furthermore, HE staining demonstrated decreased tumor metastasis area in lung tissues for
the FAM210B overexpressing group compared to the control group (Fig. 3H, right panel). These results indicated that FAM210B inhibited metastasis ability of LUAD. FAM210B ACTIVATED THE
STAT1/IRF9/IFIT3 SIGNAL PATHWAY THROUGH IFN-Α/Β UPREGULATION To explore the mechanism underlying FAM210B’s functions, we performed an RNA-seq assay with H1299 cells in which FAM210B was
either overexpressed or knocked down. A total of 113 differentially expressed genes (DEGs) were identified based on their opposite differential expression in FAM210B-overexpressed and
-knocked-down cells (Table S3, _P_ < 0.01). Strikingly, most of the top 15 GO-BP terms associated with these genes concerned IFN-mediated signal pathway, IFN production, and innate immune
response (Fig. 4A). IFN-α/β is well known to activate JAK1/STAT1/IRF9 signal pathway to promote the transcription of ISG, such as IFIT1-3 [4, 5, 26, 27] (Fig. 4B). Our data of RNA-seq assay
showed that within this pathway, the mRNA expression levels of 12 key factors were altered by FAM210B overexpression and knockdown (Fig. 4C). Four representative genes of this pathway,
STAT1, IRF9, IFIT1, and IFIT3, along with two key upstream cytokines, IFN-α and IFN-β, were identified to be positively regulated by FAM210B, as demonstrated by qRT-PCR assays in
FAM210B-overexpressed and FAM210B-knockdown cells (Fig. 4D, E). Western blotting assays revealed a similar trend of regulation for IFN-α, IFN-β, STAT1, and IRF9 by FAM210B, as well as the
positive regulation of p-STAT1 by FAM210B (Fig. 4F). Moreover, treating LUAD cells with 60 ng/mL IFN-α or IFN-β stimulated the mRNA expression of STAT1, IRF9, and IFIT3 in a time-dependent
manner (Fig. 4G, H). These results suggested that FAM210B promotes the expression of IFN-α/β and activates the IFN-α/β mediated STAT1/IRF9/IFIT3 signaling pathway. FAM210B INHIBITED THE
PROLIFERATION AND MIGRATION OF LUAD CELLS VIA STAT1/IRF9/IFIT3 AXIS We further explored whether the IFN-α/β-activated STAT1/IRF9/IFIT3 axis participated in FAM210B’s biological functions.
Knocking down STAT1, IRF9, or IFIT3 promoted colony formation and more importantly countered the effect of FAM210B overexpression (Fig. 5A–C). Similar results were also obtained in MTT
assays (Fig. 5D–F). Finally, transwell assays also demonstrated that knockdown of STAT1, IRF9, or IFIT3 partially reversed the downregulation of invasion and migration induced by FAM210B
overexpression (Fig. 5G–L, Fig. S3A–I). Taken together, these results suggested that the function of FAM210B was mediated by STAT1/IRF9/IFIT3 axis in LUAD cells. FAM210B INDUCED THE
EXPRESSION OF IFN-Α/Β THROUGH INTERACTING WITH TOM70 AND ACTIVATING IRF3 It has been reported that transcription factor IRF3 plays a crucial role in the transcription of IFN-α/β, so we
examined if FAM210B regulated IFN-α/β expression through IRF3. Our results revealed that overexpressing FAM210B increased the levels of phosphorylated IRF3 and total IRF3, and facilitated
the nuclear localization of phosphorylated IRF3 as examined by nuclear-cytoplasmic separation and IF assays (Fig. 6A–C). Consistently, knocking down IRF3 lowered the transcriptional
expression of IFN-α/β and suppressed FAM210B-induced upregulation of IFN-α/β mRNA level (Fig. 6D). These findings indicated that FAM210B indeed stimulated the transcriptional expression of
IFN-α/β through regulation of IRF3 expression, phosphorylation and nuclear import. It has been reported that FAM210B was localized in the mitochondrial membrane [15], and mitochondrial
proteins were crucial for producing IFN-α/β [28]. Using immunoprecipitation of the overexpressed FLAG-FAM210B in H1299 cells (Fig. 6E) followed by Mass Spectrometry analysis, we have
identified TOM70 (Translocase of outer mitochondrial membrane 70, also named as TOMM70), a mitochondrial outer membrane protein as a potential partner of FAM210B (Table S4). This interaction
was further confirmed by conventional co-IP assay (Fig. 6F). IF assay also demonstrated the co-localization of FAM210B with mitochondria, as well as its co-localization with TOM70 in the
mitochondria of LUAD cells (Fig. 6G). Interestingly, TOM70 silencing exhibited promoting effect on proliferation and migration of LUAD cells and reversed the inhibitory effect of FAM210B
overexpression on these phonotypes (Fig. 6H–J). Consistently, TOM70 knockdown inhibited the increased expression of IFN-α/β in OE-FAM210B cells (Fig. 6K). Our data suggested that the
interaction between FAM210B and TOM70 promoted the expression of IFN-α/β and thereby inhibited the proliferation and migration of LUAD cells. Interestingly, we found that TOM70 knockdown led
to a reduction of FAM210B expression in cells overexpressing OE-FAM210B, whereas the overexpression of FAM210B did not affect TOM70 levels (Fig. 6K). Further, we evaluated the effects of
STAT1, IRF9, and IRF3, three downstream effectors of IFN-α/β signaling, on the expression of TOM70 and FAM210B. qRT-PCR assays showed that knockdown of STAT1, IRF9, or IRF3 caused a
reduction of the expression of FAM210B, whereas knockdown of STAT1, IRF9, or IRF3 caused little or no effects on TOM70 expression (Fig. S4A–C). DISCUSSION LUAD is the most prevalent
histological subtype of lung cancer, characterized by high morbidity and fatality rates and a poor prognosis [1, 2]. Discovering new molecular mechanisms that contribute to LUAD
tumorigenesis would provide valuable insight into identifying potential drug targets for LUAD therapy. In this study, we demonstrated both in vitro and in vivo that FAM210B significantly
inhibited the growth and metastasis of LUAD cells, suggesting that FAM210B might be a promising suppressor of LUAD progression. Through analysis of transcriptional profiles, type I IFN
(IFN-α/β) signal pathway was identified as the top biological process regulated by FAM210B. Our data demonstrated that FAM210B stimulated the expression of IFN-α/β in LUAD cells and in
subcutaneous tumor tissues from mice receiving OE-FAM210B cells (Fig. S5), suggesting that elevated expression of IFN-α/β might be responsible for the repressive function of FAM210B in LUAD
cells. IFN-α/β has been used as a form of cytokine therapy for melanoma treatment [26, 29, 30]. Previous studies mainly focused on its anti-tumor activities through facilitating the
anti-tumor immune responses by increasing immune cell infiltration of CD4+ and CD8+ T-cells [31, 32]. As comparison, the autocrine signaling of IFN-α/β was less explored [6,7,8,9,10,11]. Our
research revealed a FAM210B-promoted IFN-α/β autocrine signaling pathway in LUAD cells, leading to the suppression of LUAD development. The primary function of IFN-α/β is mediated through
the activation of the JAK-STAT signal pathway [26, 27]. The role of the activated STAT1 as either a tumor suppressor or a tumor promoter has been reported [33,34,35,36]. Similarly, IRF9,
whose overexpression has been reported to cause resistance of tumor cells to drugs [37, 38] and radiation [39] and exert oncogenic effects [40, 41], has also been shown to be a key regulator
for mediating the antiproliferative effect of IFN-α in tumor cells elsewhere [27, 42]. In this study, we showed that the inhibitory effects of STAT1/IRF9 axis mediated the suppressive
function of FAM210B on the growth, mobility, and invasion of LUAD cells, while also being involved in regulating FAM210B mRNA expression. Additionally, previous studies have shown that
targeting the delivery of IFN-α via Tie2-expressing monocytes (TEMs) inhibits tumor angiogenesis [43]. Similarly, endogenous IFN-β has been reported to suppress tumor angiogenesis by
downregulating pro-angiogenic factors such as VEGF and MMP9, as well as homing factors in neutrophils [44]. Moreover, STAT1 has also been shown to negatively regulate pro-angiogenic
molecules like bFGF, significantly inhibiting tumorigenicity, angiogenesis, and metastasis, thereby serving as a negative regulator of tumor growth and metastasis [33]. In our findings,
FAM210B overexpression promoted the expression of IFN-α/β and STAT1, inhibited EMT, a key process in tumor cell metastasis, and reduced the expression of the endothelial marker CD31 in mouse
tumor tissues. Taken together, all these data might hint on the possible inhibitory role of FAM210B on tumor angiogenesis and metastasis in LUAD through IFN-α/β/ STAT1 signal axis. In this
study, we identified TOM70, a 70-kDa mitochondrial membrane-anchored adapter, as an interacting partner of FAM210B. TOM70 plays a crucial role in innate immunity by acting as an adaptor to
link mitochondrial antiviral signaling protein (MAVS) to TANK-binding kinase 1 (TBK1)/IRF3 activation [28], which is essential for the induction of type I IFN [45,46,47]. We observed that
TOM70 and FAM210B promoted IFN-α/β production and inhibited LUAD cell proliferation and migration. These findings depicted a previously unknown signaling pathway in which TOM70 interacts
with FAM210B to participate in the upregulation of IFN-α/β expression, and characterized FAM210B as a novel regulator of innate immune response (Fig. 7). Considering the absence of MAVS in
the FAM210B-interactor list in our interactome study, a direct and stable interaction between FAM210B and MAVS might be excluded. However, given our findings that FAM210B not only stimulated
the expression of two upstream regulators of MAVS, namely MDA-5 and RIG-I (Fig. S6A, B) but also interacted with TOM70, a binding protein of MAVS, cooperative functional links could exist
between FAM210B and MAVS in the signaling transduction, which, with the complexity provided by the participation of TOM70 and other factors, might need further investigation in the future to
be elucidated. Our data also indicated that both FAM210B overexpression and the treatment of LUAD cells with purified IFN-α protein enhanced the expression of MDA-5 and RIG-I which are
upstream regulators in the innate immune pathway (Fig. S6A–D). Moreover, knockdown of MDA-5 in FAM210B-overexpressing cells abolished the upregulation of IFN-α/β caused by FAM210B
overexpression (Fig. S6E). Based on these findings, we hypothesize that a positive feedback loop may take place: FAM210B promotes the expression of IFN-α/β, which in turn induces the
expression of MDA-5 and RIG-I. The increased expression of MDA-5 and RIG-I then stimulates the innate immune pathway, in which FAM210B participates to further promote the expression of
IFN-α/β (Fig. 7). Such mechanism might have important implication for future therapeutic applications based on the anti-tumor properties of FAM210B. CONCLUSION In summary, we demonstrated
the relevance of the novel protein FAM210B to LUAD development, supported by the positive correlation between FAM210B expression and the overall survival of LUAD patients. FAM210B was found
to inhibit LUAD growth and metastasis both in vitro and in vivo. Mechanistically, FAM210B interacted with TOM70, leading to the upregulation of IFN-α/β expression and activation of the
innate immune pathway in LUAD cells. The IFN-α/β-activated STAT1/IRF9/IFIT3 axis played a crucial role in suppressing the proliferation and metastasis of LUAD cells. These findings offer new
insights into LUAD pathogenesis and suggest that targeting the FAM210B/IFN-α/β/STAT1/IRF9/IFIT3 axis may hold promise as a therapeutic strategy for LUAD. DATA AVAILABILITY All data
generated and analyzed during this study are included in this article and its Supplementary files. The data and materials used in this study are available upon reasonable request to the
corresponding author. REFERENCES * Lai XN, Li J, Tang LB, Chen WT, Zhang L, Xiong LX. MiRNAs and LncRNAs: dual roles in TGF-beta signaling-regulated metastasis in lung cancer. Int J Mol Sci.
2020;21:1193. Article CAS PubMed PubMed Central Google Scholar * Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535–54. Article PubMed Google
Scholar * Yuan S, Yu Z, Liu Q, Zhang M, Xiang Y, Wu N, et al. GPC5, a novel epigenetically silenced tumor suppressor, inhibits tumor growth by suppressing Wnt/beta-catenin signaling in lung
adenocarcinoma. Oncogene. 2016;35:6120–31. Article CAS PubMed Google Scholar * Uze G, Schreiber G, Piehler J, Pellegrini S. The receptor of the type I interferon family. Curr Top
Microbiol Immunol. 2007;316:71–95. CAS PubMed Google Scholar * Xue C, Yao Q, Gu X, Shi Q, Yuan X, Chu Q, et al. Evolving cognition of the JAK-STAT signaling pathway: autoimmune disorders
and cancer. Signal Transduct Target Ther. 2023;8:204. Article PubMed PubMed Central Google Scholar * Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. Innate immune signaling and
regulation in cancer immunotherapy. Cell Res. 2017;27:96–108. Article CAS PubMed Google Scholar * Dagher OK, Schwab RD, Brookens SK, Posey AD Jr. Advances in cancer immunotherapies.
Cell. 2023;186:1814–e1. Article CAS PubMed Google Scholar * Dou Z, Ghosh K, Vizioli MG, Zhu J, Sen P, Wangensteen KJ, et al. Cytoplasmic chromatin triggers inflammation in senescence and
cancer. Nature. 2017;550:402–6. Article CAS PubMed PubMed Central Google Scholar * Gresser I, Bourali C. Antitumor effects of interferon preparations in mice. J Natl Cancer Inst.
1970;45:365–76. CAS PubMed Google Scholar * Lu C, Liu Y, Ali NM, Zhang B, Cui X. The role of innate immune cells in the tumor microenvironment and research progress in anti-tumor therapy.
Front Immunol. 2022;13:1039260. Article CAS PubMed Google Scholar * Musella M, Manic G, De Maria R, Vitale I, Sistigu A. Type-I-interferons in infection and cancer: unanticipated
dynamics with therapeutic implications. Oncoimmunology. 2017;6:e1314424. Article PubMed PubMed Central Google Scholar * Gao X, Lian Q, Guan B, Liu QY, Meng M, Chen Y, et al. ZSWIM1
promotes the proliferation and metastasis of lung adenocarcinoma cells through the STK38/MEKK2/ERK1/2 axis. J Proteome Res. 2023;22:1080–91. Article CAS PubMed Google Scholar * Wang Y,
Zhang J, Zheng CC, Huang ZJ, Zhang WX, Long YL, et al. C20orf24 promotes colorectal cancer progression by recruiting Rin1 to activate Rab5-mediated mitogen-activated protein
kinase/extracellular signal-regulated kinase signalling. Clin Transl Med. 2022;12:e796. Article CAS PubMed PubMed Central Google Scholar * Kondo A, Fujiwara T, Okitsu Y, Fukuhara N,
Onishi Y, Nakamura Y, et al. Identification of a novel putative mitochondrial protein FAM210B associated with erythroid differentiation. Int J Hematol. 2016;103:387–95. Article CAS PubMed
Google Scholar * Sun S, Liu J, Zhao M, Han Y, Chen P, Mo Q, et al. Loss of the novel mitochondrial protein FAM210B promotes metastasis via PDK4-dependent metabolic reprogramming. Cell
Death Dis. 2017;8:e2870. Article CAS PubMed PubMed Central Google Scholar * Zhou Y, Pan X, Liu Y, Li X, Lin K, Zhu J, et al. Loss of the Novel Mitochondrial Membrane Protein FAM210B Is
Associated with Hepatocellular Carcinoma. Biomedicines. 2023;11:1232. Article CAS PubMed PubMed Central Google Scholar * Friedlander P, Wood K, Wassmann K, Christenfeld AM, Bhardwaj N,
Oh WK. A whole-blood RNA transcript-based gene signature is associated with the development of CTLA-4 blockade-related diarrhea in patients with advanced melanoma treated with the checkpoint
inhibitor tremelimumab. J Immunother Cancer. 2018;6:90. Article PubMed PubMed Central Google Scholar * Plasterer C, Tsaih SW, Peck AR, Chervoneva I, O’Meara C, Sun Y, et al. Neuronatin
is a modifier of estrogen receptor-positive breast cancer incidence and outcome. Breast Cancer Res Treat. 2019;177:77–91. Article CAS PubMed PubMed Central Google Scholar * Zhang HT,
Zeng Q, Wu B, Lu J, Tong KL, Lin J, et al. TRIM21-regulated Annexin A2 plasma membrane trafficking facilitates osteosarcoma cell differentiation through the TFEB-mediated autophagy. Cell
Death Dis. 2021;12:21. Article CAS PubMed PubMed Central Google Scholar * Gao X, Feng J, He Y, Xu F, Fan X, Huang W, et al. hnRNPK inhibits GSK3beta Ser9 phosphorylation, thereby
stabilizing c-FLIP and contributes to TRAIL resistance in H1299 lung adenocarcinoma cells. Sci Rep. 2016;6:22999. Article CAS PubMed PubMed Central Google Scholar * Liu W, Xiang L,
Zheng T, Jin J, Zhang G. TranslatomeDB: a comprehensive database and cloud-based analysis platform for translatome sequencing data. Nucleic Acids Res. 2018;46:D206–D12. Article CAS PubMed
Google Scholar * Li D, Lu S, Liu W, Zhao X, Mai Z, Zhang G. Optimal settings of mass spectrometry open search strategy for higher confidence. J Proteome Res. 2018;17:3719–29. Article CAS
PubMed Google Scholar * Zeng QZ, Liu WT, Lu JL, Liu XH, Zhang YF, Liu LX, et al. YWHAZ binds to TRIM21 but is not involved in TRIM21-stimulated osteosarcoma cell proliferation. Biomed
Environ Sci. 2018;31:186–96. CAS PubMed Google Scholar * Liu X, Li L, Geng C, Wen S, Zhang C, Deng C, et al. DDX17 promotes the growth and metastasis of lung adenocarcinoma. Cell Death
Discov. 2022;8:425. Article CAS PubMed PubMed Central Google Scholar * Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, et al. Comprehensive molecular profiling of
lung adenocarcinoma. Nature. 2014;511:543–50. Article CAS Google Scholar * Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. Interferon: cellular executioner or white knight? Curr Med
Chem. 2007;14:1279–89. Article CAS PubMed Google Scholar * Tsuno T, Mejido J, Zhao T, Schmeisser H, Morrow A, Zoon KC. IRF9 is a key factor for eliciting the antiproliferative activity
of IFN-alpha. J Immunother. 2009;32:803–16. Article CAS PubMed PubMed Central Google Scholar * Liu XY, Wei B, Shi HX, Shan YF, Wang C. Tom70 mediates activation of interferon regulatory
factor 3 on mitochondria. Cell Res. 2010;20:994–1011. Article CAS PubMed Google Scholar * Aoyagi S, Hata H, Homma E, Shimizu H. Sequential local injection of low-dose interferon-beta
for maintenance therapy in stage II and III melanoma: a single-institution matched case-control study. Oncology. 2012;82:139–46. Article CAS PubMed Google Scholar * Satomi H, Wang B,
Fujisawa H, Otsuka F. Interferon-beta from melanoma cells suppresses the proliferations of melanoma cells in an autocrine manner. Cytokine. 2002;18:108–15. Article CAS PubMed Google
Scholar * Fujimura T, Okuyama R, Ohtani T, Ito Y, Haga T, Hashimoto A, et al. Perilesional treatment of metastatic melanoma with interferon-beta. Clin Exp Dermatol. 2009;34:793–9. Article
CAS PubMed Google Scholar * Uehara J, Ohkuri T, Kosaka A, Ishibashi K, Hirata Y, Ohara K, et al. Intratumoral injection of IFN-beta induces chemokine production in melanoma and augments
the therapeutic efficacy of anti-PD-L1 mAb. Biochem Biophys Res Commun. 2017;490:521–7. Article CAS PubMed Google Scholar * Huang S, Bucana CD, Van Arsdall M, Fidler IJ. Stat1 negatively
regulates angiogenesis, tumorigenicity and metastasis of tumor cells. Oncogene. 2002;21:2504–12. Article CAS PubMed Google Scholar * Zhang Y, Liu Z, Yang X, Lu W, Chen Y, Lin Y, et al.
H3K27 acetylation activated-COL6A1 promotes osteosarcoma lung metastasis by repressing STAT1 and activating pulmonary cancer-associated fibroblasts. Theranostics. 2021;11:1473–92. Article
CAS PubMed PubMed Central Google Scholar * Zhang Y, Liu Z. STAT1 in cancer: friend or foe? Discov Med. 2017;24:19–29. PubMed Google Scholar * Yan M, Sun L, Li J, Yu H, Lin H, Yu T, et
al. RNA-binding protein KHSRP promotes tumor growth and metastasis in non-small cell lung cancer. J Exp Clin Cancer Res. 2019;38:478. Article CAS PubMed PubMed Central Google Scholar *
Luker KE, Pica CM, Schreiber RD, Piwnica-Worms D. Overexpression of IRF9 confers resistance to antimicrotubule agents in breast cancer cells. Cancer Res. 2001;61:6540–7. CAS PubMed Google
Scholar * Khodarev NN, Beckett M, Labay E, Darga T, Roizman B, Weichselbaum RR. STAT1 is overexpressed in tumors selected for radioresistance and confers protection from radiation in
transduced sensitive cells. Proc Natl Acad Sci USA. 2004;101:1714–9. Article CAS PubMed PubMed Central Google Scholar * Kolosenko I, Fryknas M, Forsberg S, Johnsson P, Cheon H,
Holvey-Bates EG, et al. Cell crowding induces interferon regulatory factor 9, which confers resistance to chemotherapeutic drugs. Int J Cancer. 2015;136:E51–61. Article CAS PubMed Google
Scholar * Brunn D, Turkowski K, Gunther S, Weigert A, Muley T, Kriegsmann M, et al. Interferon regulatory factor 9 promotes lung cancer progression via regulation of versican. Cancers.
2021;13:208. Article CAS PubMed PubMed Central Google Scholar * Sharma BR, Karki R, Sundaram B, Wang Y, Vogel P, Kanneganti TD. The transcription factor IRF9 promotes colorectal cancer
via modulating the IL-6/STAT3 signaling axis. Cancers. 2022;14:919. Article CAS PubMed PubMed Central Google Scholar * Erb HH, Langlechner RV, Moser PL, Handle F, Casneuf T, Verstraeten
K, et al. IL6 sensitizes prostate cancer to the antiproliferative effect of IFNalpha2 through IRF9. Endocr Relat Cancer. 2013;20:677–89. Article CAS PubMed PubMed Central Google Scholar
* De Palma M, Mazzieri R, Politi LS, Pucci F, Zonari E, Sitia G, et al. Tumor-targeted interferon-α delivery by Tie2-expressing monocytes inhibits tumor growth and metastasis. Cancer Cell.
2008;14:299–311. Article PubMed Google Scholar * Jablonska J, Leschner S, Westphal K, Lienenklaus S, Weiss S. Neutrophils responsive to endogenous IFN-β regulate tumor angiogenesis and
growth in a mouse tumor model. J Clin Investig. 2010;120:1151–64. Article CAS PubMed PubMed Central Google Scholar * Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber
GN, et al. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J Virol. 2002;76:5532–9. Article
CAS PubMed PubMed Central Google Scholar * Gao X, Zhu K, Qin B, Olieric V, Wang M, Cui S. Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host
interactions. Nat Commun. 2021;12:2843. Article CAS PubMed PubMed Central Google Scholar * Wei B, Cui Y, Huang Y, Liu H, Li L, Li M, et al. Tom70 mediates Sendai virus-induced apoptosis
on mitochondria. J Virol. 2015;89:3804–18. Article CAS PubMed PubMed Central Google Scholar Download references FUNDING This work was supported by Guangdong Basic and Applied Basic
Research Foundation (2023A1515010248; 2023A1515011221), National Natural Science Foundation of China (82071714), Huadu District Basic and Applied Basic Research Joint Funded Project (No.
23HDQYLH02). Open projects supported by the Key Laboratory of Tumor Molecular Biology of the Ministry of Education of Jinan University (202201, 202203). AUTHOR INFORMATION Author notes *
These authors contributed equally: Xuejuan Gao, Donglan Huang, Ying Liu, Gui Zhang. AUTHORS AND AFFILIATIONS * MOE Key Laboratory of Tumor Molecular Biology and State Key Laboratory of
Bioactive Molecules and Druggability Assessment, Institute of Life and Health Engineering, College of Life Science and Technology, Jinan University, Guangzhou, China Xuejuan Gao, Donglan
Huang, Ying Liu, Gui Zhang, Xiaofen Zheng, Baiye Guan, Aiwen Chen, Jiayao Wu, Luxuan Chen, Xiaohui Liu, Jingjie Jin, Xingfeng Yin, Zhenghua Sun, Gong Zhang, Wanting Liu & Langxia Liu *
Guangzhou Key Laboratory of Metabolic Diseases and Reproductive Health, Guangdong-Hong Kong Metabolism & Reproduction Joint Laboratory, Reproductive Medicine Center, Guangdong Second
Provincial General Hospital, Guangzhou, Guangdong, China Shi-Ming Luo * Department of Biomedical Engineering, College of Life Science and Technology, Jinan University, Guangzhou, China
Zonghua Liu * Department of Nephrology, People’s Hospital of Huadu District; The Third School of Clinical Medicine, Southern Medical University, Guangzhou, China Yunfang Zhang & Meizhi
Lu Authors * Xuejuan Gao View author publications You can also search for this author inPubMed Google Scholar * Donglan Huang View author publications You can also search for this author
inPubMed Google Scholar * Ying Liu View author publications You can also search for this author inPubMed Google Scholar * Gui Zhang View author publications You can also search for this
author inPubMed Google Scholar * Xiaofen Zheng View author publications You can also search for this author inPubMed Google Scholar * Baiye Guan View author publications You can also search
for this author inPubMed Google Scholar * Aiwen Chen View author publications You can also search for this author inPubMed Google Scholar * Jiayao Wu View author publications You can also
search for this author inPubMed Google Scholar * Shi-Ming Luo View author publications You can also search for this author inPubMed Google Scholar * Zonghua Liu View author publications You
can also search for this author inPubMed Google Scholar * Luxuan Chen View author publications You can also search for this author inPubMed Google Scholar * Xiaohui Liu View author
publications You can also search for this author inPubMed Google Scholar * Jingjie Jin View author publications You can also search for this author inPubMed Google Scholar * Xingfeng Yin
View author publications You can also search for this author inPubMed Google Scholar * Zhenghua Sun View author publications You can also search for this author inPubMed Google Scholar *
Yunfang Zhang View author publications You can also search for this author inPubMed Google Scholar * Meizhi Lu View author publications You can also search for this author inPubMed Google
Scholar * Gong Zhang View author publications You can also search for this author inPubMed Google Scholar * Wanting Liu View author publications You can also search for this author inPubMed
Google Scholar * Langxia Liu View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS DH, YL, and GZ: Methodology, Investigation, Writing - original
draft. XZ, BG, AC, and JW: Methodology, Investigation. SML: Conceptualization, Funding acquisition. ZL: Resources, Supervision. LC, XL, JJ, XY, and ZS: Formal analysis, Investigation. YZ:
Conceptualization, Funding acquisition. XG, ML, GZ, WL, and LL: Conceptualization, Funding acquisition, Project administration, Writing - original draft, Writing review and editing. All
authors read and approved the final manuscript. CORRESPONDING AUTHORS Correspondence to Xuejuan Gao, Meizhi Lu, Gong Zhang, Wanting Liu or Langxia Liu. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and
institutional affiliations. Edited by Yufang Shi SUPPLEMENTARY INFORMATION SUPPLEMENTARY FILE 1 SUPPLEMENTARY TABLE S1 SUPPLEMENTARY TABLE S2 SUPPLEMENTARY TABLE S3 SUPPLEMENTARY TABLE S4
ORIGINAL DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution
and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if
changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the
material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to
obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Gao, X., Huang, D., Liu, Y. _et al._ FAM210B activates STAT1/IRF9/IFIT3 axis by upregulating IFN-α/β expression to impede the progression of lung adenocarcinoma. _Cell Death Dis_
16, 63 (2025). https://doi.org/10.1038/s41419-025-07375-9 Download citation * Received: 26 May 2024 * Revised: 20 December 2024 * Accepted: 21 January 2025 * Published: 03 February 2025 *
DOI: https://doi.org/10.1038/s41419-025-07375-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is
not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative