- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
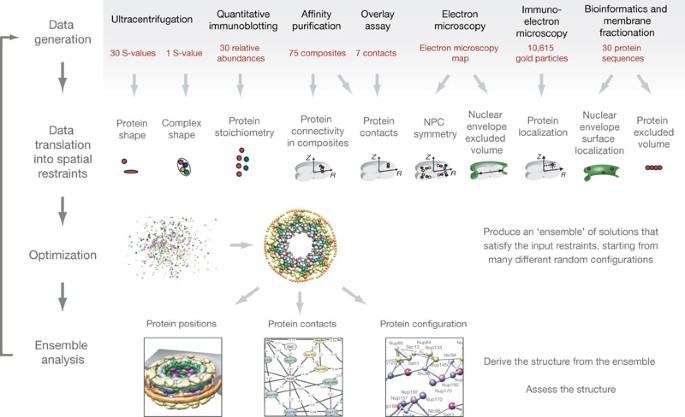
ABSTRACT To understand the workings of a living cell, we need to know the architectures of its macromolecular assemblies. Here we show how proteomic data can be used to determine such
structures. The process involves the collection of sufficient and diverse high-quality data, translation of these data into spatial restraints, and an optimization that uses the restraints
to generate an ensemble of structures consistent with the data. Analysis of the ensemble produces a detailed architectural map of the assembly. We developed our approach on a challenging
model system, the nuclear pore complex (NPC). The NPC acts as a dynamic barrier, controlling access to and from the nucleus, and in yeast is a 50 MDa assembly of 456 proteins. The resulting
structure, presented in an accompanying paper, reveals the configuration of the proteins in the NPC, providing insights into its evolution and architectural principles. The present approach
should be applicable to many other macromolecular assemblies. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS
OPTIONS Access through your institution Subscribe to this journal Receive 51 print issues and online access $199.00 per year only $3.90 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS INTEGRATIVE STRUCTURAL MODELING OF MACROMOLECULAR COMPLEXES USING ASSEMBLINE
Article 29 November 2021 A MULTI-SCALE MAP OF CELL STRUCTURE FUSING PROTEIN IMAGES AND INTERACTIONS Article 24 November 2021 A QUANTITATIVE MAP OF NUCLEAR PORE ASSEMBLY REVEALS TWO DISTINCT
MECHANISMS Article Open access 04 January 2023 REFERENCES * Sali, A., Glaeser, R., Earnest, T. & Baumeister, W. From words to literature in structural proteomics. _Nature_ 422, 216–225
(2003) Article ADS CAS PubMed Google Scholar * Rout, M. P. et al. The yeast nuclear pore complex: composition, architecture, and transport mechanism. _J. Cell Biol._ 148, 635–651 (2000)
Article CAS PubMed PubMed Central Google Scholar * Macara, I. G. Transport into and out of the nucleus. _Microbiol. Mol. Biol. Rev._ 65, 570–594 (2001) Article CAS PubMed PubMed
Central Google Scholar * Weis, K. Nucleocytoplasmic transport: cargo trafficking across the border. _Curr. Opin. Cell Biol._ 14, 328–335 (2002) Article CAS PubMed Google Scholar *
Yang, Q., Rout, M. P. & Akey, C. W. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. _Mol. Cell_ 1, 223–234 (1998)
Article CAS PubMed Google Scholar * Beck, M., Lucic, V., Förster, F., Baumeister, E. & Medalia, O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography.
_Nature_ 449, 611–615 (2007) Article ADS CAS PubMed Google Scholar * Devos, D. et al. Simple fold composition and modular architecture of the nuclear pore complex. _Proc. Natl Acad.
Sci. USA_ 103, 2172–2177 (2006) Article ADS CAS PubMed PubMed Central Google Scholar * Havel, T. F. & Wüthrich, K. A distance geometry program for determining the structures of
small proteins and other macromolecules from nuclear magnetic resonance measurements of intramolecular 1H–1H proximities in solution. _Bull. Math. Biol._ 46, 673–698 (1984) CAS MATH Google
Scholar * Malhotra, A., Tan, R. K. & Harvey, S. C. Prediction of the three-dimensional structure of _Escherichia coli_ 30S ribosomal subunit: a molecular mechanics approach. _Proc.
Natl Acad. Sci. USA_ 87, 1950–1954 (1990) Article ADS CAS PubMed PubMed Central Google Scholar * Denning, D. P., Patel, S. S., Uversky, V., Fink, A. L. & Rexach, M. Disorder in the
nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. _Proc. Natl Acad. Sci. USA_ 100, 2450–2455 (2003) Article ADS CAS PubMed PubMed Central Google
Scholar * Lim, R. Y. et al. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. _Proc. Natl Acad. Sci. USA_ 103, 9512–9517 (2006) Article ADS
CAS PubMed PubMed Central Google Scholar * Devos, D. et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. _PLoS Biol._ 2, e380 (2004)
Article PubMed PubMed Central CAS Google Scholar * Siniossoglou, S. et al. Structure and assembly of the Nup84p complex. _J. Cell Biol._ 149, 41–54 (2000) Article CAS PubMed PubMed
Central Google Scholar * Lutzmann, M., Kunze, R., Buerer, A., Aebi, U. & Hurt, E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. _EMBO J._ 21,
387–397 (2002) Article CAS PubMed PubMed Central Google Scholar * Strambio-de-Castillia, C., Blobel, G. & Rout, M. P. Isolation and characterization of nuclear envelopes from the
Yeast _Saccharomyces_ . _J. Cell Biol._ 131, 19–31 (1995) Article CAS PubMed Google Scholar * Miller, A. L. et al. Cytoplasmic inositol hexakisphosphate production is sufficient for
mediating the Gle1-mRNA export pathway. _J. Biol. Chem._ 279, 51022–51032 (2004) Article CAS PubMed Google Scholar * Solsbacher, J., Maurer, P., Vogel, F. & Schlenstedt, G. Nup2p, a
yeast nucleoporin, functions in bidirectional transport of importin alpha. _Mol. Cell. Biol._ 20, 8468–8479 (2000) Article CAS PubMed PubMed Central Google Scholar * Marelli, M.,
Aitchison, J. D. & Wozniak, R. W. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p. _J. Cell Biol._ 143,
1813–1830 (1998) Article CAS PubMed PubMed Central Google Scholar * Archambault, V. et al. Genetic and biochemical evaluation of the importance of Cdc6 in regulating mitotic exit. _Mol.
Biol. Cell_ 14, 4592–4604 (2003) Article CAS PubMed PubMed Central Google Scholar * Archambault, V. et al. Targeted proteomic study of the cyclin-Cdk module. _Mol. Cell_ 14, 699–711
(2004) Article CAS PubMed Google Scholar * Tackett, A. J. et al. I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. _J. Proteome Res._ 4,
1752–1756 (2005) Article CAS PubMed Google Scholar * Cristea, I. M., Williams, R., Chait, B. T. & Rout, M. P. Fluorescent proteins as proteomic probes. _Mol. Cell. Proteomics_ 4,
1933–1941 (2005) Article CAS PubMed Google Scholar * Niepel, M., Strambio-de-Castillia, C., Fasolo, J., Chait, B. T. & Rout, M. P. The nuclear pore complex-associated protein, Mlp2p,
binds to the yeast spindle pole body and promotes its efficient assembly. _J. Cell Biol._ 170, 225–235 (2005) Article CAS PubMed PubMed Central Google Scholar * Cristea, I. M. et al.
Tracking and elucidating alphavirus-host protein interactions. _J. Biol. Chem._ 281, 30269–30278 (2006) Article CAS PubMed Google Scholar * Zhang, W. & Chait, B. T. ProFound: an
expert system for protein identification using mass spectrometric peptide mapping information. _Anal. Chem._ 72, 2482–2489 (2000) Article CAS PubMed Google Scholar * Krutchinsky, A. N.,
Kalkum, M. & Chait, B. T. Automatic identification of proteins with a MALDI-quadrupole ion trap mass spectrometer. _Anal. Chem._ 73, 5066–5077 (2001) Article CAS PubMed Google Scholar
* Stelter, P. et al. Molecular basis for the functional interaction of dynein light chain with the nuclear-pore complex. _Nature Cell Biol._ 9, 788–796 (2007) Article CAS PubMed Google
Scholar * Murphy, R., Watkins, J. L. & Wente, S. R. GLE2, a _Saccharomyces cerevisiae_ homologue of the _Schizosaccharomyces pombe_ export factor RAE1, is required for nuclear pore
complex structure and function. _Mol. Biol. Cell_ 7, 1921–1937 (1996) Article CAS PubMed PubMed Central Google Scholar * Murphy, R. & Wente, S. R. An RNA-export mediator with an
essential nuclear export signal. _Nature_ 383, 357–360 (1996) Article ADS CAS PubMed Google Scholar * Lutzmann, M. et al. Reconstitution of Nup157 and Nup145N into the Nup84 complex.
_J. Biol. Chem._ 280, 18442–18451 (2005) Article CAS PubMed Google Scholar * Bailer, S. M. et al. Nup116p associates with the Nup82p-Nsp1p-Nup159p nucleoporin complex. _J. Biol. Chem._
275, 2354–23548 (2000) Article Google Scholar * Grandi, P., Doye, V. & Hurt, E. C. Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore
protein NIC96. _EMBO. J._ 12, 3061–3071 (1993) Article CAS PubMed PubMed Central Google Scholar * Shen, M. Y. & Sali, A. Statistical potential for assessment and prediction of
protein structures. _Protein Sci._ 15, 2507–2524 (2006) Article CAS PubMed PubMed Central Google Scholar * Harding, S. E. Determination of macromolecular homogeneity, shape, and
interactions using sedimentation velocity analytical ultracentrifugation. _Methods Mol. Biol._ 22, 61–73 (1994) CAS PubMed Google Scholar * Krogh, A., Larsson, B., von Heijne, G. &
Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. _J. Mol. Biol._ 305, 567–580 (2001) Article CAS PubMed Google
Scholar * Alber, F., Kim, M. F. & Sali, A. Structural characterization of assemblies from overall shape and subcomplex compositions. _Structure_ 13, 435–445 (2005) Article CAS PubMed
Google Scholar * Alber, F. et al. The molecular architecture of the nuclear pore complex. _Nature_ doi: 10.1038/nature06405 (this issue). * Akey, C. W. & Radermacher, M. Architecture
of the _Xenopus_ nuclear pore complex revealed by three-dimensional cryo-electron microscopy. _J. Cell Biol._ 122, 1–19 (1993) Article CAS PubMed Google Scholar * Stoffler, D. et al.
Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. _J. Mol. Biol._ 328, 119–130 (2003) Article CAS PubMed
Google Scholar * Kiseleva, E. et al. Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. _J. Struct. Biol._ 145, 272–288 (2004) Article CAS PubMed Google Scholar
* Hinshaw, J. E., Carragher, B. O. & Milligan, R. A. Architecture and design of the nuclear pore complex. _Cell_ 69, 1133–1141 (1992) Article CAS PubMed Google Scholar * Beck, M.
et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. _Science_ 306, 1387–1390 (2004) Article ADS CAS PubMed Google Scholar * Pante, N. & Kann, M.
Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. _Mol. Biol. Cell_ 13, 425–434 (2002) Article CAS PubMed PubMed Central Google Scholar * Drin, G.
et al. A general amphipathic α-helical motif for sensing membrane curvature. _Nature Struct. Mol. Biol._ 14, 138–146 (2007) Article CAS Google Scholar * Schurmann, G., Haspel, J., Grumet,
M. & Erickson, H. P. Cell adhesion molecule L1 in folded (horseshoe) and extended conformations. _Mol. Biol. Cell_ 12, 1765–1773 (2001) Article CAS PubMed PubMed Central Google
Scholar Download references ACKNOWLEDGEMENTS We thank H. Shio for performing the electron microscopic studies; J. Fanghänel, M. Niepel and C. Strambio-de-Castillia for help in developing
the affinity purification techniques; M. Magnasco for discussions and advice; A. Kruchinsky for assistance with mass spectrometry; M. Topf, D. Korkin, F. Davis, M.-Y. Shen, F. Foerster, N.
Eswar, M. Kim, D. Russel, B. Peterson and B. Webb for many discussions about structure characterization by satisfaction of spatial restraints; C. Johnson, S. G. Parker and C. Silva, T.
Ferrin and T. Goddard for preparation of some figures; and S. Pulapura and X. J. Zhou for their help with the design of the conditional diameter restraint. We are grateful to J. Aitchison
for discussion and insightful suggestions. We also thank all other members of the Chait, Rout and Sali laboratories for their assistance. We acknowledge support from an Irma T. Hirschl
Career Scientist Award (M.P.R.), a Sinsheimer Scholar Award (M.P.R.), a grant from the Rita Allen Foundation (M.P.R.), a grant from the American Cancer Society (M.P.R.), the Sandler Family
Supporting Foundation (A.S.), the Human Frontier Science Program (A.S., L.M.V.), NSF (A.S.), and grants from the National Institutes of Health (B.T.C., M.P.R., A.S.), as well as computer
hardware gifts from R. Conway, M. Homer, Intel, Hewlett-Packard, IBM and Netapp (A.S.). AUTHOR INFORMATION Author notes * Svetlana Dokudovskaya, Liesbeth M. Veenhoff, Julia Kipper, Damien
Devos, Adisetyantari Suprapto & Orit Karni-Schmidt Present address: Present addresses: Laboratory of Nucleocytoplasmic Transport, Institut Jacques Monod, 2 place Jussieu, Tour 43, Paris
75251, France (S.D.); Department of Biochemistry, University of Groningen, Nijenborgh 4, 9747 AG Groningen, The Netherlands (L.M.V.); German Aerospace Center (PT-DLR), Heinrich-Konen-Strasse
1, D-53227 Bonn, Germany (J.K.); Structural Bioinformatics, EMBL, Meyerhofstrasse 1, D-69117 Heidelberg, Germany (D.D.); Office of Technology Transfer, The Rockefeller University, 1230 York
Avenue, New York, New York 10065, USA (A.S.); Herbert Irving Comprehensive Cancer Center, Columbia University, 1130 St Nicholas Avenue, New York, New York 10032, USA (O.K.-S.)., New York *
Frank Alber, Svetlana Dokudovskaya and Liesbeth M. Veenhoff: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Bioengineering and Therapeutic Sciences,
Department of Pharmaceutical Chemistry, and California Institute for Quantitative Biosciences, Byers Hall, Suite 503B, 1700 4th Street, University of California at San Francisco, San
Francisco, California 94158-2330, USA, California Frank Alber, Damien Devos & Andrej Sali * Laboratory of Cellular and Structural Biology, and,, California Svetlana Dokudovskaya,
Liesbeth M. Veenhoff, Julia Kipper, Adisetyantari Suprapto, Orit Karni-Schmidt, Rosemary Williams & Michael P. Rout * Laboratory of Mass Spectrometry and Gaseous Ion Chemistry, The
Rockefeller University, 1230 York Avenue, New York, New York 10065, USA , New York Wenzhu Zhang & Brian T. Chait Authors * Frank Alber View author publications You can also search for
this author inPubMed Google Scholar * Svetlana Dokudovskaya View author publications You can also search for this author inPubMed Google Scholar * Liesbeth M. Veenhoff View author
publications You can also search for this author inPubMed Google Scholar * Wenzhu Zhang View author publications You can also search for this author inPubMed Google Scholar * Julia Kipper
View author publications You can also search for this author inPubMed Google Scholar * Damien Devos View author publications You can also search for this author inPubMed Google Scholar *
Adisetyantari Suprapto View author publications You can also search for this author inPubMed Google Scholar * Orit Karni-Schmidt View author publications You can also search for this author
inPubMed Google Scholar * Rosemary Williams View author publications You can also search for this author inPubMed Google Scholar * Brian T. Chait View author publications You can also search
for this author inPubMed Google Scholar * Michael P. Rout View author publications You can also search for this author inPubMed Google Scholar * Andrej Sali View author publications You can
also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Brian T. Chait, Michael P. Rout or Andrej Sali. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
INFORMATION The file contains Supplementary Methods, Supplementary Figures 1-27, Supplementary Tables 1-10 and additional references. (PDF 8230 kb) RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Alber, F., Dokudovskaya, S., Veenhoff, L. _et al._ Determining the architectures of macromolecular assemblies. _Nature_ 450, 683–694 (2007).
https://doi.org/10.1038/nature06404 Download citation * Received: 30 August 2007 * Accepted: 22 October 2007 * Published: 29 November 2007 * Issue Date: 29 November 2007 * DOI:
https://doi.org/10.1038/nature06404 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently
available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative