- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT During range expansions, even low levels of interbreeding can lead to massive introgression of local alleles into an invader's genome. Nonetheless, this pattern is not always
observed in human populations. For instance, European Americans in North America are barely introgressed by Amerindian genes in spite of known contact and admixture. With coalescent
spatially explicit simulations, we examined the impact of long-distance dispersal (LDD) events on introgression of local alleles into the invading population using a set of different
demographic scenarios applicable to a diverse range of natural populations and species. More specifically, we consider two distinct LDD models: one where LDD events originate in the range
core and targets only the expansion front and a second one where LDD events can occur from any area to any other. We find that LDD generally prevents introgression, but that LDD events
specifically targeting the expansion front are most efficient in suppressing introgression. This is likely due to the fact that LDD allows for the presence of a larger number of invader
alleles at the wave front, where effective population size is thus increased and local introgressed alleles are rapidly outnumbered. We postulate that the documented settlement of pioneers
directly on the wave front in North America has contributed to low levels of Amerindian admixture observed in European Americans and that this phenomenon may well explain the lack of
introgression after a range expansion in natural populations without the need to evoke other mechanisms such as natural selection. SIMILAR CONTENT BEING VIEWED BY OTHERS CONSTRAINTS TO GENE
FLOW INCREASE THE RISK OF GENOME EROSION IN THE NGORONGORO CRATER LION POPULATION Article Open access 21 April 2025 CLIMATE-INDUCED RANGE SHIFTS DRIVE ADAPTIVE RESPONSE VIA SPATIO-TEMPORAL
SIEVING OF ALLELES Article Open access 25 February 2023 MULTIPLE SOURCE LOCATIONS AND LONG-DISTANCE DISPERSAL EXPLAIN THE RAPID SPREAD OF A RECENT AMPHIBIAN INVASION Article Open access 16
May 2025 INTRODUCTION When a species expands its range and colonizes a new habitat, it can encounter new environmental conditions and potentially interact with other species. Although new
environments can impose novel selective pressure over the invading species, the contact with another species may lead to a reinforcement of the reproductive isolation, introgression of
adaptive, deleterious, or neutral alleles, and even hybrid speciation (Barton, 2001). In absence of selection against the invaders, it has been shown that interbreeding between a local and
an invading species can result in massive introgression of local genes into the invading gene pool (Currat et al., 2008). This asymmetric introgression is observed even in the face of a very
low (<2%) rate of interbreeding (Currat and Excoffier, 2011). This is partially caused by the ‘surfing’ of introgressed alleles at the wave front, where the effective population size of
the invading species is usually lower than that of the local population (Klopfstein et al., 2006; Currat et al., 2008). Humans have had a complex demographic history including range
expansions with admixture even between remotely related human populations (Jobling, 2012; Hellenthal et al., 2014; Homburger et al., 2015) as well as interbreeding with archaic hominids
(Sankararaman et al., 2014). A few hundred years ago, Europeans settled in America and an intricate process of admixture took place, involving autochthonous groups, European settlers and
African slaves (Ruiz-Linares et al., 2014; Bryc et al., 2015; Homburger et al., 2015). This resulted in an admixed population with varying degrees of genetic contribution from these
continental groups; for instance, while the proportion of Amerindian alleles in certain regions of South America is substantial (Wang et al., 2008; Galanter et al., 2012; Ruiz-Linares et
al., 2014; Salzano and Sans, 2014; Homburger et al., 2015; Salazar-Flores et al., 2015), there is little evidence of Amerindian genetic contribution in European Americans in North America
(Halder et al., 2009; Lao et al., 2010; Lisabeth et al., 2011; Bryc et al., 2015). The latter observation is at odds with the expected high level of introgression of local genes into the
invaders’ gene pool mentioned above (Currat et al., 2008) because, in spite of the known strong reproductive isolation between Europeans and Amerindians, interbreeding still might have
occurred at low levels (Meinig, 1986). Thus, other mechanisms that prevent introgression of local genes may be required to explain the limited genetic contribution of Amerindians to North
American populations of European descent. A factor that could have suppressed introgression of Amerindian alleles is the pattern of colonial expansion involving long-distance dispersal (LDD)
of Europeans. LDD during range expansions has been shown to lead to patchy allelic distributions (Nichols and Hewitt, 1994; Ibrahim et al., 1996), to boost genetic exchange between distant
populations (Bialozyt et al., 2006; Ray and Excoffier, 2010), and to increase diversity along the expansion range, limiting allele surfing and the effect of drift, and therefore maximizing
adaptive potential during range expansions (Berthouly-Salazar et al., 2013). In fact, during the expansion of European colonialists in North America, a peculiar form of LDD took place, where
migrants arriving from Europe settled directly in the expansion front (Meinig, 1993). Strikingly, the effects of this specific kind of LDD over the genetic diversity of the colonizing
species or population have not yet been evaluated. In this study, we simulated range expansions with and without LDD, using two different LDD models that reflect two distinct dispersal
dynamics in order to compare their impact on levels of introgression of local alleles in the invading population throughout the colonized range. MATERIALS AND METHODS SIMULATION OF RANGE
EXPANSIONS WITH COMPETITION We simulated two subsequent range expansions with a modified version of the software SPLATCHE2 (Ray et al., 2010). This software considers a two-step simulation
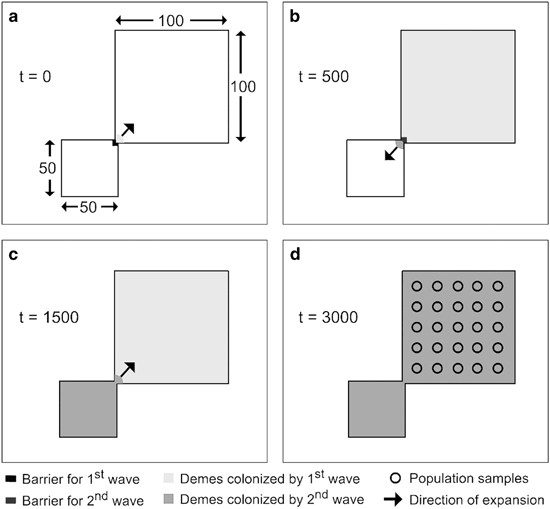
process: (1) forward demographic simulations; and (2) backward coalescent genetic simulations. In each simulation, the first step begins with a first expansion wave (locals) starting at 3000
generations before present (_t_=0; Figure 1a) and colonizes an area of 100 × 100 demes in about 500 generations (Figure 1b). The square area at the lower left, defined as a zone of 50 × 50
demes, is only colonized during the second wave to represent a source of LDD. This second wave then invades the range colonized by the first-wave 1500 generations before present (_t_=1 500
generations) from a deme at the lower-left corner of the large area (Figure 1c). Note that, even though the first step consists in a forward demographic simulation, the simulated populations
consist of haploid individuals. Thus, in this context, migration events correspond to the dispersal of genetic material (that is, gene copies at a given locus). The simulation of genetic
processes such as interbreeding, admixture and gene flow is described below in more detail. Both range expansions (of local and invading populations) are modeled according to a
Poisson-distributed stochastic nearest-neighbor migration model, with _N × m_ haploid emigrants (which can also be thought of as gene copies, as mentioned above) being sent at each
generation to surrounding demes, where _N_ is the current local population size and _m_ is the migration rate, here set to 0.1. Once a deme is occupied by one or more migrants, logistic
growth starts with intrinsic rate _r_=0.8 until the carrying capacity _K_ is reached. In our simulations, we used _K_=50 for the first wave (locals) and _K_=500 for the second wave
(invaders) and any individual colonizing an empty deme could establish a new population by itself in any scenario. Note that these parameters were arbitrarily chosen. Nonetheless, they allow
the colonization of the simulated world to occur in the timeframe chosen and are relatively realistic and in keeping with previous modeling of range expansions (Currat and Excoffier, 2004;
Currat and Excoffier, 2011). When the invaders colonize an already occupied deme, they compete with locals for local resources following a standard Lotka–Volterra model (Currat et al., 2004;
Currat and Excoffier, 2011), and they admix at a rate that is controlled by an interbreeding success rate _γ_ as follows: Where _N__i_ and _N__j_ are the haploid population size of the
local and the invader populations respectively. _A__ji_ is then used to update local population densities as follows: In this regard, a _γ_ value of 1 implies random mating between
populations, and a value of 0 implies reproductive isolation between populations (Currat and Excoffier, 2011). A more detailed explanation on this process is given in Currat and Excoffier,
2004. This density-dependent competition eventually results in the extinction of the local population due to its lower carrying capacity and it restricts interbreeding to occur only on the
invaders’ colonization front. In this regard, it is worth noting that the absolute values for carrying capacity _K_ were also chosen arbitrarily. However, the important point here is that
_K_ should be lower for locals than for colonizers such that density-dependent competition leads to the disappearance of the local population, which, in the context of the settlement of the
Americas by Europeans, models the decline in Amerindian population densities at the time of European colonization (Mulligan et al., 2004). MODELING LONG-DISTANCE DISPERSAL LDD was
implemented in two different ways. The first model, hereafter called ‘generalized LDD’, is described in more detail by Ray and Excoffier, 2010. Briefly, during the second range expansion
(invaders), a given proportion _δ_ of emigration events is chosen to be LDD events, and the remaining proportion (1-_δ_) are short-distance (nearest-neighbor) migration events as mentioned
above. For each LDD event, a migration direction and a dispersal distance are chosen at random. The dispersal distance is drawn from a gamma distribution with shape parameter _α_=0.5 and
scale parameter _β_=0.05, which leads to an average dispersal distance of _α_/_β_=10 demes for LDD events. We also arbitrarily imposed a maximum dispersal distance of 10 demes, such that the
LDD kernel is a truncated Gamma distribution. Because it has been shown that the shape of the dispersal kernel can influence the colonization dynamics (Bohrer et al., 2005; Fayard et al.,
2009), we performed additional simulations with an average LDD dispersal distance set to 5 or 20 demes. In the second model, hereafter called ‘front LDD’, LDD events occur from an
arbitrarily defined source population and exclusively target the wave front. In our simulations, the LDD source area is modeled as a zone of 50x50 demes adjacent to the 100x100 study area
(Figure 1c), but the exact location of this source should not affect our results on rate of introgression. During the second range expansion in the study area (Figure 1c), demes on the wave
front receive a certain proportion _ɛ_ of haploid migrants (that is, gene copies) from the LDD source population. This model is thus directly inspired by the colonization of North America by
European migrants who settled directly on the colonization wave front, as described by Meinig, 1993. In the front LDD model, the wave front is defined as those demes having a population
density _N_⩽0.5 _K_, which implies that all demes that have not reached 50% of their carrying capacity _K_ will eventually receive LDD directly from the source population (Supplementary
Figure S1). The parameter _ɛ_ specifies at each generation _t_ how much of the _K_—_N(t)_ available room in the front-deme will be filled by LDD migrants, which are then drawn from a random
deme of the LDD source population. Note that the proportion of front LDD migrants (_ɛ_) is very different from _δ_ defined in the generalized LDD model; _ɛ_ specifies how much of the empty
space in the front-deme will be filled by LDD migrants drawn from the source population, whereas _δ_ is the proportion of migratory events that are LDD events in the first model. We
considered four values for _γ_ (0.02, 0.04, 0.06 and 0.08) and five values (0, 0.0001, 0.001, 0.01 and 0.1) for each LDD parameter (_δ_ for the generalized LDD model and _ɛ_ for the front
LDD model). The combination of these parameters yields 20 different scenarios for each model of LDD, including the possibility of no LDD when _δ_ or _ɛ_ are equal to 0. ESTIMATES OF
INTROGRESSION LEVEL For each of these 40 tested scenarios, we performed 1000 independent forward simulations equivalent to the simulation of 1000 independent loci. At the end of each
simulation, we drew population samples belonging to 25 demes distributed over a regular grid in the study area (Figure 1d). We used a backward coalescent approach (corresponding to the
second step of the simulation process) to infer each gene copy’s origin (first or second wave) and recorded it for estimating introgression proportions (that is, the proportion of gene
copies in the invader population coming from the local population) in the 25 sampled demes in the study area over the 1000 simulations. We also recorded the number of generations between the
start of the invasion and the colonization of the whole map for each simulation (colonization time), as well as the number of generations during which locals and invaders coexisted in a
given position of the simulated grid (cohabitation time), averaged over demes and simulations. Interbreeding events were quantified as the number of haploid individuals exchanged between
locals and invaders in a given deme averaged over the whole map and over all simulations. RESULTS The dynamics of the colonization process over the large (100 × 100 demes) square world is
illustrated in Figure 2 for different LDD models. Although the front-LDD (Figure 2d) model yields a colonization pattern very similar to the case of no LDD (Figure 2a), where dispersal
occurs as a single, contiguous wave, under the generalized LDD model (Figure 2b and d), LDD events lead to independent expansion waves in the habitat occupied by the local population. The
number of these growing points is directly proportional to the proportion _δ_ of LDD events (Figure 2b and c). Levels of introgression increase with interbreeding rates _γ_ but are
negatively correlated with the proportion _δ_ of long-distance events (Figure 3a and b). Indeed, when 10% of all migration events are generalized LDD, introgression levels are reduced by 27%
and up to 61% depending on interbreeding rates (_γ_=0.08 to 0.02, respectively). However, generalized LDD is sufficient neither to completely suppress local introgression in the invading
population (Figure 3b) nor to prevent gradients of introgression along the expansion axis (Figure 3a). Contrastingly, we find that LDD events from a non-admixed population targeting directly
the wave front—the front LDD model—can very strongly inhibit introgression (Figure 3c and d). Indeed, if 10% of individuals living on the wave front (_ɛ_=0.1) come directly from the source
population, introgression is virtually no longer detectable at the end of the expansion (rightmost columns of both Figure 3c and d). This occurs even when interbreeding is relatively common
between local and invading individuals (that is, _γ_=0.08), as depicted in the lower right corner of Figure 3c. Interestingly, the gradient of introgression that is still visible with
generalized LDD (Figure 3a) is also suppressed when there is direct LDD to the wave front (Figure 3c). Although introgression levels shown in Figure 3a and c are averaged over 1000 loci,
individual loci display a much more spatially heterogeneous distribution of introgression as shown in Supplementary Figure S2. It is possible to see clear sectors of different introgression
levels for colonization without LDD (Supplementary Figure S2a), and to some extent also in the generalized LDD model (Supplementary Figure S2b), but not with front LDD (Supplementary Figure
S2c). Although there is a negative correlation between _δ_ and introgression in the generalized LDD model (Figure 3b), there is an increase in the number of interbreeding events and in the
period of cohabitation between locals and invaders when there is a larger proportion of long-distance events (Table 1). On the other hand, front LDD causes both interbreeding and
cohabitation time to decrease (Table 2). Colonization time decreases with both generalized and front LDD (Tables 1 and 2). In addition, very low levels of LDD lead to a larger variance in
the dynamics of the colonization process, particularly in the generalized LDD model given the random nature of LDD events. Note that cohabitation time and interbreeding may be slightly
underestimated for the front LDD model, since the source zone is modeled differently in the front and generalized LDD models. Indeed, the source zone is included in the computation of
cohabitation times and interbreeding levels in the case of the front LDD model, but not for the generalized LDD model. Nevertheless, values within each table are comparable and trends for
each case can be trusted. Finally, using different average dispersal distances of LDD events (5 or 20 demes; Supplementary Figure S3), we obtain results very similar to when the average LDD
distance is 10 demes, thus suggesting that the effect of LDD on admixture does not depend much on the dispersal kernel. DISCUSSION As previously described (Currat et al., 2008),
introgression levels are expected to increase along the expansion axis due to recurrent interbreeding on the wave front, where surfing of introgressed alleles may occur as a consequence of
the lower population density of the invaders relative to the locals. Interbreeding during range expansions, even at low frequency, can thus lead to high levels of introgression in the
growing invading populations. However, as we show here, the presence of LDD events during the colonization lowers the introgression of local genes into the invaders gene pool, because genes
migrating to the front from the core directly compete with local introgressed genes that are thus rapidly outnumbered. In addition, we show LDD can even completely suppress introgression
when LDD events target exclusively the wave front (Figure 3). Indeed, the effect of front LDD is much more drastic than the effect of generalized LDD. For instance, with relatively high
levels of interbreeding (_γ=_0.08) we have over 96% of introgression in absence of LDD, but if as little as 0.1% of the vacant space on the wave front is filled by migrants from the source
population (_ɛ_=0.001), introgression levels drops to 43.2% (Figure 3c). Contrastingly, under the generalized LDD model, a similar introgression proportion is reached (42.6%) with much more
extreme parameter settings, namely high proportion of LDD (_δ=_0.1) and a much lower interbreeding rate (_γ=_0.04). These results are consistent with the protective effect of intraspecific
migrations against introgression (Currat et al., 2008; Petit and Excoffier, 2009). An important implication of this effect is that, for species that are able to disperse over long distances,
such as some plants and birds, the absence of introgression after a range expansion may not necessarily be due to selection against hybrids (Barton and Bengtsson, 1986; Currat and
Excoffier, 2004) or to selection against local alleles in the invader (Excoffier et al., 2009). Strikingly, under the generalized LDD model, the number of interbreeding events increases with
larger levels of LDD events (Table 1), in contrast to the introgression levels, which decrease with increasing LDD (Figure 3a). It implies that these interbreeding events are not as
effective as in the absence of LDD. This seemingly paradoxical behavior occurs because demes colonized ahead of the wave front by LDD events take more time to reach their carrying capacity
than demes on the wave front, as they do not receive migrants from the wake of the wave. This leads to a longer cohabitation time between locals and invaders (Table 1) and hence more time
for introgression events to happen. On the other side, these introgression events are not as successful as in absence of LDD, because colonization by LDD events limits the dilution of the
invader gene pool and prevents introgressed alleles to surf (Bialozyt et al., 2006; Ray and Excoffier, 2010; Berthouly-Salazar et al., 2013) and increase in frequency during the expansion.
Since the number of interbreeding events per deme in case of front LDD is not much smaller than in the case of generalized LDD, it suggests that the much lower introgression level observed
in the case of front LDD is not due to a lowering of the amount of interbreeding, but indeed to the prevention of the surfing of introgressed genes on the wave front. Some loci show regions
of high and low introgression (Supplementary Figure S2) that look similar to sectors of low allelic diversity created by gene surfing during range expansions (Currat et al., 2006;
Hallatschek et al., 2007; Excoffier and Ray, 2008). Introgressed alleles, like any mutation, can thus potentially surf on the wave of advance and create sectors with high admixture levels.
Nonetheless direct LDD to the wave front efficiently prevents the occurrence of these sectors of introgression. We notice that the choice of demographic parameters for the simulations was
done somehow arbitrarily, but the chosen parameter values are nonetheless relatively realistic. For instance, migration and growth rates, as well as carrying capacities are within the range
of values explored previously for the analysis of modern and archaic humans evolutionary history (Currat and Excoffier 2004; Currat and Excoffier 2011). Changes in migration and growth rates
would mainly affect the speed of the colonization but could also affect introgression levels. Indeed, as mentioned earlier, higher levels of gene flow between demes of the invading
populations should prevent introgression (Petit and Excoffier, 2009) and higher growth rates should favor surfing (Klopfstein et al. 2006) and therefore have a positive effect on
introgression. However, the main conclusion of our study, which is that LDD can drastically lower introgression, should be robust to alternative (but reasonable) values of the expansion
model parameters. On a related note, we identified no effect of changes in the shape of the dispersal kernel on the patterns of admixture (Supplementary Figure S3). This is surprising given
that it has been shown that this parameter influences the colonization dynamics (Bohrer et al., 2005; Fayard et al., 2009). We notice, however, that the key feature of our simulations is to
show that heavy-tailed distributions, as the ones we considered for the generalized LDD model, can indeed preserve initial genetic diversity during range expansions, as previously shown
(Fayard et al., 2009; Goodsman et al., 2014; Alves et al., 2016), and that LDD directed to the wave front is more efficient in doing that, in agreement with an empirical study on starlings
(Berthouly-Salazar et al., 2013). Although our simulations may not be truly realistic by setting the source of LDD after the first wave, it is important to acknowledge that the exact
location and exact settlement time of this source does not matter here as this source only acts as a reservoir of non-admixed migrants. Other factors like a high ratio of local to invader
density or different interbreeding levels contribute more to final introgression levels (Currat et al., 2008). The invasion of human populations into already occupied territories has led to
a varying degree of admixture throughout the different parts of the world. For instance, the extent of Amerindian admixture in individuals of European descent is usually higher in South
America than in North America (Wang et al., 2008; Halder et al., 2009; Lao et al., 2010; Lisabeth et al., 2011; Galanter et al. 2012; Ruiz-Linares et al., 2014; Salzano and Sans, 2014; Bryc
et al., 2015; Homburger et al., 2015; Salazar-Flores et al., 2015). Our simulation results suggest that, among the various evolutionary, demographic and cultural mechanisms that may explain
these varying levels of introgression, different modes of migration could also have an important role in such cases. In fact, the comparably efficient transportation system along rivers,
roads, and, later, along railroads, facilitated migrations over large distances in the United States since early stages of colonialism, such that more than 30% of settlers were European-born
in some areas (Meinig, 1993). Earlier studies suggested that admixture levels in South America varied according to local Amerindian population density (Wang et al., 2008; Rubi-Castellanos
et al., 2009), and pre-Colombian Amerindian population densities in North America were lower than those in Central or South America (Lange et al., 2006). Interbreeding in South America was
mainly between European men and Native women resulting in a higher introgression of Amerindian alleles on the X-chromosome than the Y-chromosome (Bedoya et al., 2006; Bryc et al., 2010).
Although mainly male colonialists arrived in South America, the situation was different in North America, where whole European families migrated together (Meinig 1986, 1993). Strong
assortative mating in North America and lower population density could thus explain the absence of introgression. However, even with an interbreeding success rate of only 4% (_γ_=0.04), we
would expect to see ~80% introgression after a range expansion without LDD (Figure 3). The absence of introgression of local alleles in North Americans thus suggests that this could be at
least in part a result of the mode of colonization where many pioneers arrived directly to the front from Europe. We notice that our modeling of assortative mating may not capture all the
complex aspects of admixture in humans. However, what we show is that even if there was a very strong disassortative mating or if hybrids were strongly rejected (which is captured by setting
_γ_ to 2%), one would still expect more than 30% of introgression in absence of LDD, and still more than 10% when generalized LDD is present (Figure 3). Allowing LDD to occur directly
between the source and the wave front, as in our front LDD model, reduces this introgression rate dramatically: less than 1% introgression when only 1% of the individuals of the front come
directly from the source with low levels of interbreeding (_γ_=2%). This implies that either extremely low levels of disassortative mating were tolerated (_γ_<<2%), or that some levels
of direct gene flow from the source to the front indeed occurred. Since the latter is documented (Meinig, 1993), it is likely that current very low Amerindian introgression levels are
partially due to the migratory behavior of Europeans during the colonization of North America. It appears difficult to estimate the fraction of LDD events in a given species (Nathan et al.,
2003), but our simulations of a wide range of _δ_ values (from 0.01 up to 10%) may be suitable for explaining the observed genetic diversity of a variety of different organisms. Although
generalized LDD may be realistic for many species, direct LDD from source to the front probably only occurs in humans or in highly mobile species like birds and some plants for which there
may be seed dispersal by wind. In any case, it is remarkable that _ɛ_ values lower than 1% already have a marked effect on introgression, which shows that the establishment of even very few
non-admixed individuals at the wave front can thus have a large impact on final introgression levels. DATA ARCHIVING This article does not report new empirical data or software. The input
files as well as the executables necessary to perform the simulations are available through the Dryad electronic repository (http://datadryad.org/) under the following DOI:
10.5061/dryad.t281k. REFERENCES * Alves I, Arenas M, Currat M, Sramkova-Hanulova A, Sousa VC, Ray N _et al_. (2016). Long-distance dispersal shaped patterns of human genetic diversity in
Eurasia. _Mol Biol Evol_ 33: 946–958. Article CAS PubMed Google Scholar * Barton N, Bengtsson BO . (1986). The barrier to genetic exchange between hybridising populations. _Heredity_ 57:
357–376. Article PubMed Google Scholar * Barton NH . (2001). The role of hybridization in evolution. _Mol Ecol_ 10: 551–568. Article CAS PubMed Google Scholar * Bedoya G, Montoya P,
García J, Soto I, Bourgeois S, Carvajal L _et al_. (2006). Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate. _Proc Natl Acad
Sci USA_ 103: 7234–7239. Article CAS PubMed PubMed Central Google Scholar * Berthouly-Salazar C, Hui C, Blackburn TM, Gaboriaud C, van Rensburg BJ, van Vuuren BJ _et al_. (2013).
Long-distance dispersal maximizes evolutionary potential during rapid geographic range expansion. _Mol Ecol_ 22: 5793–5804. Article PubMed Google Scholar * Bialozyt R, Ziegenhagen B,
Petit RJ . (2006). Contrasting effects of long distance seed dispersal on genetic diversity during range expansion. _J Evol Biol_ 19: 12–20. Article CAS PubMed Google Scholar * Bohrer G,
Nathan R, Volis S . (2005). Effects of long-distance dispersal for metapopulation survival and genetic structure at ecological time and spatial scales. _J Ecol_ 93: 1029–1040. Article
Google Scholar * Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL . (2015). The genetic ancestry of African Americans, Latinos, and European Americans across the United States. _Am J
Hum Genet_ 96: 37–53. Article CAS PubMed PubMed Central Google Scholar * Bryc K, Velez C, Karafet T, Moreno-Estrada A, Reynolds A, Auton A _et al_. (2010). Genome-wide patterns of
population structure and admixture among Hispanic/Latino populations. _Proc Natl Acad Sci USA_ 107: 8954–8961. Article CAS PubMed PubMed Central Google Scholar * Currat M, Excoffier L .
(2004). Modern humans did not admix with Neanderthals during their range expansion into Europe. _Plos Biol_ 2: 2264–2274. Article CAS Google Scholar * Currat M, Excoffier L . (2011).
Strong reproductive isolation between humans and Neanderthals inferred from observed patterns of introgression. _Proc Natl Acad Sci USA_ 108: 15129–15134. Article CAS PubMed PubMed
Central Google Scholar * Currat M, Excoffier L, Maddison W, Otto SP, Ray N, Whitlock MC _et al_. (2006). Comment on ‘ongoing adaptive evolution of ASPM, a brain size determinant in _Homo
sapiens_’ and ‘microcephalin, a gene regulating brain size, continues to evolve adaptively in humans’. _Science_ 313: 172. Article CAS PubMed Google Scholar * Currat M, Ray N, Excoffier
L . (2004). SPLATCHE: a program to simulate genetic diversity taking into account environmental heterogeneity. _Mol Ecol Notes_ 4: 139–142. Article Google Scholar * Currat M, Ruedi M,
Petit RJ, Excoffier L . (2008). The hidden side of invasions: Massive introgression by local genes. _Evolution_ 62: 1908–1920. PubMed Google Scholar * Excoffier L, Foll M, Petit RJ .
(2009). Genetic consequences of range expansions. _Annu Rev Ecol Evol Syst_ 40: 481–501. Article Google Scholar * Excoffier L, Ray N . (2008). Surfing during population expansions promotes
genetic revolutions and structuration. _Trends Ecol Evol_ 23: 347–351. Article PubMed Google Scholar * Fayard J, Klein EK, Lefevre F . (2009). Long distance dispersal and the fate of a
gene from the colonization front. _J Evol Biol_ 22: 2171–2182. Article CAS PubMed Google Scholar * Galanter JM, Fernandez-Lopez JC, Gignoux CR, Barnholtz-Sloan J, Fernandez-Rozadilla C,
Via M _et al_. (2012). Development of a panel of genome-wide ancestry informative markers to study admixture throughout the Americas. _PLoS Genet_ 8: e1002554. Article CAS PubMed PubMed
Central Google Scholar * Goodsman DW, Cooke B, Coltman DW, Lewis MA . (2014). The genetic signature of rapid range expansions: how dispersal, growth and invasion speed impact
heterozygosity and allele surfing. _Theor Popul Biol_ 98: 1–10. Article PubMed Google Scholar * Halder I, Yang BZ, Kranzler HR, Stein MB, Shriver MD, Gelernter J . (2009). Measurement of
admixture proportions and description of admixture structure in different US populations. _Hum Mutat_ 30: 1299–1309. Article CAS PubMed PubMed Central Google Scholar * Hallatschek O,
Hersen P, Ramanathan S, Nelson DR . (2007). Genetic drift at expanding frontiers promotes gene segregation. _Proc Natl Acad Sci USA_ 104: 19926–19930. Article CAS PubMed PubMed Central
Google Scholar * Hellenthal G, Busby GB, Band G, Wilson JF, Capelli C, Falush D _et al_. (2014). A genetic atlas of human admixture history. _Science_ 343: 747–751. Article CAS PubMed
PubMed Central Google Scholar * Homburger JR, Moreno-Estrada A, Gignoux CR, Nelson D, Sanchez E, Ortiz-Tello P _et al_. (2015). Genomic Insights into the ancestry and demographic history
of South America. _PLoS Genet_ 11: e1005602. Article PubMed PubMed Central Google Scholar * Ibrahim KM, Nichols RA, Hewitt GM . (1996). Spatial patterns of genetic variation generated by
different forms of dispersal during range expansion. _Heredity_ 77: 282–291. Article Google Scholar * Jobling MA . (2012). The impact of recent events on human genetic diversity. _Philos
Trans R Soc Lond B Biol Sci_ 367: 793–799. Article CAS PubMed PubMed Central Google Scholar * Klopfstein S, Currat M, Excoffier L . (2006). The fate of mutations surfing on the wave of
a range expansion. _Mol Biol Evol_ 23: 482–490. Article CAS PubMed Google Scholar * Lange M, Mahoney J, vom Hau M . (2006). Colonialism and development: a comparative analysis of Spanish
and British colonies. _Am J Sociology_ 111: 1412–1462. Article Google Scholar * Lao O, Vallone PM, Coble MD, Diegoli TM, van Oven M, van der Gaag KJ _et al_. (2010). Evaluating
self-declared ancestry of US Americans with autosomal, Y-chromosomal and mitochondrial DNA. _Hum Mutat_ 31: E1875–E1893. Article PubMed PubMed Central Google Scholar * Lisabeth LD,
Morgenstern LB, Burke DT, Sun YV, Long JC . (2011). Ancestral heterogeneity in a biethnic stroke population. _Ann Hum Genet_ 75: 508–515. Article CAS PubMed PubMed Central Google Scholar
* Meinig DW . (1986) _The Shaping of America: A Geographical Perspective on 500 Years of History, Volume 1: Atlantic America_. New Haven and London: Yale University Press, 1492–1800.
Google Scholar * Meinig DW . (1993) _The Shaping of America: A Geographical Perspective on 500 Years of History, Volume 2: Continental America_. New Haven and London: Yale University Press,
1800–1867. Google Scholar * Mulligan CJ, Hunley K, Cole S, Long JC . (2004). Population genetics, history, and health patterns in native Americans. _Annu Rev Genom Hum Genet_ 5: 295–315.
Article CAS Google Scholar * Nathan R, Perry G, Cronin JT, Strand AE, Cain ML . (2003). Methods for estimating long-distance dispersal. _Oikos_ 103: 261–273. Article Google Scholar *
Nichols RA, Hewitt GM . (1994). The genetic consequences of long distance dispersal during colonization. _Heredity_ 72: 312–317. Article Google Scholar * Petit RJ, Excoffier L . (2009).
Gene flow and species delimitation. _Trends Ecol Evol_ 24: 386–393. Article PubMed Google Scholar * Ray N, Currat M, Foll M, Excoffier L . (2010). SPLATCHE2: a spatially explicit
simulation framework for complex demography, genetic admixture and recombination. _Bioinformatics_ 26: 2993–2994. Article CAS PubMed Google Scholar * Ray N, Excoffier L . (2010). A first
step towards inferring levels of long-distance dispersal during past expansions. _Mol Ecol Resour_ 10: 902–914. Article PubMed Google Scholar * Rubi-Castellanos R, Martinez-Cortes G,
Munoz-Valle JF, Gonzalez-Martin A, Cercla-Flores RM, Anaya-Palafox M _et al_. (2009). Pre-Hispanic Mesoamerican demography approximates the present-day ancestry of Mestizos throughout the
territory of Mexico. _Am J Phys Anthropol_ 139: 284–294. Article PubMed Google Scholar * Ruiz-Linares A, Adhikari K, Acuña-Alonzo V, Quinto-Sanchez M, Jaramillo C, Arias W _et al_.
(2014). Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. _PLoS Genet_ 10: e1004572. Article PubMed PubMed
Central Google Scholar * Salazar-Flores J, Zuñiga-Chiquette F, Rubi-Castellanos R, Álvarez-Miranda JL, Zetina-Hérnandez A, Martínez-Sevilla VM _et al_. (2015). Admixture and genetic
relationships of Mexican Mestizos regarding Latin American and Caribbean populations based on 13 CODIS-STRs. _Homo_ 66: 44–59. Article CAS PubMed Google Scholar * Salzano FM, Sans M .
(2014). Interethnic admixture and the evolution of Latin American populations. _Genet Mol Biol_ 27: 151–170. Article Google Scholar * Sankararaman S, Mallick S, Dannemann M, Prüfer K,
Kelso J, Pääbo S _et al_. (2014). The genomic landscape of Neanderthal ancestry in present-day humans. _Nature_ 507: 354–357. Article CAS PubMed PubMed Central Google Scholar * Wang S,
Ray N, Rojas W, Parra MV, Bedoya G, Gallo C _et al_. (2008). Geographic patterns of genome admixture in Latin American mestizos. _Plos Genet_ 4: e1000037. Article PubMed PubMed Central
Google Scholar Download references ACKNOWLEDGEMENTS We thank Miguel Arenas-Busto and Mathias Currat for SPLATCHE support and discussions. We are grateful to Jeffrey Long for his useful
comments on admixture in North America and to Joshua Schraiber and two other anonymous reviewers for their valuable comments on the manuscript. This work was supported by the Swiss National
Science Foundation (grants No 3100-126074 and 310030B-166605 to LE). CEGA is a fellow of the Science Without Borders program from CAPES foundation (BEX 8279/11-0) and CNPq, Brazil (PDE
201145/2015-4). AUTHOR INFORMATION Author notes * C E G Amorim and T Hofer: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Biological Sciences,
Columbia University, New York, NY, USA C E G Amorim * CAPES Foundation, Ministry of Education of Brazil, Brasília, Distrito Federal, Brazil C E G Amorim * Computational and Molecular
Population Genetics Lab, Institute of Ecology and Evolution, University of Bern, Bern, Switzerland T Hofer & L Excoffier * Swiss Institute of Bioinformatics, Lausanne, Switzerland T
Hofer & L Excoffier * EnviroSPACE Laboratory, Institute for Environmental Sciences, University of Geneva, Geneva, Switzerland N Ray * Genetic Cancer Susceptibility Group, International
Agency for Research on Cancer, Lyon, France M Foll * Department of Genetics, Evolution and Environment, University College London, London, UK A Ruiz-Linares Authors * C E G Amorim View
author publications You can also search for this author inPubMed Google Scholar * T Hofer View author publications You can also search for this author inPubMed Google Scholar * N Ray View
author publications You can also search for this author inPubMed Google Scholar * M Foll View author publications You can also search for this author inPubMed Google Scholar * A Ruiz-Linares
View author publications You can also search for this author inPubMed Google Scholar * L Excoffier View author publications You can also search for this author inPubMed Google Scholar
CORRESPONDING AUTHOR Correspondence to C E G Amorim. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information
accompanies this paper on Heredity website SUPPLEMENTARY INFORMATION SUPPLEMENTARY FIGURE S1 (JPG 426 KB) SUPPLEMENTARY FIGURE S2 (JPG 3903 KB) SUPPLEMENTARY FIGURE S3 (JPG 2880 KB) RIGHTS
AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Amorim, C., Hofer, T., Ray, N. _et al._ Long-distance dispersal suppresses introgression of local alleles during
range expansions. _Heredity_ 118, 135–142 (2017). https://doi.org/10.1038/hdy.2016.68 Download citation * Received: 14 January 2016 * Revised: 15 July 2016 * Accepted: 18 July 2016 *
Published: 31 August 2016 * Issue Date: February 2017 * DOI: https://doi.org/10.1038/hdy.2016.68 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative