- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Since initial identification in China, the widespread geographical occurrence of plasmid-mediated colistin resistance gene _mcr-1_ in _Enterobacteriaceae_ has been of great concern.
In this study, a total of 22 _Salmonella enterica_ were resistant to colistin, while only five isolates which belonged to ST34 _Salmonella enterica_ serovar Typhimurium (_S._ Typhimurium)
were _mcr-1_ positive. Four of them shared nearly identical PFGE type, although they were from different host species and diverse geographical locations. All the _mcr-1_-positive _S._
Typhimurium exhibited multi-resistant phenotypes including ampicillin, streptomycin, gentamicin, florfenicol, nalidixic acid, tetracycline, trimethoprim-sulfamethox, in addition to colistin.
The _oqxAB_ and _aac_(_6_′)_-Ib-cr_ genes were present alone or in combination in four (80.0%) and five (100%) isolates, respectively. The _mcr-1_ gene was located on a transferable IncI2
plasmid in the four genetically related strains. In the other one strain, _mcr-1_ was located on an approximately 190 kb IncHI2 plasmid. In conclusion, we report five _mcr-1-_positive _S._
Typhimurium/ST34 isolates. Both clonal expansion and horizontal transmission of IncI2-type plasmids were involved in the spread of the _mcr-1_ gene in _Salmonella enterica_ from
food-producing animals in China. There is a great need to monitor the potential dissemination of the _mcr-1_ gene. SIMILAR CONTENT BEING VIEWED BY OTHERS OCCURRENCE OF _MCR_-MEDIATED
COLISTIN RESISTANCE IN _SALMONELLA_ CLINICAL ISOLATES IN THAILAND Article Open access 08 July 2021 MULTIDRUG RESISTANCE PLASMIDS UNDERLIE CLONAL EXPANSIONS AND INTERNATIONAL SPREAD OF
_SALMONELLA ENTERICA_ SEROTYPE 1,4,[5],12:I:- ST34 IN SOUTHEAST ASIA Article Open access 03 October 2023 WHOLE GENOME SEQUENCE ANALYSIS OF THE FIRST REPORTED ISOLATE OF _SALMONELLA_ AGONA
CARRYING BLACTX-M-55 GENE IN BRAZIL Article Open access 09 February 2023 INTRODUCTION Salmonellosis is one of important global public health zoonoses, causing life-threatening infections.
Each year, there are an estimated 1.0 million _Salmonella_ infections in the United States1. By contrast, 30 millions of infections every year in China, approximately 75% of the food-borne
diseases, are attributed to this bacterium2. Unpublished data from the China CDC surveillance system indicated that the carriage rate of human salmonellosis is 549 per 100,000 people in
2013, which is higher than that in the USA in 2012 (16.4 per 100,000)3. _Salmonella enterica_, especially non-typhoidal _Salmonella_ (NTS), is a leading cause of food-borne disease of humans
and livestock worldwide3,4. Various animal species, such as poultry, pigs, cattle, and reptiles, are reservoirs for NTS. Human NTS infections are frequently due to the consumption of
contaminated cooked or raw meat, milk, eggs, seafood, and other fresh products derived from animals5. In recently year, emerging fluoroquinolone resistance prevalence has been identified in
several _Salmonella_ serovars and the resistance rate to fluoroquinolone has increased dramatically both in clinical and food-borne _Salmonella_ isolates around the world6. Resistance toward
quinolone and fluoroquinolone antimicrobials is mainly attributed to mutations of quinolone resistance-determining regions (QRDRs) and plasmid-mediated quinolone resistance (PMQR) mechanism
including Qnr peptides (QnrA, QnrB, QnrS, QnrD and QnrC), AAC(6′)-Ib-cr and the efflux pumps QepA and OqxAB7. In our previous study, we have characterized a high prevalence of _oqxAB_
(31.7%) in _Salmonella enterica_ serotype Typhimurium (_S._ Typhimurium) isolated from food-producing animals in China. The _oqxAB_ gene was present alone or in combination with other PMQR
genes such as _aac_(_6_′)-_Ib-cr_ and _qnrS1_ genes. Interestingly, the _S._ Typhimurium isolates carrying _oqxAB_ were clonally related as determined by PFGE and also defined as ST34 by
MLST type7. A high prevalence of _oqxAB_-positive _S._ Typhimurium/ST34 was also detected in human clinical and food samples in Hong Kong in the same period8. It aroused a possibility that
the _oqxAB_-positive S. Typhimurium/ST34 transmitted from food animals to humans via food chain. More recently, _Salmonella_ species have been isolated that carried PMQR, can occur as a
multiple drug resistance phenotype, which has caused international concern because it brought an even greater challenge for clinical treatment9,10. Colistin (polymyxin E) is a cationic,
multi-component, lipopeptide antibacterial agent discovered in the 1940s with significant activity against Gram-negative bacteria. Colistin has been used both in human and veterinary
medicine for more than 50 years, although their parenteral usage in humans has been limited because of concerns about nephrotoxicity and neurotoxicity11. However, with a global increase in
Gram-negative bacteria that are multidrug-resistant (MDR), extensively drug-resistant (XDR) and pandrug-resistant (PDR), colistin has been re-introduced as a last-resort drug for infections
with such bacteria, which are frequently the cause of healthcare-associated infections12,13. In veterinary medicine, colistin is more widely used, mainly to treat Gram-negative infections of
the intestinal tract14. In addition, colistin is used as a growth promoter in some countries duo to its great growth performance in pig and poultry production14,15. Resistance to colistin
in Gram-negative bacteria has been characterized by chromosomal mutations and was generally thought non-transferable by mobile genetic elements11. Specific regions mutations like _pmrAB_ and
_phoPQ_, which were related to structural changes (ParR-ParS two-component system) of the LPS at both the cytosol and periplasmic site of the cell membrane, could decrease colistin activity
in _Klebsiella, E. coli_ and _Salmonella enterica_14. Recently, a plasmid-mediated colistin resistance gene (_mcr-1_) was firstly reported in food animals, food and humans in China15. The
_mcr-1_ gene was proved subsequently to be disseminated worldwide, mainly found in _E. coli, K. pneumonia and Salmonella spp_15,16. In China, _mcr-1_ was dominantly identified in _E. coli,
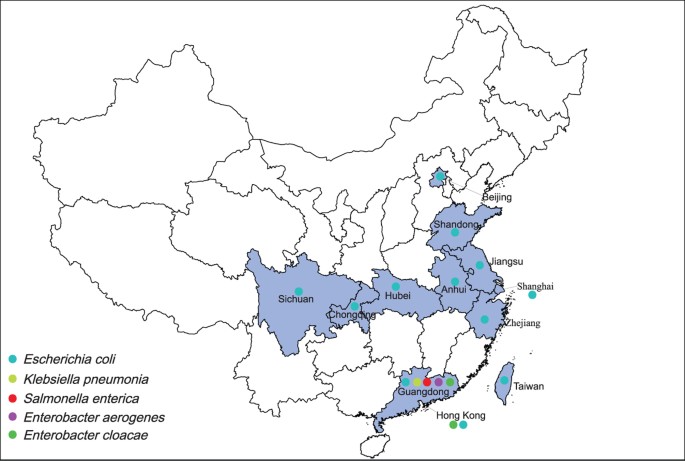
K. pneumonia, Enterobacter aerogenes_, and _Enterobacter cloacae_ in many regions (Fig. 1), mainly in Guangdong15,17,18, Shanghai15, Zhejiang, Hubei, Jiangsu19, Sichuan20, Shandong, Anhui21,
Chongqing22, Hong Kong23 and Taiwan24, whereas data on the transmission of _mcr-1_-mediated colistin resistance in _Salmonella spp._ are lacking. Although, during the preparation of this
study, the prevalence of _mcr-1_ among ESBL-positive _Salmonella spp._ isolates was investigated, but the presence of this gene in particular successful resistant clone has not been
demonstrated25. In this study, we did a retrospective study to examine the emergence of the _mcr-1_ gene in _Salmonella enterica_ isolates from food-producing animals during 2007 to 2015.
RESULTS ANTIMICROBIAL SUSCEPTIBILITY AND DETECTION OF RESISTANCE GENES These isolates showed the minimal inhibitory concentrations (MIC) for colistin of 0.25 to 16 mg/L. A total of 22
_Salmonella_ isolates were resistant to colistin with MIC ≥ 8 mg/L. The _mcr-1_ gene was detected in only five colistin-resistant _S._ Typhimurium isolates with diverse origins (Table 1).
Susceptibility testing showed that all the _mcr-1_-positive isolates in this study were resistant to nalidixic acid, olaquindox, ampicillin, streptomycin, gentamicin, florfenicol,
tetracycline and trimethoprim-sulfamethox. All the five _mcr-1-_containing strains carried _aac_(_6_′)_-Ib-cr_, and four of them also carried _oqxAB_ and _floR_ (Table 1). Interestingly, all
strains showed increased MIC values and exhibited intermediate resistance phenotype (strain GDS 79, GDS82, and GDS141) or resistance phenotype (strain S01) but except strain GDS78 retained
sensitivity to ciprofloxacin according to the cutoff of CLSI, although most of them harboured one or more PMQR genes (Table 1). TRANSFER OF _MCR-1_ GENE Four transconjugants were
successfully obtained from the five _mcr-1_-positive isolates. Conjugation and transformation tests were not successful for strain S01 despite repeated attempts. All transconjugants showed
32-fold increase in the MICs of colistin, in comparison with the recipient _E. coli_ C600 (0.125 mg/L). However, other antibiotic-resistant phenotypes could not be co-transfered with
colistin except for strain GDS78. The transconjugant of GDS78 was multidrug-resistant and showed resistance to more than five antibiotics, in addition to colistin (Table 1). PLASMID ANALYSIS
PCR-based replicon typing (PBRT) including IncHI2, IncI2, IncFIB and IncFII were detected in the original strains. While, only IncI2 was found in the three transconjugants. For GDS78T, both
IncI2 and IncFII were identified (Table 1). _S1_-PFGE and Southern blot showed that _mcr-1_ in all the four transconjugants was located in an IncI2 plasmid with size of 70 kb,
approximately. In strain S01, the _mcr-1_ was located in a round 190 kb IncHI2 plasmid (Fig. 2A). Restriction Fragment Length Polymorphism (RFLP) indicated that pattern of GDS79T was
different from GDS82T and GDS141T (Fig. 2B). After 14 days of passage without colistin, the plasmids carrying _mcr-1_ were stable both in the parent strains and transconjugants. PCR-mapping
and sequencing showed that an approximately 2,600 bp long fragment designed as the _mcr-1_ cassette26, was inserted between _nikB_ and _ydfA_ on the backbone of IncI2 plasmid in the four
transconjugants. The _mcr-1_ cassette encompasses the likely promoter sequences, the _mcr-1_ gene and a hypothetical protein26. Nevertheless, compared with pHNSHP45 (KP347127), the firstly
identified _mcr-1_-carrying IncI2 plasmid, the IS_Apl1_ is absent from the upstream of the _mcr-1_ gene (Table 2). In the original strain S01, we then confirmed that a 3,679 bp length of
_ISApl1-mcr-1_ fragment was inserted in approximately 8,500 bp downstream of _terY2_ on pHNSHP45-2 (KU341381), which is in accordance with pMR0516mcr (KX276657) (Fig. 3). MOLECULAR TYPING
All of the five isolates were successfully typed by pulsed-field gel electrophoresis (PFGE), and two different PFGE clusters designated A and B were obtained (Fig. 4). Cluster A contains
four isolates, three of which were isolated from different pig farms in 2008 and 2009, while the remaining one was isolated from duck in 2010. The single strain of cluster B was isolated
from chicken in 2007. Multi-locus sequence typing showed that the five _mcr-1_-positive _S_. Typhimurium belongs to ST34, though they were classified into two PFGE clusters (Fig. 4).
DISCUSSION Since initial identification in China, the _mcr-1_ gene has been detected in _Enterobacteria_ from almost 30 countries on five continents16. The _mcr-1_ gene was frequently
detected in _Enterobacteriaceae_, including _E. coli_15,27,28, _Enterobacter aerogenes_ and _Enterobacter cloacae_18, _Klebsiella pneumonia_15, _Shigella somnei_29, as well as _Salmonella
enterica_30. Among them, _Salmonella enterica_ has attracted much attention owing to it as an important food-borne pathogen. _Salmonella enterica_ harbours a number of serovars, but the
_mcr-1_ is not restricted to a certain serovar, which has been identified in _Salmonella enterica_ serotype Anatum, Derby, 1,4,[5],12:i:2,31 Java, Paratyphi B, Rissen, Schwartzengrund,
Typhimurium and Virchow16. Moreover, several reports confirmed the _mcr-1_-positive _Salmonella enterica_ were multidrug resistance strains, which usually carry other resistance genes
including ESBL genes and quinolone resistance genes25,31,32. More recently, the _mcr-1_ gene was found in the multidrug resistant and copper-tolerant _Salmonella spp._ from pigs33. Notably,
the _mcr-1_-positive _Salmonella_ isolates were strongly associated with the particular successful MDR clonal lineages, including _S._ 1,4,[5],12:i:/ST34 and _S._ Rissen/ST469, which had
widely spread epidemically in European countries33. Recently, _mcr-1_ was detected in MDR _S._ Typhimurium/ST34 and _S._ Typhimurium/ST36 in England and Wales32. Here, all the five
_mcr-1_-positive _Salmonella_ strains belong to _S._ Typhimurium/ST34, which is the predominant ST of _S._ Typhimurium in Guangdong, China34. In our previous study, we have characterized a
high prevalence of MDR _S._ Typhimurium/ST34 carrying PMQR8, which was also frequently detected in human clinical and food samples in Hong Kong in the same period8. The dissemination of this
clone carrying _mcr-1_ from food-producing animals to humans via food chain might expedite colistin resistance in _Salmonella_ strains. The _mcr-1_ gene was so far associated with diverse
plasmids belonging to the IncI2, IncHI2, IncP, IncX4, IncFII and IncF replicon types15,35,36,37,38,39. Among them, IncHI2 and IncX4 rather than IncI2 plasmids, which were firstly identified
to harbour the _mcr-1_ gene in _E. coli_, were the most common replicons in _Salmonella spp_10,32,36. However, the _mcr-1_ gene was located on the IncI2 plasmid in all _S._ Typhimurium/ST34
strains except strain S01 in this study, which indicated that the IncI2 plasmid harbouring the _mcr-1_ gene has circulated between _E. coli_ and _Salmonella spp_. Interestingly, the
_mcr-1_-positive IncI2 plasmid was co-occurrence with the other plasmids with diverse replicon types such as IncHI2 and IncFII in the four clonal _mcr-1_-positive strains. Unlike the
original _E. coli_ SHP45, which carried two _mcr-1_-positive plasmids including an IncI2 type pHNSHP45 and an IncHI2 type pHNSHP45-215,40, the _mcr-1_ gene was only found on IncI2 plasmid in
this study. This indicated that other mobile elements were probably involved in the mobilization of the _mcr-1_ gene, in addition to plasmids. Indeed, IS_Apl1_ was present upstream of the
_mcr-1_ gene on both _mcr-1_-carrying plasmids in _E. coli_ SHP45, while it was absent on all the _mcr-1_ positive IncI2 plasmids in this study. Only five _mcr-1_-positive strains were
identified in 276 isolates, indicating that the _mcr-1_-positive _Salmonella_ isolates are sporadic in animals in China. It is possible that the transferable _mcr-1_ gene here appears in the
form lacking the insertion sequence IS_Apl1_41. In addition, all the _mcr-1_-positive _Salmonella enterica_ were isolated from the disease animals, whereas most of the _mcr-1_-negative
colistin resistant strains were from healthy animals. Although the prescription was not recorded during the sampling, existing evidence suggested that exposure to colistin was closely
related to its resistance rates14. The high rate of colistin resistance and low _mcr-1_ positive rates showed that the plasmid-mediated colistin resistance was not the main way conferred
colistin resistance among _Salmonella_ isolates. It is of possibility that other colistin-resistant mechanisms or even novel _mcr-1_ type exisit35, which needs to be evaluated in future
study. In conclusion, we reported five _mcr-1_-positive _Salmonella_ isolates from animals in China between 2007 and 2015. Clonal spread of PMQR-carrying ST34 _Salmonella_ isolates and
horizontal transmission of IncI2 plasmids were the main way to disseminate _mcr-1_ gene in this study. Colistin should be used more prudently in food-producing animals to prevent _mcr-1_
gene spreading between different specials. MATERIALS AND METHODS BACTERIAL STRAINS A total of 276 nonduplicate _Salmonella enterica_ isolates (246 from avian and 30 from swine), isolated
from faecal swabs of healthy or sick animals at poultry farms, swine farms and two diagnostic laboratories in Guangdong and Shandong province in China among 2007 and 2015, were used in this
study (Table 3). Among them 127 were _S._ Typhimurium, 79 were _Salmonella enterica_ serotype Indiana, six were _Salmonella enterica_ serotype Enteritidis, two were _Salmonella enterica_
serotype Meleagridis, two were _Salmonella enterica_ serotype Bredeney, one was _Salmonella enterica_ serotype Abaetetuba and 59 were non-typeble. The details of these strains have been
described in our previous works7,42. SCREENING OF THE _MCR-1_ GENE AND SUSCEPTIBILITY TESTING MICs of Colistin were determined by the agar dilution method according to the guidelines of the
Clinical and Laboratory Standards Institution43,44. Colistin-resistant isolates were screened for the presence of _mcr-1_ by PCR with the primers as described by Liu _et al_.15. All the
_mcr-1_-positive isolates were detected by other resistance determinants including PMQR genes, _rmtB, armA_ and _floR_. Susceptibility testing was also assessed in the _mcr-1_-positive
isolates using the following antimicrobial agents: ampicillin (AMP), cefotaxime (CTX), cefoxitin (FOX), meropenem (MEM), streptomycin (STR), amikacin (AMK), gentamicin (GEN), florfenicol
(FFC), tetracycline (TET), nalidixic acid (NAL), ciprofloxacin (CIP), olaquindox (OQX), trimethoprim-sulfamethox (SMZ/TMP), and fosfomycin (FOS). _E. coli_ ATCC 25922 was used as a quality
control strain. CONJUGATION AND TRANSFORMATION ANALYSIS _E. coli_ C600 was used as the recipient for the conjugation experiment of MCR-producing _Salmonella_ isolates. The transconjugants
were selected on MacConkey agar containing colistin (2 mg/L) and streptomycin (2,000 mg/L), and finally confirmed by PCR and ERIC-PCR45. Plasmids that are not transferable by conjugation
were studied by transformation assay. Plasmid DNA was extracted using a QIAGEN Prep Plasmid Midi Kit. Purified plasmids were used in electroporation experiments with _E. coli_ DH5α following
the manufacturer’s instructions. Transformants were incubated at 37 °C for 1 h and were then selected on LB agar containing 2 mg/L colistin. PLASMID CHARACTERIZATION Incompatibility (Inc)
groups were assigned by PBRT. To analyze the location of the _mcr-1_ gene, _S1_ nuclease-PFGE and Southern blot analysis were performed. Briefly, whole-cell DNA of the donor strains and the
transconjugants harbouring _mcr-1_ were extracted and embedded in agarose gel plugs. Subsequently, the agarose gel plugs were treated with _S1_ nuclease (TaKaRa, Dalian, China) and the DNA
fragments were separated by PFGE. Southern blot hybridization was then performed with DNA probes specific for the _mcr-1_ gene, which was non-radioactively labeled with a DIG High Prime DNA
labeling and detection kit (Roche Diagnostics, Mannheim, Germany). Transconjugants containing one plasmid were extracted and analyzed by RFLP using _ApaLI_ (TaKaRa, Dalian, China) digestion.
The genetic context surrounding the _mcr-1_ gene was investigated by PCR mapping and sequencing according to the plasmids sequences which had been submitted to GenBank. The primers used to
determine the regions upstream and downstream of the _mcr-1_ gene are listed in Table 2. To assess the stability of _mcr-1_-positive plasmids, the original strains and transconjugants were
cultured by in daily serial passages with the absence of colistin for two weeks. MOLECULAR TYPING Genomic DNA of the _mcr-1_-positive isolates was analyzed by PFGE following digestion with
_XbaI_46. _Salmonella enterica_ serotype Braenderup H9812 standard was used as size marker. Comparison of PFGE patterns was performed by BioNumerics®v6.6 (Applied Maths, Ghent, Belgium) with
a cut-off at 90% of the similarity values to indicate identical PFGE types. Multi-locus sequence typing (MLST) was performed by using the primers and protocol specified at the _Salmonella
enterica_ MLST website (http://mlst.warwick.ac.uk/mlst/dbs/Senterica). ADDITIONAL INFORMATION HOW TO CITE THIS ARTICLE: Li, X.-P. _et al_. Clonal spread of _mcr-1_ in PMQR-carrying ST34
_Salmonella_ isolates from animals in China. _Sci. Rep._ 6, 38511; doi: 10.1038/srep38511 (2016). PUBLISHER'S NOTE: Springer Nature remains neutral with regard to jurisdictional claims
in published maps and institutional affiliations. REFERENCES * National Center for Emerging and Zoonotic Infectious Diseases. Pathogens causing US foodborne illnesses, hospitalizations, and
deaths, 2000–2008. Available at: http://www.cdc.gov/salmonella/index.html (2012). * Wu, H. et al. Prevalence of extended-spectrum β-lactamase-producing _Salmonella_ on retail chicken in six
provinces and two national cities in the People’s Republic of China. J Food Prot 76, 2040–2044 (2013). Article CAS PubMed Google Scholar * Zhang, W. H. et al. CTX-M-27 producing
_Salmonella enterica_ serotypes Typhimurium and Indiana are prevalent among food-producing animals in China. Front Microbiol 7, 436 (2016). Article PubMed PubMed Central Google Scholar *
Liang, Z. et al. Serotypes, seasonal trends, and antibiotic resistance of non-typhoidal _Salmonella_ from human patients in Guangdong Province, China, 2009–2012. BMC Infect Dis 15, 53
(2015). Article PubMed PubMed Central Google Scholar * Foley, S. L. & Lynne, A. M. Food animal-associated _Salmonella_ challenges: pathogenicity and antimicrobial resistance. J Anim
Sci 86, E173–187 (2008). Article CAS PubMed Google Scholar * Li, L. et al. Spread of _oqxAB_ in _Salmonella enterica_ serotype Typhimurium predominantly by IncHI2 plasmids. J Antimicrob
Chemother 68, 2263–2268 (2013). Article CAS PubMed Google Scholar * Wong, M. H., Chan, E. W., Liu, L. Z. & Chen, S. PMQR genes _oqxAB_ and _aac_(_6_′)_Ib-cr_ accelerate the
development of fluoroquinolone resistance in _Salmonella_ typhimurium. Front Microbiol 5, 521 (2014). Article PubMed PubMed Central Google Scholar * Wong, M. H. et al. Expansion of
_Salmonella_ Typhimurium ST34 clone carrying multiple resistance determinants in China. Antimicrob Agents Chemother 57, 4599–4601 (2013). Article PubMed PubMed Central CAS Google Scholar
* Jones-Dias, D. et al. QnrS1- and Aac(6′)-Ib-cr-producing _Escherichia coli_ among Isolates from animals of different sources: susceptibility and genomic characterization. Front Microbiol
7, 671 (2016). PubMed PubMed Central Google Scholar * Campos, J. et al. Clinical _Salmonella_ Typhimurium ST34 with metal tolerance genes and an IncHI2 plasmid carrying
_oqxAB-aac_(_6_′)_-Ib-cr_ from Europe. J Antimicrob Chemother 71, 843–845 (2016). Article CAS PubMed Google Scholar * Landman, D., Georgescu, C., Martin, D. A. & Quale, J. Polymyxins
revisited. Clin Microbiol Rev 21, 449–465 (2008). Article PubMed PubMed Central CAS Google Scholar * Olaitan, A. O., Morand, S. & Rolain, J.-M. Mechanisms of polymyxin resistance:
acquired and intrinsic resistance in bacteria. Front Microbiol 5, 643 (2014). Article PubMed PubMed Central Google Scholar * Nation, R. L. & Li, J. Colistin in the 21st century. Curr
Opin Infect Dis 22, 535–543 (2009). Article PubMed PubMed Central CAS Google Scholar * Catry, B. et al. Use of colistin-containing products within the European Union and European
Economic Area (EU/EEA): development of resistance in animals and possible impact on human and animal health. Int J Antimicrob Agents 46, 297–306 (2015). Article CAS PubMed Google Scholar
* Liu, Y.-Y. et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect
Dis 16, 161–168 (2015). Article CAS PubMed Google Scholar * Schwarz, S. & Johnson, A. P. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71,
2066–2070 (2016). Article PubMed Google Scholar * Sun, J. et al. Complete nucleotide sequence of an IncI2 plasmid coharboring _bla_CTX-M-55 and _mcr-1_. Antimicrob Agents Chemother 60,
5014–5017 (2016). Article PubMed PubMed Central CAS Google Scholar * Zeng, K. J., Doi, Y., Patil, S., Huang, X. & Tian, G. B. Emergence of the Plasmid-Mediated _mcr-1_ Gene in
Colistin-resistant _Enterobacter aerogenes_ and _Enterobacter cloacae_. Antimicrob Agents Chemother 60, 3862–3863 (2016). Article PubMed PubMed Central CAS Google Scholar * Li, Z., Tan,
C., Lin, J. & Feng, Y. Diversified variants of the _mcr-1_-carrying plasmid reservoir in the swine lung microbiota. _Sci China_ Life Sci (2016). * Yu, H. et al. Detection of the _mcr-1_
colistin resistance gene in carbapenem-resistant Enterobacteriaceae from different hospitals in china. Antimicrob Agents Chemother 60, 5033–5035 (2016). Article PubMed PubMed Central CAS
Google Scholar * Zheng, B. et al. Coexistence of MCR-1 and NDM-1 in clinical _Escherichia coli_ Isolates. Clin Infect Dis (2016). * Bai, L. et al. Characterisation of multidrug-resistant
Shiga toxin-producing _Escherichia coli_ cultured from pigs in China: co-occurrence of extended-spectrum beta-lactamase- and _mcr-1_-encoding genes on plasmids. Int J Antimicrob Agents
(2016). * Sally, C. Y. et al. Colistin-resistant Enterobacteriaceae carrying the _mcr-1_ gene among patients in Hong Kong. Emerg Infect Dis 22, 1667–1669 (2016). Article Google Scholar *
Kuo, S. C. et al. Colistin resistance gene _mcr-1_ in _Escherichia coli_ isolates from humans and retail meats, Taiwan. J Antimicrob Chemother 71, 2327–2329 (2016). Article CAS PubMed
Google Scholar * Yang, Y. Q. et al. Co-occurrence of _mcr-1_ and ESBL on a single plasmid in _Salmonella enterica_. J Antimicrob Chemother 71, 2336–2338 (2016). Article CAS PubMed Google
Scholar * Poirel, L. K. N. et al. Genetic features of MCR-1-producing colistin-resistant _Escherichia coli_ isolates in South Africa. Antimicrob Agents Chemother 60, 4394–4397 (2016).
Article PubMed PubMed Central CAS Google Scholar * Zhang, H., Seward, C. H., Wu, Z., Ye, H. & Feng, Y. Genomic insights into the ESBL and MCR-1-producing ST648 _Escherichia coli_
with multi-drug resistance. Sci Bull (Beijing) 61, 875–878 (2016). Article CAS Google Scholar * Ye, H. et al. Diversified _mcr-1_-harbouring plasmid reservoirs confer resistance to
colistin in human gut microbiota. mBio 7, e00177–00116 (2016). Article PubMed PubMed Central CAS Google Scholar * Pham Thanh, D. et al. Inducible colistin resistance via a disrupted
plasmid-borne _mcr-1_ gene in a 2008 Vietnamese _Shigella sonnei_ isolate. J Antimicrob Chemother 71, 2314–2317 (2016). Article PubMed PubMed Central Google Scholar * Hu, Y., Liu, F.,
Lin, I. Y., Gao, G. F. & Zhu, B. Dissemination of the _mcr-1_ colistin resistance gene. Lancet Infect Dis 16, 146–147 (2016). Article PubMed Google Scholar * Figueiredo, R. et al.
Detection of an _mcr-1_-encoding plasmid mediating colistin resistance in _Salmonella enterica_ from retail meat in Portugal. J Antimicrob Chemother 71, 2338–2340 (2016). Article CAS
PubMed Google Scholar * Doumith, M. et al. Detection of the plasmid-mediated _mcr-1_ gene conferring colistin resistance in human and food isolates of _Salmonella enterica_ and
_Escherichia coli_ in England and Wales. J Antimicrob Chemother 71, 2300–2305 (2016). Article CAS PubMed Google Scholar * Campos, J., Cristino, L., Peixe, L. & Antunes, P. MCR-1 in
multidrug-resistant and copper-tolerant clinically relevant _Salmonella_ 1,4,[5],12:i:- and _S._ Rissen clones in Portugal, 2011 to 2015. Euro Surveill 21 (2016). * Sun, J. F. et al. The
molecular epidemiological characteristics and genetic diversity of _Salmonella_ Typhimurium in Guangdong, China, 2007–2011. PLoS One 9, e113145 (2014). Article ADS PubMed PubMed Central
Google Scholar * Malhotra-Kumar, S. et al. Colistin resistance gene _mcr-1_ harboured on a multidrug resistant plasmid. Lancet Infect Dis 16, 283–284 (2016). Article CAS PubMed Google
Scholar * Webb, H. E. et al. Dissemination of the _mcr-1_ colistin resistance gene. Lancet Infect Dis 16, 144–145 (2016). Article MathSciNet PubMed Google Scholar * Tse, H. & Yuen,
K. Y. Dissemination of the _mcr-1_ colistin resistance gene. Lancet Infect Dis 16, 145–146 (2016). Article PubMed Google Scholar * McGann, P. et al. _Escherichia coli_ harboring _mcr-1_
and _bla_CTX-M on a novel IncF plasmid: first report of _mcr-1_ in the United States. Antimicrob Agents Chemother 60, 4420–4421 (2016). Article PubMed PubMed Central CAS Google Scholar
* Xavier, B. B., Lammens, C., Butaye, P., Goossens, H. & Malhotra-Kumar, S. Complete sequence of an IncFII plasmid harbouring the colistin resistance gene _mcr-1_ isolated from Belgian
pig farms. J Antimicrob Chemother 71, 2342–2344 (2016). Article CAS PubMed Google Scholar * Zhi, C., Lv, L., Yu, L.-F., Doi, Y. & Liu, J.-H. Dissemination of the _mcr-1_ colistin
resistance gene. Lancet Infect Dis 16, 292–293 (2016). Article PubMed Google Scholar * Petrillo, M., Angers-Loustau, A. & Kreysa, J. Possible genetic events producing colistin
resistance gene mcr-1. Lancet Infect Dis 16, 280 (2016). CAS PubMed Google Scholar * Li, L. et al. Co-spread of _oqxAB_ and _bla_CTX-M-9G in non-Typhi _Salmonella enterica_ isolates
mediated by ST2-IncHI2 plasmids. Int J Antimicrob Agents 44, 263–268 (2014). Article CAS PubMed Google Scholar * CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
CLSI document M100-S25. Clinical and Laboratory Standards Institute Wayne, PA (2015). * CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacterial
Isolated from Animals. Approved Standard-Fourth Edition and Supplement. CLSI documents VET01A4E and VET01S3E. Clinical and Laboratory Standards Institute Wayne, PA (2015). * James
Versalovic, T. K. a. J. R. L. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res 19, 6823–6831 (1991). Article
Google Scholar * Seifert, H. et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of _Acinetobacter baumannii_. J
Clin Microbiol 43, 4328–4335 (2005). Article PubMed PubMed Central CAS Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the National Natural Science
Foundation and the Natural Science Foundation of Guangdong Province, China (grant U1201214), the Program for Changjiang Scholars and Innovative Research Team at the University of the
Ministry of Education of China (grant IRT13063), and the Natural Science Foundation of Guangdong Province (grant S2012030006590), and the Basic Research Program of China (2016YFC1200100).
Dr. Feng is a recipient of the “Young 1000 Talents” Award. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * National Risk Assessment Laboratory for Antimicrobial Resistance of Animal Original
Bacteria, South China Agricultural University, Guangzhou, P.R. China Xing-Ping Li, Liang-Xing Fang, Jia-Qi Song, Jing Xia, Wei Huo, Jin-Tao Fang, Xiao-Ping Liao, Ya-Hong Liu & Jian Sun *
Guangdong Provincial Key Laboratory of Veterinary Pharmaceutics Development and Safety Evaluation, South China Agricultural University, Guangzhou, P.R. China Xing-Ping Li, Liang-Xing Fang,
Jia-Qi Song, Jing Xia, Wei Huo, Jin-Tao Fang, Xiao-Ping Liao, Ya-Hong Liu & Jian Sun * Department of Medical Microbiology and Parasitology, Zhejiang University School of Medicine,
Zhejiang, P.R. China Youjun Feng Authors * Xing-Ping Li View author publications You can also search for this author inPubMed Google Scholar * Liang-Xing Fang View author publications You
can also search for this author inPubMed Google Scholar * Jia-Qi Song View author publications You can also search for this author inPubMed Google Scholar * Jing Xia View author publications
You can also search for this author inPubMed Google Scholar * Wei Huo View author publications You can also search for this author inPubMed Google Scholar * Jin-Tao Fang View author
publications You can also search for this author inPubMed Google Scholar * Xiao-Ping Liao View author publications You can also search for this author inPubMed Google Scholar * Ya-Hong Liu
View author publications You can also search for this author inPubMed Google Scholar * Youjun Feng View author publications You can also search for this author inPubMed Google Scholar * Jian
Sun View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS X.-P.L. performed experiments, analyzed the data and wrote the main manuscript;
L.-X.F. and Y.F. edited the manuscript; J.-Q.S., W.H. and J.-T.F. performed experiments; J.X. analyzed the data; X.-P.L. and Y.-H.L. coordinated the whole project; J.S. designed this
project, analyzed the data and revised the article. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. RIGHTS AND PERMISSIONS This work is licensed
under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless
indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the
material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Li, XP., Fang, LX., Song, JQ. _et
al._ Clonal spread of _mcr-1_ in PMQR-carrying ST34 _Salmonella_ isolates from animals in China. _Sci Rep_ 6, 38511 (2016). https://doi.org/10.1038/srep38511 Download citation * Received: 06
September 2016 * Accepted: 09 November 2016 * Published: 05 December 2016 * DOI: https://doi.org/10.1038/srep38511 SHARE THIS ARTICLE Anyone you share the following link with will be able
to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative




:max_bytes(150000):strip_icc():focal(734x339:736x341)/spike-lee-glenn-close-ab1259d23d1448af8942a6dccda2e318.jpg)