- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
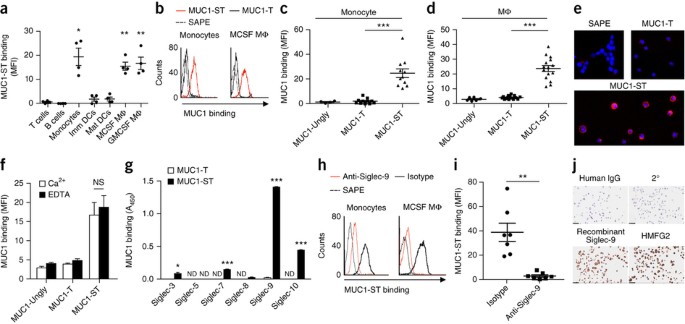
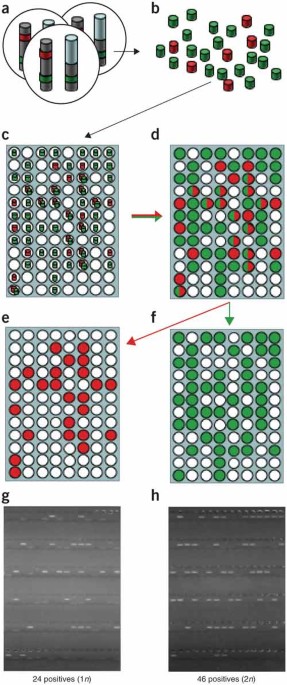
Human cancers and some congenital traits are characterized by cytogenetic aberrations including translocations, amplifications, duplications or deletions that can involve gain or loss of
genetic material. We have developed a simple method to precisely delineate such regions with known or cryptic genomic alterations. Molecular copy-number counting (MCC) uses PCR to
interrogate miniscule amounts of genomic DNA and allows progressive delineation of DNA content to within a few hundred base pairs of a genomic alteration. As an example, we have located the
junctions of a recurrent nonreciprocal translocation between chromosomes 3 and 5 in human renal cell carcinoma, facilitating cloning of the breakpoint without recourse to genomic libraries.
The analysis also revealed additional cryptic chromosomal changes close to the translocation junction. MCC is a fast and flexible method for characterizing a wide range of chromosomal
aberrations.
*Note: Correspondence should be addressed to M. Thangavelu ([email protected]) instead of T. H. Rabbitts. The error has been corrected in the PDF version of the article.
Correspondence should be addressed to M. Thangavelu ([email protected]) instead of T. H. Rabbitts. The error has been corrected in the PDF version of the article.
Albertson, D.G., Collins, C., McCormick, F. & Gray, J.W. Chromosome aberrations in solid tumors. Nat. Genet. 34, 369–376 (2003).
This work was supported by the Medical Research Council. G.C. was partly supported by the Kay Kendall Leukemia Fund. We thank L. Old for generously supplying the renal cell line SK-RC-9 and
N. Copeland for providing an inverse PCR protocol.
Present address: Institute of Human Genetics, Johannes Gutenberg University, Langenbeckstr. 1, D-55131, Mainz, Germany
Present address: MRC Cancer Cell Unit, Hills Road, Cambridge, CB2 2XZ, UK
Present address: Department of Anatomical and Cellular Pathology, Prince of Wales Hospital, The Chinese University of Hong Kong, Hong Kong
Present address: Prime Health, L1, St Georges Tce, Perth, 6000, Australia
Angelika Daser and Madan Thangavelu: These authors contributed equally to this work.
MRC Laboratory of Molecular Biology, Hills Road, Cambridge, CB2 2QH, UK
Angelika Daser, Madan Thangavelu, Richard Pannell, Alan Forster, Grace Chung, Paul H Dear & Terence H Rabbitts
A.D., M.T., R.P., A.F. and G.C. conducted the experimental procedures; A.D., M.T., P.H.D. and T.H.R. devised the project; A.D., P.H.D. and T.H.R. wrote the manuscript.
Identification of micro-deletion in SK-RC-9 chromosome t(3;5). (PDF 33 kb)
Agarose gel fractionation of PCR products for round one MCC of SK-RC-12 DNA. (PDF 99 kb)
Filter hybridization of SK-RC-12 DNA to confirm genomic alteration detected by MCC. (PDF 66 kb)
Anyone you share the following link with will be able to read this content:


![[Withdrawn] Islamic Trust (Maidenhead) - GOV.UK](https://www.gov.uk/assets/static/govuk-opengraph-image-03837e1cec82f217cf32514635a13c879b8c400ae3b1c207c5744411658c7635.png)