- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Myosin light-chain kinase (_MYLK_) is a gene known to be significantly associated with severe asthma in African Americans. Here we further examine the molecular function of a
single-nucleotide polymorphism (SNP), located in the non-muscle myosin light-chain kinase isoform (nmMLCK), in asthma susceptibility and pathobiology. We identified nmMLCK variant (reference
SNP: rs9840993, NM_053025: 721C>T, c.439C>T) with a distinct mRNA secondary structure from the other variants. The nmMLCK variant (721C) secondary structure exhibits increased
stability with an elongated half-life in the human endothelial cell, and greater efficiency in protein translation initiation owing to an increased accessibility to translation start site.
Finally, nmMLCK expression of 721C- and 721T-containing _MYLK_ transgenes were compared in nmMLCK−/− mice and confirmed deleterious effects of nmMLCK expression on asthmatic indices and
implicated the augmented influence of _MYLK_ 721C>T (c.439C>T) SNP on asthma severity. The confirmation of the novel mechanism of the regulation of asthmatic inflammation by a _MYLK_
advances knowledge of the genetic basis for asthma disparities, and further suggests the potential of nmMLCK as a therapeutic target. Our study suggests that in addition to altering protein
structure and function, non-synonymous SNPs may also lead to phenotypic disparity by altering protein expression. SIMILAR CONTENT BEING VIEWED BY OTHERS ALTERNATIVE SPLICING IN LUNG
INFLUENCES COVID-19 SEVERITY AND RESPIRATORY DISEASES Article Open access 04 October 2023 EXPRESSION QUANTITATIVE TRAIT LOCI FOR _ETV4_ AND _MEOX1_ ARE ASSOCIATED WITH ADULT ASTHMA IN
JAPANESE POPULATIONS Article Open access 22 September 2021 GENETIC REGULATION OF GENE EXPRESSION OF MIF FAMILY MEMBERS IN LUNG TISSUE Article Open access 12 October 2020 INTRODUCTION Within
a rising US and world-wide prevalence of asthma, there is strong evidence for ethnic disparities in asthma susceptibility and severity with greater mortality,1 more severe obstruction and
greater number of severe attacks occurring in asthmatics of African descent (AD).2 We recently explored _MYLK_, a gene encoding the Ca2+/calmodulin-dependent myosin light-chain kinase
(MLCK), as a candidate gene for asthma susceptibility3 taking advantage of extensive gene re-sequencing in samples from Europeans and African Americans.4 We identified the significant
association of an African-specific non-synonymous variant in _MYLK_ (reference single-nucleotide polymorphism (SNP): rs9840993, NM_053025: 721C>T, p.Pro147Ser or c.439C>T) with severe
asthma.3 The human _MYLK_ gene encodes three isoforms including non-muscle MLCK isoform (nmMLCK), smooth muscle isoform (smMLCK) and telokin (KRP), a small myosin filament-binding protein.
Both smMLCK and nmMLCK phosphorylate myosin light chains to regulate cellular contraction and relaxation.5 smMLCK has been well studied in the pathogenesis of asthma as a key contributor to
airway smooth muscle contractile function remodeling, characteristic of the asthmatic phenotype.6 In contrast, limited information is known about the role of the nmMLCK isoform in asthma
pathobiology. The identified asthma susceptibility _MYLK_ variant (NM_053025: 721C>T, p.Pro147Ser or c.439C>T) within the unique N terminus of nmMLCK, residing at significant distance
from the smMLCK start site at 922aa.3 Consistent with a potential role for nmMLCK in asthma pathobiology, our structure/function studies in non-muscle tissues, such as gastrointestinal
epithelium and lung vascular endothelium, have underscored a key role for nmMLCK in inflammatory responses wherein nmMLCK regulates vascular integrity (via interplay of cell contractile
forces and cell–cell/cell–matrix contacts) and leukocyte influx into lung tissues.7,8 We recently reported that protein levels of nmMLCK correlate with experimental asthma susceptibility and
severity.9 In this report, we analyzed the functionality of the SNP 721C>T (c.439C>T), which is strongly associated with severe asthma in African Americans, and identified a novel
aspect of mRNA secondary structure. MATERIALS AND METHODS We calculated RNA folding energies (Δ_G_) using the RNAfold10,11 and mfold program.12 The recombinant nmMLCK1 constructs (721T or
721C) were transfected into human endothelial cells for mRNA decay and protein expression analysis. In addition, these plasmids were used to transfect the mouse lung in a model of
OVA-induced experimental asthma. Asthmatic inflammatory parameters were analyzed as described previously.9 Detailed methods are in online methods session. RESULTS AND DISCUSSION Within the
four identified coding SNPs in nmMLCK in our previous study (Supplementary Table 1),3 rs9840993 (NM_053025: 721C>T, c.439C>T) is the only SNP significantly associated with severe
asthma (the corresponding variant information has been deposited in the LOVD 3.0 database: http://databases.lovd.nl/shared/variants/0000040191). This is also an AD-specific SNP3 with very
low minor allelic frequencies in European decent individuals. To analyze the global and local mRNA stability of each variant, we calculated RNA folding energies (Δ_G_) using both the
_RNAfold_ program in the Vienna package10,11 and _mfold_ program12 based on NCBI reference sequence NM_053025 (ancestral allele of each SNP was used to define the wild-type (WT) mRNA
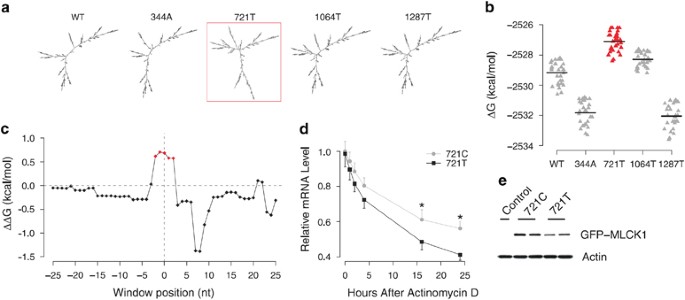
sequence). _In silico_ prediction by _RNAfold_ demonstrated that compared with other _MYLK_ variants, the 721T variant greatly changes the global mRNA secondary structure of minimum free
energy (MFE; Figure 1a and Supplementary Figure S1). The MFE of 721T is higher than all other variants (WT, 344A, 1064T and 1287T; Supplementary Figure S2). As expected, a similar trend was
obtained when the _mfold_ program is applied. We used _mfold_ to predict the top 30 optimal and suboptimal foldings of each variant. The free energy of the optimal and suboptimal foldings of
721T is significantly higher than that of all the other variants (_t_-test: _P_<10−10) (Figure 1b and Supplementary Figure S3). In comparison with MFE structure, RNA centroid structure
of the ensemble was reported to produce 30% fewer prediction errors. Here we also computed the centroid structures of WT and 721T by _RNAfold_13 and found that the folding energy of the
centroid structure of 721T is higher than that of WT. The free energy gap (ΔΔ_G_) between 721T and WT is >4 kcal/mol. These results suggest that the global structure of 721T variant is
the most unstable compared with other _MYLK_ variants and thus is more likely to be degraded.14, 15, 16 Increasing evidences suggest that reduced mRNA local stability near the translation
initiation region may lead to increased translation efficiency.17, 18, 19 Therefore, we further analyzed the mRNA local accessibility (free energy required to open local structure) around
the translation start codon using a sliding window of three nucleotides in length and one nucleotide in step. Interestingly, we found that there are five clustered windows with lower
accessibility in 721T comparing with 721C (Figure 1c). The ΔΔ_G_ between 721T and 721C is >0.5 kcal/mol for each of the five continuous windows, which can be rarely observed along the
nmMLCK mRNA (Supplementary Figure S4). We also checked the probability of observing five windows with the sum of ΔΔ_G_ larger than that of the five clustered windows around start codon by
randomly selecting the nucleotides within nmMLCK mRNA. Among the 10 000 time randomization, we failed to observe such a pattern (Supplementary Figure S5), indicating that the clustered
windows with decreased accessibility around start codon in 721T do not reflect random chance but rather suggest that the translation efficiency of 721C is potentially higher than that of
721T. To further validate our findings, we assayed and compared mRNA stability and translational efficiency of variants of 721C and 721T. EGFP-labeled nmMLCK constructs with 721C or 721T
were ectopically transfected and expressed in human endothelial cells to compare the rate of mRNA decay. Twenty four hours after the transfection, transcription was inhibited with
actinomycin D (5 _μ_g/ml), and the levels of nmMLCK-GFP transcripts at 0–24 h were determined by real-time PCR. Actinomycin D significantly reduced mRNA levels by both constructs, while the
relative mRNA level by 721C variant is significantly higher than that of 721T (Figure 1d). These observations indicate that 721C variant mediates an increased mRNA stability and reduced
intracellular decay. Protein levels of nmMLCK-GFP were determined in these ECs without actinomycin D treatment (Figure 1e). Our results demonstrated that mutating nmMLCK at position 721 from
T to C recapitulates higher expression levels of protein, indicating that protein translation is more efficient with the variant 721C. To verify the functional effects of 721C variant
(147Pro) in contributing to asthmatic susceptibility and severity, we reversely expressed human nmMLCK1 in the pulmonary endothelium of nmMLCK−/− mice utilizing the ACE antibody-tagged
liposome delivery system used to deliver MLCK expressing plasmids. This gene delivery system is consistently targeting pulmonary endothelium with to overexpress nmMLCK1 in mouse lung
endothelium (Supplementary Figure S6). nmMLCK1 overexpression in mouse lung tissues with the nmMLCK variant 721C (147Pro) exhibited higher expression efficiency than that of the 721T variant
(147Ser or WT; Figure 2a and b), consistent with the findings in computation analysis and endothelial cell _in vitro_ assays. As expected, nmMLCK1 overexpression augmented asthmatic
inflammatory parameters including airway hyper-responsiveness (Figure 2c), with the disease-associated 721C variant eliciting stronger allergic inflammation than the _MYLK_ variant 721T
(Figure 2c). Similar effects were observed in other inflammatory parameters such as airway inflammatory leukocyte infiltration (Figure 2d–f) and BAL protein leakage (Figure 2g). These data
are consistent with findings of upregulation of nmMLCK transcripts in both human or murine asthmatic subjects (Supplementary Figure S7). These studies represent the first characterization of
a biological function of a _MYLK_ SNP _in vivo_. In summary, our studies underscore the contributory role of _MYLK_ genetic variants to asthma susceptibility in populations of African
ancestry, and highlight the role of nmMLCK expression, especially in lung endothelium, to asthma pathogenesis. Although the link between _MYLK_ variants, expression of specific MLCK isoforms
and asthma appears to be certain, additional studies are required to establish a mechanistic relationship between genetic variants and functional alterations of this interesting gene
product. Moreover, although _MYLK_ variants may account for severe symptoms in African American asthmatics, further association studies, analyzing additional nearby SNPs and independent
samples are required. REFERENCES * Masoli M, Fabian D, Holt S, Beasley R : The global burden of asthma: executive summary of the GINA Dissemination Committee report. _Allergy_ 2004; 59:
469–478. Article Google Scholar * El-Ekiaby A, Brianas L, Skowronski ME _et al_. Impact of race on the severity of acute episodes of asthma and adrenergic responsiveness. _Am J Respir Crit
Care Med_ 2006; 174: 508–513. Article Google Scholar * Flores C, Ma SF, Maresso K, Ober C, Garcia JG : A variant of the myosin light chain kinase gene is associated with severe asthma in
African Americans. _Genet Epidemiol_ 2007; 31: 296–305. Article Google Scholar * Gao L, Grant A, Halder I _et al_. Novel polymorphisms in the myosin light chain kinase gene confer risk for
acute lung injury. _Am J Respir Cell Mol Biol_ 2006; 34: 487–495. Article Google Scholar * Dudek SM, Garcia JG : Cytoskeletal regulation of pulmonary vascular permeability. _J Appl
Physiol_ 2001; 91: 1487–1500. Article CAS Google Scholar * Benayoun L, Druilhe A, Dombret MC, Aubier M, Pretolani M : Airway structural alterations selectively associated with severe
asthma. _Am J Respir Crit Care Med_ 2003; 167: 1360–1368. Article Google Scholar * Clayburgh DR, Rosen S, Witkowski ED _et al_. A differentiation-dependent splice variant of myosin light
chain kinase, MLCK1, regulates epithelial tight junction permeability. _J Biol Chem_ 2004; 279: 55506–55513. Article CAS Google Scholar * Garcia JG, Verin AD, Herenyiova M, English D :
Adherent neutrophils activate endothelial myosin light chain kinase: role in transendothelial migration. _J Appl Physiol_ 1998; 84: 1817–1821. Article CAS Google Scholar * Wang T,
Moreno-Vinasco L, Ma SF _et al_. Nonmuscle myosin light chain kinase regulates murine asthmatic inflammation. _Am J Respir Cell Mol Biol_ 2014; 50: 1129–1135. Article Google Scholar *
Hofacker IL, Stadler PF : Memory efficient folding algorithms for circular RNA secondary structures. _Bioinformatics_ 2006; 22: 1172–1176. Article CAS Google Scholar * Hofacker IL,
Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P : Fast folding and comparison of RNA secondary structures. _Monatshefte für Chemie_ 1994; 125: 167–188. Article CAS Google
Scholar * Zuker M, David H, Mathews DH, Turner DH . Algorithms and Thermodynamics for RNA Secondary Structure Prediction: a Practical Guide. _RNA Biochemistry and Biotechnology_. Dordrecht,
NL, USA: Kluwer Academic Publishers, 1999, pp 11–43. Chapter Google Scholar * Ding Y, Chan CY, Lawrence CE : RNA secondary structure prediction by centroids in a Boltzmann weighted
ensemble. _RNA_ 2005; 11: 1157–1166. Article CAS Google Scholar * Nackley AG, Shabalina SA, Tchivileva IE _et al_. Human catechol-O-methyltransferase haplotypes modulate protein
expression by altering mRNA secondary structure. _Science_ 2006; 314: 1930–1933. Article CAS Google Scholar * Chamary JV, Hurst LD : Evidence for selection on synonymous mutations
affecting stability of mRNA secondary structure in mammals. _Genome Biol_ 2005; 6: R75. Article CAS Google Scholar * Qiu L, Moreira A, Kaplan G _et al_. Degradation of hammerhead
ribozymes by human ribonucleases. _Mol Gen Genet_ 1998; 258: 352–362. Article CAS Google Scholar * Gingold H, Pilpel Y : Determinants of translation efficiency and accuracy. _Mol Syst
Biol_ 2011; 7: 481. Article Google Scholar * Kudla G, Murray AW, Tollervey D, Plotkin JB : Coding-sequence determinants of gene expression in Escherichia coli. _Science_ 2009; 324:
255–258. Article CAS Google Scholar * Gu W, Zhou T, Wilke CO : A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. _PLoS Comput
Biol_ 2010; 6: e1000664. Article Google Scholar Download references ACKNOWLEDGEMENTS We are grateful to the superb technical support on the liposome preparation by Drs Yulia Epshtein and
Alicia Rizzo. We also thank Dr Julian Solway for providing human lung biopsy samples. This study is supported National Institute of Health grants HL 91899 (JGNG) and HL 58064 (JGNG). AUTHOR
INFORMATION Author notes * Ting Wang and Tong Zhou: These authors contributed equally to this work. AUTHORS AND AFFILIATIONS * Arizona Respiratory Center and Department of Medicine,
University of Arizona, Tucson, AZ, USA Ting Wang, Tong Zhou, Laleh Saadat & Joe GN Garcia Authors * Ting Wang View author publications You can also search for this author inPubMed Google
Scholar * Tong Zhou View author publications You can also search for this author inPubMed Google Scholar * Laleh Saadat View author publications You can also search for this author inPubMed
Google Scholar * Joe GN Garcia View author publications You can also search for this author inPubMed Google Scholar CORRESPONDING AUTHOR Correspondence to Joe GN Garcia. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no conflict of interest. ADDITIONAL INFORMATION Supplementary Information accompanies this paper on European Journal of Human Genetics website
SUPPLEMENTARY INFORMATION SUPPLEMENTARY METHODS (DOC 58 KB) SUPPLEMENTARY INFORMATION (PDF 967 KB) RIGHTS AND PERMISSIONS This work is licensed under a Creative Commons Attribution 3.0
Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the
material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit
http://creativecommons.org/licenses/by/3.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Wang, T., Zhou, T., Saadat, L. _et al._ A MYLK variant regulates asthmatic
inflammation via alterations in mRNA secondary structure. _Eur J Hum Genet_ 23, 874–876 (2015). https://doi.org/10.1038/ejhg.2014.201 Download citation * Received: 17 March 2014 * Revised:
20 August 2014 * Accepted: 26 August 2014 * Published: 01 October 2014 * Issue Date: June 2015 * DOI: https://doi.org/10.1038/ejhg.2014.201 SHARE THIS ARTICLE Anyone you share the following
link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature
SharedIt content-sharing initiative





:max_bytes(150000):strip_icc():focal(319x0:321x2)/people_social_image-60e0c8af9eb14624a5b55f2c29dbe25b.png)