- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
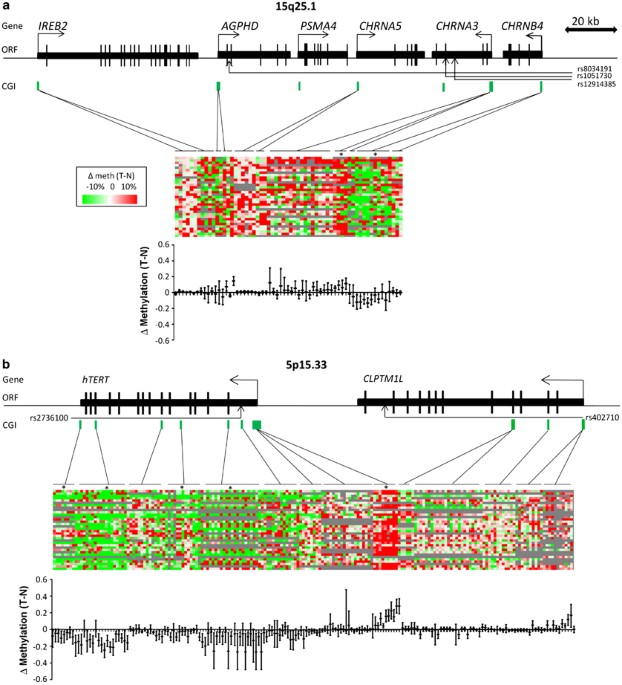
ABSTRACT Genome-wide association studies have highlighted three major lung cancer susceptibility regions at 15q25.1, 5p15.33 and 6p21.33. To gain insight into the possible mechanistic
relevance of the genes in these regions, we investigated the regulation of candidate susceptibility gene expression by epigenetic alterations in healthy and lung tumor tissues. For genes up
or downregulated in lung tumors, the influence of genetic variants on DNA methylation was investigated and _in vitro_ studies were performed. We analyzed 394 CpG units within 19 CpG islands
in the susceptibility regions in a screening set of 34 patients. Significant findings were validated in an independent patient set (_n_=50) with available DNA and RNA. The most consistent
overall DNA methylation difference between tumor and adjacent normal tissue on 15q25 was tumor hypomethylation in the promoter region of _CHRNB4_ with a median difference of 8%
(_P_<0.001), which resulted in overexpression of the transcript in tumors (_P_<0.001). Confirming previous studies, we also found hypermethylation in _CHRNA3_ and _telomerase reverse
transcriptase_ (_TERT_) with significant expression changes. Decitabine treatment of H1299 cells resulted in reduced methylation levels in gene promoters, elevated transcript levels of
_CHRNB4_ and _CHRNA3_, and a slight downregulation of _TERT_ demonstrating epigenetic regulation of lung cancer cells. Single-nucleotide polymorphisms rs421629 on 5p15.33 and rs1948,
rs660652, rs8040868 and rs2036527 on 15q25.1, previously identified as lung cancer risk or nicotine-addiction modifiers, were associated with tumor DNA methylation levels in the promoters of
_TERT_ and _CHRNB4_ (_P_<0.001), respectively, in two independent sample sets (_n_=82; _n_=150). In addition, _CHRNB4_ knockdown in two different cell lines (A549 and H1299) resulted in
reduced proliferation (_P_A549<0.05;_P_H1299<0.001) and propensity to form colonies in H1299 cells. These results suggest epigenetic deregulation of nicotinic acetylcholine receptor
subunit (_nAChR_) genes which in the case of _CHRNB4_ is strongly associated with genetic lung cancer susceptibility variants and a functional impact on tumorigenic potential. Access through
your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 50
print issues and online access $259.00 per year only $5.18 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject
to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT
BEING VIEWED BY OTHERS IDENTIFYING POLYMORPHIC CIS-REGULATORY VARIANTS AS RISK MARKERS FOR LUNG CARCINOGENESIS AND CHEMOTHERAPY RESPONSES IN TOBACCO SMOKERS FROM EASTERN INDIA Article Open
access 10 March 2023 INTEGRATIVE GENOMICS IDENTIFIES SHPRH AS A TUMOR SUPPRESSOR GENE IN LUNG ADENOCARCINOMA THAT REGULATES DNA DAMAGE RESPONSE Article Open access 18 June 2024 UNRAVELING
THE INTRICATE MOLECULAR LANDSCAPE AND POTENTIAL BIOMARKERS IN LUNG ADENOCARCINOMA THROUGH INTEGRATIVE EPIGENOMIC AND TRANSCRIPTOMIC PROFILING Article Open access 17 March 2025 REFERENCES *
Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T _et al_. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. _Nat Genet_ 2008; 40:
616–622. Article CAS Google Scholar * Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D _et al_. A susceptibility locus for lung cancer maps to nicotinic acetylcholine
receptor subunit genes on 15q25. _Nature_ 2008; 452: 633–637. Article CAS Google Scholar * Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP _et al_. A variant
associated with nicotine dependence, lung cancer and peripheral arterial disease. _Nature_ 2008; 452: 638–642. Article CAS Google Scholar * McKay JD, Hung RJ, Gaborieau V, Boffetta P,
Chabrier A, Byrnes G _et al_. Lung cancer susceptibility locus at 5p15.33. _Nat Genet_ 2008; 40: 1404–1406. Article CAS Google Scholar * Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan
J, Matakidou A _et al_. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. _Nat Genet_ 2008; 40: 1407–1409. Article CAS Google Scholar * Freedman ML, Monteiro AN, Gayther SA,
Coetzee GA, Risch A, Plass C _et al_. Principles for the post-GWAs functional characterization of cancer risk loci. _Nat Genet_ 2011; 43: 513–518. Article CAS Google Scholar * Parkin DM,
Bray F, Ferlay J, Pisani P . Global cancer statistics, 2002. _CA Cancer J Clin_ 2005; 55: 74–108. Article Google Scholar * Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ . Genomic
profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. _Nucleic Acids Res_ 2007; 35: e119. Article
Google Scholar * Herman JG, Baylin SB . Gene silencing in cancer in association with promoter hypermethylation. _N Engl J Med_ 2003; 349: 2042–2054. Article CAS Google Scholar * He L,
Hannon GJ . MicroRNAs: small RNAs with a big role in gene regulation. _Nat Rev Genet_ 2004; 5: 522–531. Article CAS Google Scholar * Dai Z, Lakshmanan RR, Zhu WG, Smiraglia DJ, Rush LJ,
Fruhwald MC _et al_. Global methylation profiling of lung cancer identifies novel methylated genes. _Neoplasia_ 2001; 3: 314–323. Article CAS Google Scholar * Shivapurkar N, Stastny V,
Xie Y, Prinsen C, Frenkel E, Czerniak B _et al_. Differential methylation of a short CpG-rich sequence within exon 1 of TCF21 gene: a promising cancer biomarker assay. _Cancer Epidemiol
Biomarkers Prev_ 2008; 17: 995–1000. Article CAS Google Scholar * Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E _et al_. Aberrant methylation of p16(INK4a)
is an early event in lung cancer and a potential biomarker for early diagnosis. _Proc Natl Acad Sci USA_ 1998; 95: 11891–11896. Article CAS Google Scholar * Belinsky SA, Liechty KC,
Gentry FD, Wolf HJ, Rogers J, Vu K _et al_. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. _Cancer Res_ 2006; 66: 3338–3344.
Article CAS Google Scholar * Brena RM, Morrison C, Liyanarachchi S, Jarjoura D, Davuluri RV, Otterson GA _et al_. Aberrant DNA methylation of OLIG1, a novel prognostic factor in non-small
cell lung cancer. _PLoS Med_ 2007; 4: e108. Article Google Scholar * Brena RM, Costello JF . Genome-epigenome interactions in cancer. _Hum Mol Genet_ 2007; 1: R96–R105 16 Spec No. Article
Google Scholar * Esteller M, Garcia A, Martinez Palones JM, Xercavins J, Reventos J . Germ line polymorphisms in cytochrome-P450 1A1 (C4887 CYP1A1) and methylenetetrahydrofolate reductase
(MTHFR) genes and endometrial cancer susceptibility. _Carcinogenesis_ 1997; 18: 2307–2311. Article CAS Google Scholar * Shames DS, Girard L, Gao B, Sato M, Lewis CM, Shivapurkar N _et
al_. A genome-wide screen for promoter methylation in lung cancer identifies novel methylation markers for multiple malignancies. _PLoS Med_ 2006; 3: e486. Article Google Scholar * Laird
PW . Principles and challenges of genome-wide DNA methylation analysis. _Nat Rev Genet_ 2010; 11: 191–203. Article CAS Google Scholar * Park JC, Chae YK, Son CH, Kim MS, Lee J, Ostrow K
_et al_. Epigenetic silencing of human T (brachyury homologue) gene in non-small-cell lung cancer. _Biochem Biophys Res Commun_ 2008; 365: 221–226. Article CAS Google Scholar * Hitchins
MP, Rapkins RW, Kwok CT, Srivastava S, Wong JJ, Khachigian LM _et al_. Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single
nucleotide variant within the 5′UTR. _Cancer Cell_ 2011; 20: 200–213. Article CAS Google Scholar * Paliwal A, Vaissiere T, Krais A, Cuenin C, Cros MP, Zaridze D _et al_. Aberrant DNA
methylation links cancer susceptibility locus 15q25.1 to apoptotic regulation and lung cancer. _Cancer Res_ 2010; 70: 2779–2788. Article CAS Google Scholar * Renaud S, Loukinov D,
Abdullaev Z, Guilleret I, Bosman FT, Lobanenkov V _et al_. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT
gene. _Nucleic Acids Res_ 2007; 35: 1245–1256. Article CAS Google Scholar * Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN _et al_. Allelic skewing of DNA
methylation is widespread across the genome. _Am J Hum Genet_ 2010; 86: 196–212. Article CAS Google Scholar * Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E _et al_. Genomic surveys
by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. _Nat Genet_ 2008; 40: 904–908. Article CAS Google Scholar * Zhang Y, Rohde C, Reinhardt
R, Voelcker-Rehage C, Jeltsch A . Non-imprinted allele-specific DNA methylation on human autosomes. _Genome Biol_ 2009; 10: R138. Article Google Scholar * Amos CI, Gorlov IP, Dong Q, Wu
X, Zhang H, Lu EY _et al_. Nicotinic acetylcholine receptor region on chromosome 15q25 and lung cancer risk among African Americans: a case-control study. _J Natl Cancer Inst_ 2011; 102:
1199–1205. Article Google Scholar * Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ _et al_. The CHRNA5/A3/B4 gene cluster variability as an important determinant of
early alcohol and tobacco initiation in young adults. _Biol Psychiatry_ 2008; 63: 1039–1046. Article CAS Google Scholar * Stephens SH, Hoft NR, Schlaepfer IR, Young SE, Corley RC, McQueen
MB _et al_. Externalizing behaviors are associated with SNPs in the CHRNA5/CHRNA3/CHRNB4 gene cluster. _Behav Genet_ 2011; 42: 402–414. Article Google Scholar * Erlich PM, Hoffman SN,
Rukstalis M, Han JJ, Chu X, Linda Kao WH _et al_. Nicotinic acetylcholine receptor genes on chromosome 15q25.1 are associated with nicotine and opioid dependence severity. _Hum Genet_ 2010;
128: 491–499. Article CAS Google Scholar * Improgo MR, Schlichting NA, Cortes RY, Zhao-Shea R, Tapper AR, Gardner PD . ASCL1 regulates the expression of the CHRNA5/A3/B4 lung cancer
susceptibility locus. _Mol Cancer Res_ 8: 194–203. Article CAS Google Scholar * Jones PA . The DNA methylation paradox. _Trends Genet_ 1999; 15: 34–37. Article CAS Google Scholar *
Allemand E, Batsche E, Muchardt C . Splicing, transcription, and chromatin: a menage a trois. _Curr Opin Genet Dev_ 2008; 18: 145–151. Article CAS Google Scholar * Luco RF, Pan Q,
Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T . Regulation of alternative splicing by histone modifications. _Science_ 2010; 327: 996–1000. Article CAS Google Scholar * Chodavarapu
RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y _et al_. Relationship between nucleosome positioning and DNA methylation. _Nature_ 2010; 466: 388–392. Article CAS Google Scholar *
McCabe MT, Lee EK, Vertino PM . A multifactorial signature of DNA sequence and polycomb binding predicts aberrant CpG island methylation. _Cancer Res_ 2009; 69: 282–291. Article CAS
Google Scholar * Gebhard C, Benner C, Ehrich M, Schwarzfischer L, Schilling E, Klug M _et al_. General transcription factor binding at CpG islands in normal cells correlates with resistance
to _de novo_ DNA methylation in cancer cells. _Cancer Res_ 2010; 70: 1398–1407. CAS PubMed Google Scholar * Herceg Z . Epigenetic information in chromatin and cancer. _Eur J Cancer_
2009; 45: 442–444. Article Google Scholar * Chen SS, Raval A, Johnson AJ, Hertlein E, Liu TH, Jin VX _et al_. Epigenetic changes during disease progression in a murine model of human
chronic lymphocytic leukemia. _Proc Natl Acad Sci USA_ 2009; 106: 13433–13438. Article CAS Google Scholar * Van den Berg RM, Brokx H, Vesin A, Field JK, Brambilla C, Meijer CJ _et al_.
Prognostic value of hTERT mRNA expression in surgical samples of lung cancer patients: the European Early Lung Cancer Project. _Int J Oncol_ 2010; 37: 455–461. CAS PubMed Google Scholar *
Schuller HM . Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? _Nat Rev Cancer_ 2009; 9: 195–205. Article CAS Google Scholar * Schuller HM, Tithof PK,
Williams M, Plummer H . The tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone is a beta-adrenergic agonist and stimulates DNA synthesis in lung adenocarcinoma via
beta-adrenergic receptor-mediated release of arachidonic acid. _Cancer Res_ 1999; 59: 4510–4515. CAS PubMed Google Scholar * Kawai H, Berg DK . Nicotinic acetylcholine receptors
containing alpha 7 subunits on rat cortical neurons do not undergo long-lasting inactivation even when up-regulated by chronic nicotine exposure. _J Neurochem_ 2001; 78: 1367–1378. Article
CAS Google Scholar * Joseph J, Niggemann B, Zaenker KS, Entschladen F . The neurotransmitter gamma-aminobutyric acid is an inhibitory regulator for the migration of SW 480 colon carcinoma
cells. _Cancer Res_ 2002; 62: 6467–6469. CAS PubMed Google Scholar * Shiraishi K, Kohno T, Kunitoh H, Watanabe S, Goto K, Nishiwaki Y _et al_. Contribution of nicotine acetylcholine
receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. _Carcinogenesis_ 2009; 30: 65–70. Article CAS Google Scholar * Liu Y, Liu P, Wen W, James MA,
Wang Y, Bailey-Wilson JE _et al_. Haplotype and cell proliferation analyses of candidate lung cancer susceptibility genes on chromosome 15q24-25.1. _Cancer Res_ 2009; 69: 7844–7850. Article
CAS Google Scholar * Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G _et al_. Quantitative high-throughput analysis of DNA methylation patterns by base-specific
cleavage and mass spectrometry. _Proc Natl Acad Sci USA_ 2005; 102: 15785–15790. Article CAS Google Scholar * Oeth P, del Mistro G, Marnellos G, Shi T, van den Boom D . Qualitative and
quantitative genotyping using single base primer extension coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY). _Methods Mol Biol_ 2009;
578: 307–343. Article CAS Google Scholar * Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C . Clonogenic assay of cells _in vitro_. _Nat Protoc_ 2006; 1: 2315–2319. Article CAS
Google Scholar Download references ACKNOWLEDGEMENTS We thank Frau Heinzmann-Groth, Birgit Jäger and all other members of the clinical and laboratory teams for help with sample and/or data
collection and archiving for the Heidelberg lung study. We are grateful to all patients at the Thoraxklinik Heidelberg, who participated in the study. We thank Ruprecht Kuner for the cell
lines, Oliver Mücke for competent technical assistance, Chris Amos for helpful comments on the manuscript and the members of the Plass Laboratory for thoughtful discussions. The scientific
development and funding of this project were in part supported by the Genetic Associations and Mechanisms in Oncology (GAME-ON): a NCI Cancer Post-GWAS Initiative. This work was in part
supported by the National Institute of Health (USA; grant number CA148127) and earlier sample collection by the Deutsche Krebshilfe (grant number 70–2387). JLB was in part supported by a
grant of the Deutsche Forschungsgemeinschaft (DFG, SFB/TRR77, project Z2). AUTHOR INFORMATION Author notes * T Muley, M Meister, H Dienemann and A Risch: Translational Lung Research Centre
Heidelberg (TLRC-H), Member of the German Center for Lung Research. AUTHORS AND AFFILIATIONS * Division of Epigenomics and Cancer Risk Factors (C010), German Cancer Research Center (DKFZ),
Heidelberg, Germany D B Scherf, N Sarkisyan, H Jacobsson, R Claus, L Gu, C Plass & A Risch * Division of Molecular Genetic Epidemiology, Institute of Medical Biometry and Informatics,
University Hospital Heidelberg, German Cancer Research Center, Heidelberg, Germany J L Bermejo & B Peil * Translational Research Unit, Thoraxklinik-Heidelberg gGmbH, University of
Heidelberg, Germany T Muley & M Meister * Department of Thoracic Surgery, Thoraxklinik-Heidelberg gGmbH, University of Heidelberg, Germany H Dienemann Authors * D B Scherf View author
publications You can also search for this author inPubMed Google Scholar * N Sarkisyan View author publications You can also search for this author inPubMed Google Scholar * H Jacobsson View
author publications You can also search for this author inPubMed Google Scholar * R Claus View author publications You can also search for this author inPubMed Google Scholar * J L Bermejo
View author publications You can also search for this author inPubMed Google Scholar * B Peil View author publications You can also search for this author inPubMed Google Scholar * L Gu View
author publications You can also search for this author inPubMed Google Scholar * T Muley View author publications You can also search for this author inPubMed Google Scholar * M Meister
View author publications You can also search for this author inPubMed Google Scholar * H Dienemann View author publications You can also search for this author inPubMed Google Scholar * C
Plass View author publications You can also search for this author inPubMed Google Scholar * A Risch View author publications You can also search for this author inPubMed Google Scholar
CORRESPONDING AUTHOR Correspondence to A Risch. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information
accompanies the paper on the Oncogene website SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (DOC 1893 KB) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Scherf, D., Sarkisyan, N., Jacobsson, H. _et al._ Epigenetic screen identifies genotype-specific promoter DNA methylation and oncogenic potential of _CHRNB4_. _Oncogene_ 32,
3329–3338 (2013). https://doi.org/10.1038/onc.2012.344 Download citation * Received: 26 January 2012 * Revised: 21 May 2012 * Accepted: 20 June 2012 * Published: 03 September 2012 * Issue
Date: 11 July 2013 * DOI: https://doi.org/10.1038/onc.2012.344 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a
shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * DNA methylation * risk factors *
non-small cell lung cancer (NSCLC) * _CHRNB4_ * _TERT_





