- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
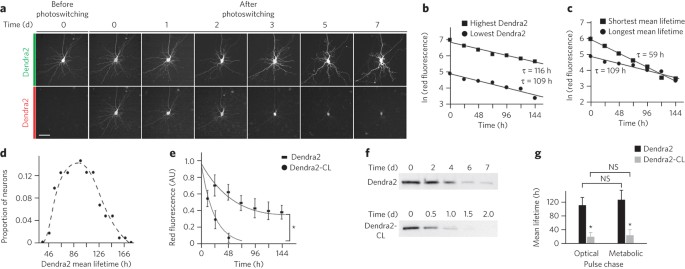
ABSTRACT In polyglutamine (polyQ) diseases, only certain neurons die, despite widespread expression of the offending protein. PolyQ expansion may induce neurodegeneration by impairing
proteostasis, but protein aggregation and toxicity tend to confound conventional measurements of protein stability. Here, we used optical pulse labeling to measure effects of polyQ
expansions on the mean lifetime of a fragment of huntingtin, the protein that causes Huntington's disease, in living neurons. We show that polyQ expansion reduced the mean lifetime of
mutant huntingtin within a given neuron and that the mean lifetime varied among neurons, indicating differences in their capacity to clear the polypeptide. We found that neuronal longevity
is predicted by the mean lifetime of huntingtin, as cortical neurons cleared mutant huntingtin faster and lived longer than striatal neurons. Thus, cell type–specific differences in turnover
capacity may contribute to cellular susceptibility to toxic proteins, and efforts to bolster proteostasis in Huntington's disease, such as protein clearance, could be neuroprotective.
Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this
journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now
Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer
support SIMILAR CONTENT BEING VIEWED BY OTHERS POLYGLUTAMINE-MEDIATED RIBOTOXICITY DISRUPTS PROTEOSTASIS AND STRESS RESPONSES IN HUNTINGTON’S DISEASE Article 13 May 2024 PERIPHERAL
SEQUESTRATION OF HUNTINGTIN DELAYS NEURONAL DEATH AND DEPENDS ON N-TERMINAL UBIQUITINATION Article Open access 18 August 2024 OSMOLYTES DYNAMICALLY REGULATE MUTANT HUNTINGTIN AGGREGATION AND
CREB FUNCTION IN HUNTINGTON’S DISEASE CELL MODELS Article Open access 23 September 2020 REFERENCES * Han, I., You, Y., Kordower, J.H., Brady, S.T. & Morfini, G.A. Differential
vulnerability of neurons in Huntington's disease: the role of cell type–specific features. _J. Neurochem._ 113, 1073–1091 (2010). CAS PubMed PubMed Central Google Scholar * Taylor,
J.P., Hardy, J. & Fischbeck, K.H. Toxic proteins in neurodegenerative disease. _Science_ 296, 1991–1995 (2002). Article CAS PubMed Google Scholar * Kayed, R. et al. Common structure
of soluble amyloid oligomers implies common mechanism of pathogenesis. _Science_ 300, 486–489 (2003). Article CAS PubMed Google Scholar * Miller, J. et al. Identifying polyglutamine
protein species _in situ_ that best predict neurodegeneration. _Nat. Chem. Biol._ 7, 925–934 (2011). Article CAS PubMed PubMed Central Google Scholar * Gidalevitz, T., Ben-Zvi, A., Ho,
K.H., Brignull, H.R. & Morimoto, R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. _Science_ 311, 1471–1474 (2006). Article CAS PubMed
Google Scholar * Mitra, S., Tsvetkov, A.S. & Finkbeiner, S. Single neuron ubiquitin-proteasome dynamics accompanying inclusion body formation in Huntington disease. _J. Biol. Chem._
284, 4398–4403 (2009). Article CAS PubMed PubMed Central Google Scholar * Tsvetkov, A.S. et al. A small-molecule scaffold induces autophagy in primary neurons and protects against
toxicity in a Huntington disease model. _Proc. Natl. Acad. Sci. USA_ 107, 16982–16987 (2010). Article CAS PubMed PubMed Central Google Scholar * Takahashi, M. & Ono, Y. Pulse-chase
analysis of protein kinase C. _Methods Mol. Biol._ 233, 163–170 (2003). CAS PubMed Google Scholar * Gurskaya, N.G. et al. Engineering of a monomeric green-to-red photoactivatable
fluorescent protein induced by blue light. _Nat. Biotechnol._ 24, 461–465 (2006). Article CAS PubMed Google Scholar * Arrasate, M. & Finkbeiner, S. Automated microscope system for
determining factors that predict neuronal fate. _Proc. Natl. Acad. Sci. USA_ 102, 3840–3845 (2005). Article CAS PubMed PubMed Central Google Scholar * Leutenegger, A. et al. It's
cheap to be colorful. Anthozoans show a slow turnover of GFP-like proteins. _FEBS J._ 274, 2496–2505 (2007). Article CAS PubMed Google Scholar * Dantuma, N.P., Lindsten, K., Glas, R.,
Jellne, M. & Masucci, M.G. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. _Nat. Biotechnol._ 18, 538–543 (2000).
Article CAS PubMed Google Scholar * Zoghbi, H.Y. & Orr, H.T. Glutamine repeats and neurodegeneration. _Annu. Rev. Neurosci._ 23, 217–247 (2000). Article CAS PubMed Google Scholar
* DiFiglia, M. Clinical Genetics, II. Huntington's disease: from the gene to pathophysiology. _Am. J. Psychiatry_ 154, 1046 (1997). Article CAS PubMed Google Scholar * Sathasivam,
K. et al. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. _Proc. Natl. Acad. Sci. USA_ 110, 2366–2370 (2013). Article CAS PubMed PubMed Central
Google Scholar * Wellington, C.L. & Hayden, M.R. Caspases and neurodegeneration: on the cutting edge of new therapeutic approaches. _Clin. Genet._ 57, 1–10 (2000). Article CAS PubMed
Google Scholar * Mangiarini, L. et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. _Cell_ 87, 493–506
(1996). Article CAS PubMed Google Scholar * Arrasate, M., Mitra, S., Schweitzer, E.S., Segal, M.R. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and
the risk of neuronal death. _Nature_ 431, 805–810 (2004). Article CAS PubMed Google Scholar * Persichetti, F. et al. Differential expression of normal and mutant Huntington's
disease gene alleles. _Neurobiol. Dis._ 3, 183–190 (1996). Article CAS PubMed Google Scholar * Dyer, R.B. & McMurray, C.T. Mutant protein in Huntington disease is resistant to
proteolysis in affected brain. _Nat. Genet._ 29, 270–278 (2001). Article CAS PubMed Google Scholar * Kaytor, M.D., Wilkinson, K.D. & Warren, S.T. Modulating huntingtin half-life
alters polyglutamine-dependent aggregate formation and cell toxicity. _J. Neurochem._ 89, 962–973 (2004). Article CAS PubMed Google Scholar * Roscic, A., Baldo, B., Crochemore, C.,
Marcellin, D. & Paganetti, P. Induction of autophagy with catalytic mTOR inhibitors reduces huntingtin aggregates in a neuronal cell model. _J. Neurochem._ 119, 398–407 (2011). Article
CAS PubMed Google Scholar * Wu, J.C. et al. The regulation of N-terminal Huntingtin (Htt552) accumulation by Beclin1. _Acta Pharmacol. Sin._ 33, 743–751 (2012). Article CAS PubMed
PubMed Central Google Scholar * Kazantsev, A., Preisinger, E., Dranovsky, A., Goldgaber, D. & Housman, D. Insoluble detergent-resistant aggregates form between pathological and
nonpathological lengths of polyglutamine in mammalian cells. _Proc. Natl. Acad. Sci. USA_ 96, 11404–11409 (1999). Article CAS PubMed PubMed Central Google Scholar * Kopito, R.R.
Aggresomes, inclusion bodies and protein aggregation. _Trends Cell Biol._ 10, 524–530 (2000). CAS PubMed Google Scholar * Hartl, F.U. & Hayer-Hartl, M. Converging concepts of protein
folding _in vitro_ and _in vivo_. _Nat. Struct. Mol. Biol._ 16, 574–581 (2009). Article CAS PubMed Google Scholar * Snell, R.G. et al. Relationship between trinucleotide repeat expansion
and phenotypic variation in Huntington's disease. _Nat. Genet._ 4, 393–397 (1993). Article CAS PubMed Google Scholar * Saudou, F., Finkbeiner, S., Devys, D. & Greenberg, M.E.
Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. _Cell_ 95, 55–66 (1998). Article CAS PubMed Google Scholar
* Matsumoto, G., Kim, S. & Morimoto, R.I. Huntingtin and mutant SOD1 form aggregate structures with distinct molecular properties in human cells. _J. Biol. Chem._ 281, 4477–4485 (2006).
Article CAS PubMed Google Scholar * Lin, C.H. et al. Neurological abnormalities in a knock-in mouse model of Huntington's disease. _Hum. Mol. Genet._ 10, 137–144 (2001). Article
CAS PubMed Google Scholar * Colby, D.W., Cassady, J.P., Lin, G.C., Ingram, V.M. & Wittrup, K.D. Stochastic kinetics of intracellular huntingtin aggregate formation. _Nat. Chem. Biol._
2, 319–323 (2006). Article CAS PubMed Google Scholar * Slow, E.J. et al. Absence of behavioral abnormalities and neurodegeneration _in vivo_ despite widespread neuronal huntingtin
inclusions. _Proc. Natl. Acad. Sci. USA_ 102, 11402–11407 (2005). Article CAS PubMed PubMed Central Google Scholar * Tsakiri, E.N. et al. Proteasome dysfunction in _Drosophila_ signals
to an Nrf2-dependent regulatory circuit aiming to restore proteostasis and prevent premature aging. _Aging Cell_ http://dx.doi.org/10.1111/acel.12111 (2013). * Riley, B.E. et al. Ubiquitin
accumulation in autophagy-deficient mice is dependent on the Nrf2-mediated stress response pathway: a potential role for protein aggregation in autophagic substrate selection. _J. Cell
Biol._ 191, 537–552 (2010). Article CAS PubMed PubMed Central Google Scholar * Zhang, Q.C. et al. A compact β model of huntingtin toxicity. _J. Biol. Chem._ 286, 8188–8196 (2011).
Article CAS PubMed PubMed Central Google Scholar * Bilimoria, P.M. & Bonni, A. Cultures of cerebellar granule neurons. _Cold Spring Harb. Protoc._
http://dx.doi.org/10.1101/pdb.prot5107 (2008). Download references ACKNOWLEDGEMENTS This work was supported by grants R01 3NS039746 and 2R01 NS045191 from the US National Institute of
Neurological Disease and Stroke; grant P01 2AG022074 from the National Institute on Aging; by the Huntington's Disease Society of America (made possible with a gift from the James E.
Bashaw Family); the Taube-Koret Center for Neurodegenerative disease and the Gladstone Institutes (S.F.); the Milton Wexler Award and a fellowship from the Hereditary Disease Foundation
(A.S.T.); a fellowship from the Hillblom Foundation (M.A.); a fellowship from California Institute for Regenerative Medicine (P.S.), and in part by DMS-0914906 from the US National Science
Foundation (B.A.S.). Gladstone Institutes received support from a US National Center for Research Resources Grant RR18928-01. We thank Y. Dabaghian, I. Kelmanson, A. Gelfand and members of
the Finkbeiner laboratory for helpful discussions. The animal care facility was partly supported by a US National Institutes of Health Extramural Research Facilities Improvement Project (C06
RR018928). K. Nelson provided administrative assistance, and G.C. Howard, A.L. Lucido and S. Ordway edited the manuscript. AUTHOR INFORMATION Author notes * Montserrat Arrasate Present
address: Present address: Division of Neuroscience, Center for Applied Medical Research, University of Navarra, Pamplona, Spain., AUTHORS AND AFFILIATIONS * Gladstone Institute of
Neurological Disease, San Francisco, California, USA Andrey S Tsvetkov, Montserrat Arrasate, Sami Barmada, Punita Sharma & Steven Finkbeiner * Taube-Koret Center for Neurodegenerative
Disease Research, San Francisco, California, USA Andrey S Tsvetkov, Montserrat Arrasate, Punita Sharma & Steven Finkbeiner * Biomedical Sciences Graduate Program, University of
California–San Francisco, San Francisco, California, USA D Michael Ando & Steven Finkbeiner * Department of Statistical Science, Duke University, Durham, North Carolina, USA Benjamin A
Shaby * Graduate Programs in Neuroscience and Biomedical Sciences, University of California–San Francisco, San Francisco, California, USA Steven Finkbeiner * Program in Biological Sciences
and Medical Scientist Training Program, University of California–San Francisco, San Francisco, California, USA Steven Finkbeiner * Department of Neurology and Physiology, University of
California–San Francisco, San Francisco, California, USA Steven Finkbeiner Authors * Andrey S Tsvetkov View author publications You can also search for this author inPubMed Google Scholar *
Montserrat Arrasate View author publications You can also search for this author inPubMed Google Scholar * Sami Barmada View author publications You can also search for this author inPubMed
Google Scholar * D Michael Ando View author publications You can also search for this author inPubMed Google Scholar * Punita Sharma View author publications You can also search for this
author inPubMed Google Scholar * Benjamin A Shaby View author publications You can also search for this author inPubMed Google Scholar * Steven Finkbeiner View author publications You can
also search for this author inPubMed Google Scholar CONTRIBUTIONS A.S.T., M.A. and S.F. designed the study. A.S.T. and S.F. wrote the manuscript. B.A.S. performed statistical analysis and
wrote the statistical analysis section of the manuscript. A.S.T., M.A., P.S., S.B. and D.M.A. wrote scripts for automated photoswitching and imaging. A.S.T. cloned all of the constructs used
in the study. A.S.T. and M.A. cultured primary neurons and performed transfections, automated microscopy, fluorescence intensity measurements and data analysis. A.S.T. performed detergent
extraction, metabolic labeling and photobleaching experiments. A.S.T. and P.S. performed survival analyses. CORRESPONDING AUTHOR Correspondence to Steven Finkbeiner. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TEXT AND FIGURES Supplementary Results and Supplementary Figures 1–6. (PDF
1333 kb) RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Tsvetkov, A., Arrasate, M., Barmada, S. _et al._ Proteostasis of polyglutamine varies among
neurons and predicts neurodegeneration. _Nat Chem Biol_ 9, 586–592 (2013). https://doi.org/10.1038/nchembio.1308 Download citation * Received: 24 September 2012 * Accepted: 25 June 2013 *
Published: 21 July 2013 * Issue Date: September 2013 * DOI: https://doi.org/10.1038/nchembio.1308 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative




:strip_icc()/i.s3.glbimg.com/v1/AUTH_63b422c2caee4269b8b34177e8876b93/internal_photos/bs/2024/A/Z/yTof2gRYALQKQgAvs2rw/foto15carr-101-accor-b2.jpg)