- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT White Spot Syndrome Virus (WSSV) is regarded as a virus with the strongest pathogenicity to shrimp. For the threshold trait such as disease resistance, marker assisted selection
(MAS) was considered to be a more effective approach. In the present study, association analyses of single nucleotide polymorphisms (SNPs) located in a set of immune related genes were
conducted to identify markers associated with WSSV resistance. SNPs were detected by bioinformatics analysis on RNA sequencing data generated by Illimina sequencing platform and Roche 454
sequencing technology. A total of 681 SNPs located in the exons of immune related genes were selected as candidate SNPs. Among these SNPs, 77 loci were genotyped in WSSV susceptible group
and resistant group. Association analysis was performed based on logistic regression method under an additive and dominance model in GenABEL package. As a result, five SNPs showed
associations with WSSV resistance at a significant level of 0.05. Besides, SNP-SNP interaction analysis was conducted. The combination of SNP loci in _TRAF6, Cu/Zn SOD_ and _nLvALF2_
exhibited a significant effect on the WSSV resistance of shrimp. Gene expression analysis revealed that these SNPs might influence the expression of these immune-related genes. This study
provides a useful method for performing MAS in shrimp. SIMILAR CONTENT BEING VIEWED BY OTHERS WHOLE-GENOME ASSOCIATION STUDY SEARCHING FOR QTL FOR _AEROMONAS SALMONICIDA_ RESISTANCE IN
RAINBOW TROUT Article Open access 08 September 2021 A MAJOR QUANTITATIVE TRAIT LOCUS AFFECTING RESISTANCE TO TILAPIA LAKE VIRUS IN FARMED NILE TILAPIA (_OREOCHROMIS NILOTICUS_) Article Open
access 14 July 2021 GENOMIC SELECTION FOR WHITE SPOT SYNDROME VIRUS RESISTANCE IN WHITELEG SHRIMP BOOSTS SURVIVAL UNDER AN EXPERIMENTAL CHALLENGE TEST Article Open access 25 November 2020
INTRODUCTION The Pacific white shrimp _Litopenaeus vannamei (L. vannamei_) is a world-wide aquaculture species. After it was firstly introduced to China in 1988, its production has increased
rapidly, and it has become the major cultured shrimp species with an annual output value of 7.4 billion US dollars. However, the huge profit is accompanied by multiple challenges like
environmental degradation and various shrimp diseases. White spot syndrome virus (WSSV) is regarded as the most severe virus for its wide spread ability and high lethality1. Up to now, there
is no effective method to prevent the shrimp from WSSV infection. Breeding of WSSV-resistant varieties should be the most effective method to solve the virus disease problem. As a threshold
trait, disease-resistance appears to have low heritability, which is easily influenced by external environments. Marker assisted selection (MAS) is considered to be an effective method in
breeding for a complex trait, especially for disease resistance. In order to conduct MAS, markers related to disease-resistance is needed. Association analysis is regarded as a powerful tool
for trait-related marker screening2,3. Through this approach, multiple quantitative trait loci (QTLs) have been successfully identified in some species4,5,6. Genome-wide association study
(GWAS) and candidate gene association study are two powerful strategies for association analyses. GWAS has been applied for mapping complex diseases traits in human beings, economic crops
and animals7,8. However, the genome reference sequence is necessary for GWAS. For species without whole genome sequences, the candidate gene strategy was more widely used due to its
advantages in specificity and accuracy for association analysis9. With the development of Single Nucleotide polymorphisms (SNPs) genotyping method, a large number of SNPs can be genotyped
simultaneously at a lower cost. Recently, SNPs in gene sets based on regulatory networks and gene pathways began to be used for association analyses10,11,12. Gene set based association
analyses can eliminate redundant information, improve the efficiency at a lower cost and detect associations with weak effects13,14. This method has been widely used in human diseases
analysis and economic traits studies of animals11,15,16. In shrimp, innate immune signaling pathways, including Toll pathway, IMD pathway and JAK/STAT pathway were involved in the immune
response of shrimp to bacterial or WSSV infection. The immune response of innate immunity includes pathogen recognition, signal transduction and production of effectors. A large number of
effectors including antimicrobial peptides (AMPs), antioxidant enzymes, hemocyanin _etc._ are produced to kill the pathogens directly. Both Toll pathway and IMD pathway were responsive to
bacteria and virus, and JAK/STAT pathway was involved in WSSV infection17. Therefore the key genes involved in the innate immunity were regarded as the basis for disease resistance of
shrimp. The generalized multifactor dimensionality reduction (GMDR) method was developed based on the multifactor dimensionality reduction (MDR) method to be applied in detecting the
interactions for gene-by-gene or gene-by-environment18,19,20,21. The genes used for GMDR analysis were usually from one pathway which showed similar functions11,15. It has been used for risk
assessment on cancer and other diseases in humans14,22,23. This method was recently introduced for genetic breeding of livestock and poultry11. In the present study, the SNPs in the gene
set of the signaling pathways regulating the innate immune response to WSSV infection were selected for association analysis. The GMDR method was used to further explore the influence of
gene-by-gene interactions on the WSSV resistance/susceptibility of shrimp. RESULTS WSSV COPY NUMBERS IN WSSV-SUSCEPTIBLE AND WSSV-RESISTANT GROUPS Before WSSV infection, the average viral
load of WSSV per ng DNA of shrimp from the experimental population was 7.46 copies/ng, so the experimental shrimp could be regarded as WSSV free. At 3 days after WSSV infection, the average
viral load in dead shrimp in shrimp reached 1.05 × 105 copies/ng DNA. After checking the virus load in shrimp from WSSV-susceptible (sus.) group or WSSV-resistant (res.) group, we found that
the viral load in shrimp from sus group was (1.286 ± 0.355) × 105 copies/ng DNA, while that in res. group was (2.793 ± 0.982) × 103 copies/ng DNA. The _P_ value from the independent t-test
between two groups was 0.001, which means that a very significant difference existed for the viral load between sus. and res. group. SNPS VALIDATION, SELECTION AND GENOTYPING Validation on
the SNPs predicted by bioinformatics analysis was shown in Table S1. The prediction accuracy on the SNPs from Illumina data was 86.5% when the quality score was set at more than 20. The
prediction accuracy for SNPs with high quality score (Q > 100) and relative low score (20 ≤ Q ≤ 100) showed no obvious difference. Meanwhile, the parameter of _duel min_ was a determinant
factor for SNPs prediction with 454 data. The prediction accuracy of SNP loci with parameter _duel min_ ≥9 reached 88.2%, while it was not accurate for loci with duel min <9. Thus SNPs
with quality score higher than 20 and duel min more than 8 were used as basic data for later SNP screening. Through bioinformatics analysis, a total of 868 loci from Illumina sequencing data
and 776 loci from 454 sequencing data, which were located in the immune related genes, were screened with the Q value higher than 20 for Illumina data and _duel min_ higher than 9 for 454
sequencing data. After blasting the related cDNA sequence of these unigenes to the assembled genome sequences established in our lab, 681 loci (593 from Illumina data, 88 from 454 data) with
high quality were obtained for further SNPs genotyping analyses in sus. and res. group. Due to the limitations of primer design for the genotyping analysis, a total of 77 SNPs from the 681
loci were chosen for the genotyping based on the SNP annotation and function. POPULATION STRATIFICATION AND ASSOCIATIONS ANALYSIS After quality control, a total of 51 SNPs were qualified for
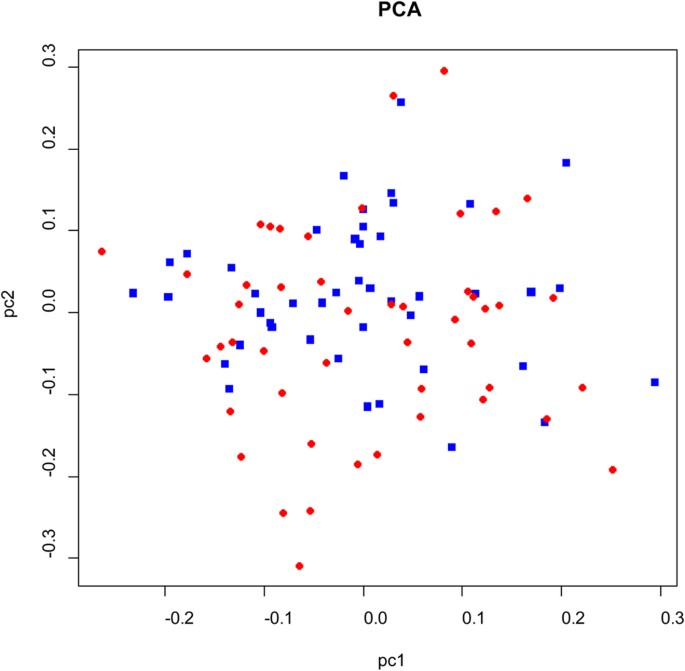
genotyping in 93 individuals of shrimp. The multidimensional scaling analysis (MDS) on these shrimp based on 51 SNPs showed that they were evenly distributed, which indicated that no
genetic stratification existed in the studied population (Fig. 1). The genome-wide degree of inflation (λ) was 1.11 and 1.18 for additive and dominance model respectively, which showed that
the structure of the studied population had a minor impact on the association analyses. Under the additive model, three SNPs located in unigene 15411 (_TLR_, sSNP), unigene 16729 (_TRAF6_,
nsSNP) and unigene 34129 (_BRAFLDRAFT_123571,_ sSNP) were associated with WSSV resistance at a significant level (P < 0.05). Under the dominance model, five SNPs located in unigene 15411
(_TLR_, sSNP), unigene 34129 (_BRAFLDRAFT_123571,_ sSNP), unigene 16729 (_TRAF6_, nsSNP), unigene 34569 (_Cu/Zn SOD_, sSNP) and unigene 30237 (_STAM_, 3′ UTR) were significantly associated
with the WSSV resistance of shrimp (P < 0.05) (Table 1, Table S3). GMDR ANALYSIS FOR SNP-SNP INTERACTION ASSOCIATED WITH WSSV RESISTANCE All loci showing associations with WSSV resistance
identified in this study and those in our previous study24,25 were used for GMDR analyses to detect the combined effects. The best interaction models from two-loci to four-loci were shown
in Table 2. Combination of three SNPs (Unigene34569, Unigene16729 and _nLvALF2_ g.2422) exhibited significant association with resistance/susceptibility of shrimp to WSSV. The interaction
among above three SNPs were shown in Fig. 2, in which the risks of shrimp to WSSV infection caused by different combinations of SNPs were displayed. Different combinations of genotypes were
classified as ‘high risk’ (hr) group and ‘low risk’ (lr) group. Individuals with low risk combinations are supposed to be more difficult to be infected than those with high risk
combinations. Therefore, shrimp with the combination of AA-TT-AG, AA-CC-AA, AG-TT-AA, AG-TT-AG, AG-CT-AG, AG-CC-AA, AG-CC-AG in _nLvALF2_ g.2422, Unigene34569, Unigene16729 were supposed to
be susceptible to WSSV, while shrimp with the combination of AA-TT-AA, AA-CT-AA, AA-CT-AG, AG-CT-AA were resistance to WSSV. VERIFICATION OF GMDR ANALYSIS Before WSSV injection, the viral
load of WSSV in shrimp were around zero, which indicated that the shrimp was WSSV free. The viral load in shrimp from the ‘high risk’ (hr) group was much higher than that in shrimp from the
‘low risk’ (lr) group at 48 hpi, However, no obvious difference was observed between these two groups at 72 hpi (Fig. 3). The expression levels of Unigene 34569 (_Cu/Zn SOD_), Unigene 16729
(_TRAF6_) and _nLvALF2_ in hr group and lr group showed significant differences (_P_ < 0.01) (Fig. 4) in shrimp without WSSV infection. During WSSV challenge, the gene expression levels
of Unigene34569 (_Cu/Zn SOD_), Unigene16729 (_TRAF6_) in lr group were higher than those in hr group at 48 h after infection, while no difference was detected for _nLvALF2_ at this time. At
72 h after WSSV infection, the expression levels of _TRAF6_ and _nLvALF2_ in lr group were higher than those in hr group, while no difference was detected for the expression level of _Cu/Zn
SOD_ between these two groups. DISCUSSION SNPs are the most abundant and widely-distributed mutations in genomes. They can be divided into coding-region SNPs (cSNPs), perigenic SNPs (pSNPs)
and intergenic SNPs (iSNPs) based on their positions in the genomes. As the SNPs may directly influence the gene translation or gene transcription, they were widely used as molecular markers
for disease genetics and pharmacogenomics studies26. The application of SNPs in association study for quantitative trait locus (QTL) mapping also have been attracting more
interests27,28,29. As enormous transcriptome data were produced by next generation sequencing, high throughput SNPs can be easily obtained by bioinformatics prediction. In our previous
study, a total of 96,040 SNPs were predicted from two transcriptomes of _L. vannamei_. In order to reduce the false-positive rate of SNPs prediction and obtain the most accurate SNPs, we
firstly analyzed the filtering parameters for different sequencing methods in the present study. The SNPs prediction accuracy was up to more than 86% when a high Q score (for Illumina
sequencing data) and _duel min_ score (for 454 sequencing data) were set30. Shrimp immune related genes were involved in the defense of shrimp against WSSV infection31. In the present study,
77 loci located in the immune related genes were chosen as candidate SNPs for WSSV resistance analyses. The JAK/STAT signaling pathway plays key roles in the antiviral immunity17. In the
present study, one locus on unigene 30237 (_STAM_, 3′ UTR SNP) was associated with the WSSV resistance of shrimp both in additive and dominance model (P < 0.05). STAM (Signal transduction
adaptor molecule), as a key gene in the JAK/STAT signaling pathway, was directly interacted with JAK to mediate the signal transduction and promote the endocytosis of WSSV32. It was
reported that SNPs located at the 3′ UTR of the gene could alter the gene transcriptional efficiency through affecting the combination between mRNA and the regulatory elements33,34. As the
identified locus located at 3′ UTR of _STAM_, the SNP in unigene 30237 (_STAM_) which showed association with WSSV resistance might affect the transcriptional efficiency of _STAM_. The SNPs
located in unigene 16729 (_TRAF6_, nsSNP) and unigene 15411 (_TLR_, sSNP) showed significant association with WSSV-resistance (P < 0.05). TLR are the upstream factor in Toll pathway, and
_TRAF6_ can regulate the activation of NF-kB and various stress kinases35. _TRAF6_ showed up-regulation at the transcriptional level after WSSV infection, and could activate the expressions
of AMPs in _L. vannamei_36. The detected SNP in _TRAF6_ was a nonsynonymous mutation which changed the amino acid sequence. Considering the crucial function of _TRAF6_ in Toll pathway, this
nonsynonymous SNP may play important roles in shrimp to defend WSSV infection. One SNPs located in genes encoding immune effectors also showed association with WSSV-resistance. Shrimp with
allele C at locus 389 of Unigene34569 (_Cu/Zn SOD_, sSNP) showed resistance to WSSV. This SNP was a synonymous SNPs. Although the synonymous SNPs can not change the protein structure
directly, it has been proved that sSNP can affect the biological process by the following three mechanisms. One was that sSNPs and some functional nsSNPs might be in strong linkage
disequilibrium, and the second was that allele-specific differences in mRNA folding might influence the splicing, transcription and regulation. The third explanation was that the codon usage
bias can affect the protein translation and folding. As mentioned previously, multiple molecules in the humoral and cellular immune system take part in defending against WSSV. Compared to
one gene, mutations in gene pathways could explain most phenotypic variance for certain traits16,37. That’s why we chose lots of SNPs from immune gene pathways in this research. Through this
strategy, more potential genes or SNPs which might affect the WSSV-resistance could be found. In some ways, development in human researches can greatly facilitate the study in animals or
non-model species. Although thousands of SNPs were detected to be associated with various human diseases, most of them can only explain part epigenetic changes of complex disease38. As we
know, the biological processes are usually accomplished by the interactions among lots of molecules or pathways. Investigation on the contribution of functionally relevant gene-gene
(SNP-SNP) interaction and gene-environment interaction to complex diseases has been proved to be useful in many cases11,13,21,22. Statistical method including logistic regression models,
multi-loci linkage disequilibrium (LD) tests were developed to analyze a large sample size data39. Using unconditional multiplicative logistic regression method, researchers have found the
interaction of SNP genotypes in several mismatch repair genes was associated with risk for breast cancer15. MDR method was the primary tool for studying genes or SNPs interaction network for
moderate sample size data18. Different versions like OR-MDR, GMDR, MB-MDR were created for different purposes19,40,41. Based on the GMDR analysis, several SNPs as well as SNP-SNP
combination from the IGF1/FoxO signal transduction pathway were reported to be associated with economical traits in pigs12. The present study applied the GMDR method in shrimp WSSV
resistance association analysis for the first time. The loci in _LvALFs_ related to WSSV resistance of shrimp found in our previous studies were also involved24,25. In the present study, a
three-factor model consisted of loci from unigene34569, unigene16729 and _nLvALF2_ showed significant association with viral resistance. According to GMDR analysis results, lr group and hr
group were divided based on the genotyping result of above three loci. The average WSSV copy numbers in hr group was apparently higher than that in lr group at 48 h after WSSV infection,
which suggested that lr group might have higher WSSV resistance than hr group. When we compare the expression level of unigene34569, unigene16729 and _nLvALF2_ in lr group and hr group, we
found that the three genes in lr group showed higher expression levels than those in the hr group. These data indicated that the high expression of these genes might affect the replication
of WSSV. Further work needs to be done to validate the combination of SNPs related to WSSV resistance of shrimp in a larger population and investigate the feasibility of these markers in
shrimp breeding. CONCLUSION This study presents the first gene set based association analyses for WSSV resistance in _L. vannamei._ A total of 5 SNPs in these immune related genes were
detected to be associated with WSSV resistance. The marker combinations associated with WSSV resistance were also identified through SNP-SNP interaction analysis. These results were useful
for understanding the genetic basis for disease resistance in shrimp and will assist the selective breeding of shrimp with WSSV-resistance. MATERIALS AND METHODS EXPERIMENTAL ANIMALS FOR
ASSOCIATION STUDY The shrimp individuals used for association analysis were generated from multiple families of Kehai No. 1 variety, which was a new selective breeding variety in China42.
Five hundred healthy shrimp, with an average body length of 6.18 ± 1.11 cm and body weight of 3.59 ± 1.57 g, were cultured in filtered sea water at 24 °C with continuous aeration in plastic
tanks. The shrimp were reared without feeding for 2 days. Then the shrimp were fed with WSSV infected shrimp tissue (average 105–106copies/ng DNA) four times (2 times per day) to simulate
the normal infection process during shrimp culture. Every shrimp was checked to ensure that they ate shrimp tissues infected with WSSV. The shrimp which did not eat would be fed with shrimp
tissues in the second day. After the infection, the shrimp were fed with artificial food pellets twice a day. All tanks were checked daily and dead shrimps were collected and marked. On the
16th day after artificial infection, the number of live shrimp was stable. Forty eight survived individuals on 19th day were regarded as WSSV-resistant samples (res group), and forty eight
individuals died at the beginning after inoculation were regarded as WSSV-susceptible samples (sus group). All experimental materials were stored at −20 °C for DNA extraction and virus
detection. SNP VALIDATION AND SELECTION SNPs were predicted based on several transcriptomic data sets through Illumina Hiseq 2500 and Roche 454 sequencing30. According to annotations, all
SNPs in 49 unigenes predicted from Illumina data and in 7 unigenes predicted from 454 data related with JAK/STAT, TOLL and IMD pathways were picked out. SNPs with quality score higher than
20 were chosen for further analysis. In order to analyze the accuracy of predicted SNPs, we randomly selected 43 loci from Illumina data and 32 loci from 454 data for validation by PCR
method (Table S2). After validation, SNPs with high quality were selected and the genomic sequence of selected SNPs were obtained by blasting the unigenes’ cDNA sequences to the genomic
database of our lab. SNP GENOTYPING Selected SNPs located in the immune related pathways were genotyped in WSSV susceptible/resistant groups using ABI Prism SNaPshot™ Multiplex System. For
SNaPshot genotyping, the SNP must satisfy the condition that no other SNPs existed in the upstream and downstream sequence of the SNP within 150 bp. Therefore, 12 SNPs located in the genes
involved in JAK/STAT pathway, 19 SNPs in TOLL pathway related genes, 12 SNPs in IMD pathway related genes, and 34 SNPs located in other immune genes were genotyped (Table 3). The SNP
information were submitted to NCBI dbSNP with submitted SNP (ss) number from 2137123241 to 2137123317. The SNP genotyping was conducted by Shanghai MapBiotech Company Limited. For easy to
describe, the name of the unigenes used in later part represented the testing SNP locus in it. DATA ANALYSIS Quality control of genotyped SNPs was performed using “_check.marker_” function
in GenABEL package43. Those SNPs with low Minor Allele Frequency (MAF), low call rate and deviation to Hardy-Weinberg Equilibrium were dropped prior to main analysis. Multidimensional
scaling were implemented on filtered SNPs to assess the population stratification in GenABEL package. As the phenotype is binary, we performed association analysis using logistic regression
method under an additive and dominance model, the body weight of each shrimp was used as covariate. This analysis was accomplished by “_mlreg_” function in GenABEL package. The genome-wide
degree of inflation (λ) was calculated to test for any hidden substructure, and the final P-value was corrected by inflation factor. Associations were deemed statistically significant at the
5% empirical significance cutoff. In order to know the potential interactions among SNPs located in different genes, SNPs associated with WSSV resistance with statistical significance (P ≤
0.05) were chosen for further gene-gene interaction analysis. Meanwhile, SNPs located in _nLvALF1_ and _nLvALF2_ (loci _nLvALF1_ g.1370, g.1419 and _nLvALF2_ g.2422, g.2466, g.2529) which
was proved to be potentially associated with WSSV resistance in previous reports24,25 were also chosen for the interaction analysis. Through the cross-validation strategy, GMDR divided
various combinations of genotypes into ‘high’ or ‘low’ risk genotypes19. In the present case, the high risk means more susceptible to WSSV, and the best two-, three-, and four-factor (SNP)
models were given. Shrimps with ‘low’ risk genotypes were considered to be high resistant to WSSV than those with ‘high’ risk genotypes. Models were considered to be significant at _P_ <
0.05. VALIDATION OF SNPS INTERACTION MODEL Two hundred health shrimp carrying no specific pathogens were chosen to verify the effect of gene-gene interaction based on the best three-factor
model given by GMDR analysis. Firstly, we separated shrimp into two groups (100 per group), then each shrimp was labeled with different combinations of four fluorescent dyes (green, red,
blue, orange) (Fig. 5) in different part of the body. After marking, two pleopods from each shrimp were taken to extract DNA for SNP examinations. Shrimp were then reared in seawater with
continuous aeration. Genomic DNA extracted from the pleopods of each shrimp were used for SNPs genotyping by Sanger sequencing. The primers used for genotyping were shown in Table 4. All PCR
products were sent to Beijing Genomics Institute for sequencing. Shrimps with predicted best combinations were grouped as ‘low’ risk (lr) group and vice versa. There were forty individuals
in lr group and hr group, respectively. Each shrimp was injected with 104 copies of WSSV (dissolved in PBS) at the last abdominal segment (set as 0 h) following the method described
previously24. Five or six shrimp were collected at 0, 48 and 72 h after WSSV injection (hpi) from lr group and hr group, separately, and these shrimps were preserved in liquid nitrogen for
further gene expression analysis. Shrimp died during the experiment were also collected for virus check. The swimming legs were picked to extract DNA for quantification of viral load. Total
RNA was extracted from cephalothoraxes. The procedures for RNA and genomic DNA extraction were the same as those described previously24. The cDNA synthesis was performed using PrimeScript RT
Reagent kit with gDNA Eraser (TaKaRa, Japan). The virus load in each shrimp was checked according to the method described by Sun _et al_.44. Transcriptional levels of the three genes
referred in the best three-factor models, including unigene34569, Unigene16729 and _nLvALF2_ were analyzed by real-time qPCR (RT-qPCR). 18 S rRNA was used as a reference gene. Relative
expression levels were calculated by 2−ΔΔCt method. The average transcriptional level of the genes in hr group and lr group at 0 h were used as controls. All these data were analyzed by
independent samples _t-test_ with a significance level of _P_ = 0.05 using SPSS 16.0. ADDITIONAL INFORMATION HOW TO CITE THIS ARTICLE: Yu, Y. _et al_. Gene set based association analyses for
the WSSV resistance of Pacific white shrimp _Litopenaeus vannamei. Sci. Rep._ 7, 40549; doi: 10.1038/srep40549 (2017). PUBLISHER'S NOTE: Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. REFERENCES * Li, F. & Xiang, J. Recent advances in researches on the innate immunity of shrimp in China.
_Developmental and Comparative Immunology_ 39, 11–26 (2013). Article Google Scholar * Kumar, L. S. DNA markers in plant improvement: An overview. _Biotechnol Adv_ 17, 143–182 (1999).
Article ADS CAS Google Scholar * Vignal, A., Milan, D., SanCristobal, M. & Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. _Genet Sel
Evol_ 34, 275–305 (2002). Article CAS Google Scholar * Werner, J. D. Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural
flowering-time variation. _Proceedings of the National Academy of Sciences of the United States of America_ 102, 2460–2465 (2005). Article ADS CAS Google Scholar * Yu, J. & Buckler,
E. S. Genetic association mapping and genome organization of maize. _Current opinion in biotechnology_ 17, 155–160 (2006). Article CAS Google Scholar * Bryant, R. et al. Genetic variation
and association mapping of silica concentration in rice hulls using a germplasm collection. _Genetica_ 139, 1383–1398 (2011). Article CAS Google Scholar * Barrett, J. C. et al.
Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. _Nature Genetics_ 41, 703–707 (2009). Article CAS Google Scholar * Timmann, C. et
al. Genome-wide association study indicates two novel resistance loci for severe malaria. _Nature_ 489, 443–446 (2012). Article ADS CAS Google Scholar * Patnala, R., Clements, J. &
Batra, J. Candidate gene association studies: a comprehensive guide to useful in silicotools. _BMC Genetics_ 14, 39 (2013). Article CAS Google Scholar * Braun, R. & Buetow, K.
Pathways of distinction analysis: a new technique for multi-SNP analysis of GWAS data. _PLoS genetics_ 7, e1002101 (2011). Article CAS Google Scholar * Wang, M., Wang, Q. & Pan, Y.
From QTL to QTN: candidate gene set approach and a case study in porcine IGF1-FoxO pathway. _PLoS One_ 8, e53452 (2013). Article ADS CAS Google Scholar * He, T., Zhong, P. S. & Cui,
Y. A set-based association test identifies sex-specific gene sets associated with type 2 diabetes. _Frontiers in Genetics_. 5, 395 (2014). PubMed PubMed Central Google Scholar * Weng, L.
et al. SNP-based pathway enrichment analysis for genome-wide association studies. _BMC bioinformatics_ 12, 99 (2011). Article Google Scholar * Yu, Y. et al. Association of genetic variants
in tachykinins pathway genes with colorectal cancer risk. _International journal of colorectal disease_ 27, 1429–1436 (2012). Article Google Scholar * Conde, J. et al. Association of
common variants in mismatch repair genes and breast cancer susceptibility: a multigene study. _BMC cancer_ 9, 344 (2009). Article Google Scholar * Raven, L. A., Cocks, B. G., Pryce, J. E.,
Cottrell, J. J. & Hayes, B. J. Genes of the RNASE5 pathway contain SNP associate with milk production traits in dairy cattle. _Genet Sel Evol_ 45, 25 (2013). Article CAS Google
Scholar * Li, F. & Xiang, J. Signaling pathways regulating innate immune responses in shrimp. _Fish & Shellfish Immunology_ 34, 973–980 (2013). Article CAS Google Scholar *
Ritchie, M. D. et al. Multifactor-Dimensionality Reduction Reveals High-Order Interactions among Estrogen-Metabolism Genes in Sporadic Breast Cancer. _The American Journal of Human Genetics_
69, 138–147 (2001). Article CAS Google Scholar * Lou, X. Y. et al. A generalized combinatorial approach for detecting gene-by-gene and gene-by-environment interactions with application
to nicotine dependence. _American journal of human genetics_ 80, 1125–1137 (2007). Article CAS Google Scholar * Lee, J. H. et al. Genetic interactions model among Eotaxin gene
polymorphisms in asthma. _Journal of human genetics_ 53, 867–875 (2008). Article CAS Google Scholar * Chen, G. B. et al. Practical and theoretical considerations in study design for
detecting gene-gene interactions using MDR and GMDR approaches. _PLoS One_ 6, e16981 (2011). Article ADS CAS Google Scholar * Onay, V. U. et al. SNP-SNP interactions in breast cancer
susceptibility. _BMC cancer_ 6, 114 (2006). Article Google Scholar * Suyama, Y. et al. Effects of six functional SNPs on the urinary 8-isoprostane level in a general Japanese population;
Shimane COHRE Study. _Disease markers_ 30, 291–298 (2011). Article CAS Google Scholar * Liu, J., Yu, Y., Li, F., Zhang, X. & Xiang, J. A new anti-lipopolysaccharide factor (ALF) gene
with its SNP polymorphisms related to WSSV-resistance of _Litopenaeus vannamei_ . _Fish Shellfish Immunol_ 39, 24–33 (2014). Article CAS Google Scholar * Liu, J., Yu, Y., Li, F., Zhang,
X. & Xiang, J. A new ALF from _Litopenaeus vannamei_ and its SNPs related to WSSV resistance. _Chin J Oceanol Limn_ 32, 1232–1247 (2014). Article CAS Google Scholar * Ragoussis, J.
Genotyping technologies for genetic research. _Annu Rev Genomics Hum Genet_ 10, 117–133 (2009). Article CAS Google Scholar * Rafalski, J. A. Novel genetic mapping tools in plants: SNPs
and LD-based approaches. _Plant Science_ 162, 329–333 (2002). Article CAS Google Scholar * Gupta, P., Rustgi, S. & Kulwal, P. Linkage disequilibrium and association studies in higher
plants: Present status and future prospects. _Plant Mol Biol_ 57, 461–485 (2005). Article CAS Google Scholar * Duran, C. et al. AutoSNPdb: an annotated single nucleotide polymorphism
database for crop plants. _Nucleic Acids Research_ 37, 951–953 (2009). Article Google Scholar * Yu, Y. et al. SNP Discovery in the Transcriptome of White Pacific Shrimp _Litopenaeus
vannamei_ by Next Generation Sequencing. _PLoS One_ 9, e87218 (2014). Article ADS Google Scholar * Chen, Y., Li, X. & He, J. Recent Advances in Researches on Shrimp Immune Pathway
Involved in White Spot Syndrome Virus Genes Regulation. _Journal of Aquaculture Research & Development_ 5, 228 (2014). Google Scholar * Lohi, O. & Lehto, V.-P. STAM/EAST/Hbp adapter
proteins – integrators of signalling pathways. _FEBS letters_ 508, 287–290 (2001). Article CAS Google Scholar * Moor, C. H., Meijer, H. & Lissenden, S. Mechanisms of translational
control by the 3′ UTR in development and differentiation. _Seminars in Cell & Developmental Biology_ 16, 49–58 (2005). Article Google Scholar * Nicoloso, M. S. et al. Single-nucleotide
polymorphisms inside microRNA target sites influence tumor susceptibility. _Cancer Research_ 70, 2789–2798 (2010). Article CAS Google Scholar * Takeuchi, O. & Akira, S. Pattern
Recognition Receptors and Inflammation. _Cell_ 140, 805–820 (2010). Article CAS Google Scholar * Wang, P. et al. _Litopenaeus vannamei_ tumor necrosis factor receptor-associated factor 6
(_TRAF6_) responds to Vibrio alginolyticus and white spot syndrome virus (WSSV) infection and activates antimicrobial peptide genes. _Dev Comp Immunol_ 35, 105–114 (2011). Article Google
Scholar * Lu, Y. et al. Polymorphisms in Wnt signaling pathway genes are significantly associated with chicken carcass traits. _Poultry science_ 91, 1299–1307 (2012). Article ADS CAS
Google Scholar * Frazer, K. A., Murray, S. S., Schork, N. J. & Topol, E. J. Human genetic variation and its contribution to complex traits. _Nat Rev Genet_ 10, 241–251 (2009). Article
CAS Google Scholar * Moore, J. H. & Williams, S. M. New strategies for identifying gene-gene interactions in hypertension. _Annals of medicine_ 34, 88–95 (2002). Article CAS Google
Scholar * Chung, Y., Lee, S. Y., Elston, R. C. & Park, T. Odds ratio based multifactor-dimensionality reduction method for detecting gene-gene interactions. _Bioinformatics_ 23, 71–76
(2007). Article CAS Google Scholar * Calle, M. L., Urrea, V., Malats, N. & Van, S. K. mbmdr: an R package for exploring gene–gene interactions associated with binary or quantitative
traits. _Bioinformatics_ 26, 2198–2199 (2010). Article CAS Google Scholar * Yu, Y. et al. Molecular markers for identifying a new selected variety of Pacific white shrimp _Litopenaeus
vannamei_ . _Chin J Oceanol Limn_ 33, 1–10 (2015). Article Google Scholar * Aulchenko, Y. S., Ripke, S., Isaacs, A. & van Duijn, C. M. GenABEL: an R package for genome-wide association
analysis. _Bioinformatics._ 23, 1294–6 (2007). Article CAS Google Scholar * Sun, Y., Li, F. & Xiang, J. Analysis on the dynamic changes of the amount of WSSV in Chinese shrimp
_Fenneropenaeus chinensis_ during infection. _Aquaculture_ 376, 124–132 (2013). Article Google Scholar Download references ACKNOWLEDGEMENTS This work is supported by National Natural
Science Foundation of China (Grant No. 31502161 and 31272683), China Agriculture Research System-47 (CARS-47), and the Scientific and Technological Innovation Project Financially Supported
by Qingdao National Laboratory for Marine Science and Technology (No. 2015ASKJ02). AUTHOR INFORMATION Author notes * Yang Yu and Jingwen Liu: These authors contributed equally to this work.
AUTHORS AND AFFILIATIONS * Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, 266071, China Yang Yu, Jingwen Liu, Fuhua Li, Xiaojun
Zhang, Chengsong Zhang & Jianhai Xiang * University of Chinese Academy of Sciences, Beijing, 100049, China Jingwen Liu * Laboratory for Marine Biology and Biotechnology, Qingdao
National Laboratory for Marine Science and Technology, Qingdao 266237, China. , Fuhua Li & Jianhai Xiang Authors * Yang Yu View author publications You can also search for this author
inPubMed Google Scholar * Jingwen Liu View author publications You can also search for this author inPubMed Google Scholar * Fuhua Li View author publications You can also search for this
author inPubMed Google Scholar * Xiaojun Zhang View author publications You can also search for this author inPubMed Google Scholar * Chengsong Zhang View author publications You can also
search for this author inPubMed Google Scholar * Jianhai Xiang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Y.Y. and J.L. conducted the
experiment and data processing. J.X. and F.L. conceived and supervised the project. X.Z. contributed to prepare the genomic DNA for SNP genotyping. C.Z. prepare and cultured the experimental
animals. Y.Y., J.L. and F.L. wrote the manuscript. All authors have read and approved the manuscript. CORRESPONDING AUTHOR Correspondence to Fuhua Li. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing financial interests. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION (PDF 177 KB) RIGHTS AND PERMISSIONS This work is licensed under a Creative
Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in
the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy
of this license, visit http://creativecommons.org/licenses/by/4.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Yu, Y., Liu, J., Li, F. _et al._ Gene set based association
analyses for the WSSV resistance of Pacific white shrimp _Litopenaeus vannamei_. _Sci Rep_ 7, 40549 (2017). https://doi.org/10.1038/srep40549 Download citation * Received: 04 July 2016 *
Accepted: 07 December 2016 * Published: 17 January 2017 * DOI: https://doi.org/10.1038/srep40549 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative



