- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Physical activity (PA) has been shown to reduce the impact of _FTO_ variation and obesity genetic risk scores (GRS) on BMI. We examined this interaction using a quantitative measure
of PA and two adiposity indexes in a longitudinal multi-ethnic study. We analyzed the impact of PA on the association between 14 obesity predisposing variants (analyzed independently and as
a GRS) and baseline/follow-up obesity measures in the multi-ethnic prospective cohort EpiDREAM (17423 participants from six ethnic groups). PA was analyzed using basic (low-moderate-high)
and quantitative measures (metabolic equivalents (METS)), while BMI and the body adiposity index (BAI) were used to measure obesity. Increased PA was associated with decreased BMI/BAI at
baseline/follow-up. _FTO_ rs1421085, _CDKAL1_ rs2206734, _TNNl3K_ rs1514176, _GIPR_ rs11671664 and the GRS were associated with obesity measures at baseline and/or follow-up. Risk alleles of
three SNPs displayed nominal associations with increased (_NTRK2_ rs1211166, _BDNF_ rs1401635) or decreased (_NPC1_ rs1805081) basic PA score independently of BMI/BAI. Both basic and
quantitative PA measures attenuated the association between _FTO_ rs1421085 risk allele and BMI/BAI at baseline and follow-up. Our results show that physical activity can blunt the genetic
effect of _FTO_ rs1421085 on adiposity by 36–75% in a longitudinal multi-ethnic cohort. SIMILAR CONTENT BEING VIEWED BY OTHERS GENOME-WIDE ASSOCIATION ANALYSES OF PHYSICAL ACTIVITY AND
SEDENTARY BEHAVIOR PROVIDE INSIGHTS INTO UNDERLYING MECHANISMS AND ROLES IN DISEASE PREVENTION Article Open access 07 September 2022 ASSOCIATION BETWEEN _PTPN1_ POLYMORPHISMS AND
OBESITY-RELATED PHENOTYPES IN EUROPEAN ADOLESCENTS: INFLUENCE OF PHYSICAL ACTIVITY Article 11 November 2022 INTERACTION OF GENETIC AND ENVIRONMENTAL FACTORS FOR BODY FAT MASS CONTROL:
OBSERVATIONAL STUDY FOR LIFESTYLE MODIFICATION AND GENOTYPING Article Open access 23 June 2021 INTRODUCTION Obesity has become a global epidemic1 and is a known risk factor for a number of
adverse health outcomes, including psychological disturbance, osteoarthritis, type 2 diabetes, hypertension, cardiovascular disease, cancer and ultimately 8–13 years shorter life expectancy
in its more severe forms2,3. Since the rising prevalence of obesity has been attributed primarily to changes in environmental exposures, such as excessive energy intake, sedentary lifestyle
and sleep debt, among others4, recent research has focused on preventative strategies to control the obesity epidemic5. Despite the global impact of these environmental changes, obesity
appears to manifest preferentially in genetically predisposed individuals and a high level of inter-individual variation has been observed among exposed populations6. Current evidence has
shown that heritability estimates for obesity-related traits can be modulated by lifestyle factors such as physical activity (PA)7. Significant gene-environment interactions (GEI) between
_FTO_ intron 1 variation and PA have been consistently found in 16 independent cross-sectional and intervention studies performed with children and adult populations of European, East Asian
and African ancestry8,9,10. A recent meta-analysis in 111 421 subjects of European ancestry confirmed a significant PA x genetic risk score (GRS) interaction using 12 obesity predisposing
SNPs and showed that this interaction was more apparent in subjects living in North America11. Although these data provide convincing evidence of an interaction between genetic
predisposition to obesity and PA, we identified several important limitations of the existing literature. First, almost all current gene x PA interaction studies in the obesity field have
been conducted using cross sectional designs, although there are exceptions12. Strengths of the cohort design include the ability to study temporal relationships, which strengthens the
confidence in statistical associations13,14. Second, most GEI analyses of PA have used imprecise self-report measurements of PA due to concerns regarding cost, feasibility and participant
burden15 and only one study has examined the impact of a quantitative metabolic equivalent (MET) score16. Third, nearly all GEI studies of obesity have used BMI as the outcome measure of
obesity, although the validity of the BMI can be compromised by different body compositions (e.g. lean vs. fat mass)17. Fourth, GEI studies have mainly been performed in European
populations, but the transferability of the conclusions of these studies to other ethnic backgrounds remains unclear18,19. Although a previous meta-analysis analyzed multiple ethnic groups,
the sample was over 95% European18. This prompted us to assess (1) the association between PA behavior and obesity and (2) the interaction between 14 obesity predisposing variants (analyzed
independently and as a GRS) and PA on obesity-related traits in the multi-ethnic longitudinal cohort EpiDREAM. We used a quantitative measure of energy expenditure (Metabolic Equivalent
Score, MET score)20 and the recently validated body adiposity index (BAI)21. The BAI may provide new insight into this interaction due to its strong correlation (r = 0.85) with body
adiposity (as measured by dual x-ray absorptiometry)21. MATERIALS AND METHODS STUDY PARTICIPANTS The data for this investigation were collected through a prospective cohort of participants
at risk for type 2 diabetes (T2D), which has been described in detail previously22,23. Briefly, EpiDREAM enrolled a total of 24 872 individuals recruited from 21 countries who were screened
for eligibility to enter the DREAM clinical trial22. Individuals who were identified as at risk for type 2 diabetes based on family history, ethnicity and abdominal adiposity were screened
using a 75-gram oral glucose tolerance test (OGTT). A subset of 1960 DREAM participants received rosiglitazone over follow-up. All participants were between the ages of 18–85 years and were
screened between July 2001 and August 2003. We focused on 17 423 subjects from six ethnic groups (South Asian, East Asian, European, African, Latin American, Native North American) having
both phenotypic and 50 K gene-centric array information in the EpiDREAM study (Supplementary Figure 1). Self-reported ethnicity has been verified in the 17 423 individuals using the
eigensoft software (http://genepath.med.harvard.edu/~reich/Software.htm) and 40 individuals were reclassified. The first 10 components from this principal components analysis were retained
to adjust for population stratification (Supplementary Figure 2). Of the 17 423 participants at baseline, our follow-up analyses included 9 228 participants with complete genotype and
phenotype data. Interim and final visits with participants occurred 12–24 months and 36–48 months (median follow-up 3.3 years) after their screening visit, respectively. Participants who
were not able to complete in-person clinic visits were contacted through telephone or mail23. The EpiDREAM study has been approved by local ethics committees and the study methodology was
carried out in accordance with the approved guidelines. Informed consent was obtained from each subject before participating in the study, in accordance with the Declaration of Helsinki.
GENOTYPING Buffy coats for DNA extraction have been collected in 19 197 participants of the EpiDREAM study (Supplementary Figure 1). DNA has been extracted by the Gentra System. Genotyping
was performed using the Illumina CVD bead chip microarray ITMAT Broad Care (IBC) array24. Genotyping was performed at the McGill University and Genome Quebec Innovation Centre using the
Illumina Bead Studio genotyping module, version 3.2. We established a list of SNPs that reached genome-wide significance (P < 5 × 10−8) with BMI or binary obesity status in populations of
European ancestry. We used three different strategies to optimize the SNP selection procedure using a key word search (e.g. BMI) on i) the National Human Genome Research Institute (NHGRI)
GWAS Catalog (www.genome.gov/gwastudies/) ii) the HuGE Navigator GWAS Integrator (www.hugenavigator.net/HuGENavigator/gWAHitStartPage.do) iii) the PubMed database
(www.ncbi.nlm.nih.gov/pubmed). Using this strategy, we ended-up with a list of 72 independent SNPs associated with obesity-related traits. From this list, 14 SNPs were available on versions
1 and 2 of the IBC 50 K SNP array (Supplementary Table 1). The SNPs selected showed no significant (P < 10−6) deviation from Hardy-Weinberg Equilibrium (HWE) in the six ethnic groups. The
call rate for each of the 14 SNPs was between 99.8–100% (Supplementary Table 1). PHENOTYPING In addition to the OGTT, participants completed a questionnaire that included demographic data,
medical history and PA behaviors at baseline and follow-up. Anthropometric measurements including height, weight, waist and hip circumference were performed using a standardized protocol22.
Height (m), weight and hip circumference (HC) (cm) were measured by trained medical staff. Standing height was measured to the nearest 0.1 cm and weight was measured to the nearest 0.1 kg in
light clothing. Hip circumference was measured in duplicate at the level of the greater trochanters using a non-flexible tape measure with an attached spring balance with a mass of 750 g.
Averages of the two measures were used in all analyses. Body mass index (BMI) was calculated as weight in kilograms divided by height in meters (m) squared. We used Body Adiposity Index
(BAI), which estimates body adiposity percentage directly based on height and hip circumference25. Specifically, BAI = [(hip circumference)/((height)(1.5)) − 18], with hip circumference and
height being expressed in centimeters and meters, respectively25. The 2003 ADA criteria were used to classify participants as having normal glucose tolerance (NGT), impaired fasting glucose
(IFG), impaired glucose tolerance (IGT), or T2D at baseline, as confirmed by an oral glucose tolerance test. Normoglycemia was defined as a fasting plasma glucose < 5.6 mmol/L, IFG was
defined as a fasting plasma glucose of 5.6 to 6.9 mmol/L, IGT was defined as a fasting plasma glucose below 7.0 mmol/L and a 2-h glucose between 7.8 and 11.0 mmol/L and diabetes was defined
if either the fasting plasma glucose was ≥ 7.0 mmol/L or the 2-h glucose was ≥ 11.1 mmol/L26. IFG and IGT were collapsed into one category and these three groups (normoglycemia, IFG/IGT,
diabetes) comprised the glycemic status variable. PA measures were based on self-reported time of participation in 41 different physical activities (Supplementary Table 2). A quantitative
measure of energy expenditure (Metabolic Equivalent score, MET score) was derived using this information and the updated compendium of physical activities20. A MET score was assigned to each
activity based on the energy cost of that activity. The energy expended on each leisure time and work-related activity was estimated by multiplying the hours/week of participation by the
corresponding MET value. These measures were summed across all activities to provide an overall estimate of energy expenditure represented as MET minutes per week. This quantitative measure
was compared to a brief, self-reported, categorical measure based on PA participation during leisure time and at work. Participants rated their PA level at work and during leisure on a scale
of one to three to form the basic PA score (1 = sedentary, 2 = moderately active, 3 = very active). STATISTICAL ANALYSES Statistical analyses were performed using SPSS (version 20, New
York, USA, IBM Corporation) and the power of our study was calculated using QUANTO (version 1.2.4, University of Southern California, Los Angeles, CA, USA). Single SNP analyses were
performed under the additive model and the obesity risk alleles previously identified for each of the 14 SNPs in literature were used as the risk allele. General linear models (GLM) were
used to examine (1) the association between energy expenditure (METS)/basic PA score at baseline and BMI/BAI at baseline and follow-up (2) the association between 14 obesity predisposing
gene variants (analyzed independently and as a GRS) and BMI/BAI at baseline and follow-up. These tests were adjusted for covariates including, sex, age, ethnicity and glycemic status.
General linear models were used, with or without the inclusion of a SNP x PA or a GRS x PA interaction term. Since previous studies have demonstrated a compelling association between each of
the SNPs analyzed and obesity traits, the effect of the SNPs on BMI/BAI was considered a confirmatory step. Therefore, all SNPs reaching a nominal level of association with BMI/BAI (_P_
< 0.05) were considered significant and followed up for interaction analysis. These tests were also adjusted for covariates including, sex, age, ethnicity and glycemic status. All
analyses with BMI/BAI follow-up or change as the dependent variable were adjusted for rosiglitazone use in addition to the aforementioned variables. A natural log transformation was used
successfully to correct the positive skew of the MET score data. Ordinal logistic regression was used to analyze the impact of the 14 obesity predisposing SNPs on the basic PA score
(adjusted for sex, age, ethnicity, glycemic status and BMI). This test was performed since a correlation between the genetic variant and environmental exposure can bias the estimates of the
main genetic effect and the gene-environment interaction27. Previous examples of these effects have been observed in cancer research, where variants on 15q25 have been linked to both smoking
behaviors and lung cancer28. The genetic predisposition score was calculated by summing the alleles of the 14 obesity predisposing SNPs so that the score could range from 0 to 28. Since
weighting has been shown to have no major impact on the effect of a GRS29, an unweighted GRS was used. We performed imputations for missing genotypic values as previously described30 using
the mean number of predisposing obesity alleles in successfully genotyped individuals. This procedure was performed separately in each ethnic group. Individuals with more than one out of 14
missing genotype were not included in the genetic risk score calculation. Although multiple SNPs included in the gene score are within the same gene, our linkage disequilibrium analyses of
this cohort indicated that it is appropriate to include all 14 SNPs in the genetic risk score (r2 < 0.25, Supplementary Table 3). Two-tailed P-values are presented in this manuscript and
_P_ < 0.05 were considered as nominally significant. After applying a Bonferroni’s correction for multiple testing, a _P_ < 0.00025 (0.05/200) was considered as significant. RESULTS
CHARACTERISTICS OF THE STUDIED COHORT The clinical and anthropometric characteristics of the EpiDREAM study are summarized in Table 1. The mean age of participants was 52.7 years and the
ethnic distribution of the cohort was 53.9% European, 18.9% Latino, 15.8% South Asian, 7.2% African, 2.9% Native American, 1.3% East Asian. The individuals in this analysis represented 17 of
the original 21 countries from which recruitment took place (Supplementary Table 4). At baseline, a mean BMI of 30.2 (SD = 6.22) kg/m2 and a mean BAI of 33.0 (SD = 7.49) were observed and
the mean energy expenditure was 320.50 MET-minutes/week (SD = 409.20). For the present analyses, we focused on 17 423 participants at baseline and 9 228 at follow-up who had complete
genotype and phenotype data. The median time between the baseline screening visit and final contact was 3.3 years. After follow-up, the mean BMI and BAI were 30.32 (SD = 5.79) and 33.78 (SD
= 7.59), respectively and the mean energy expenditure appeared to decrease slightly to 301.50 MET-minutes/week (SD = 368.04). EFFECT OF PHYSICAL ACTIVITY ON BMI/BAI At baseline, the
quantitative MET score was significantly associated with both decreased BMI and BAI (Table 2). Congruently, the basic PA score (low – moderate – high PA) was significantly associated with
lower baseline BMI and BAI. Similar associations were found between the baseline MET score and BMI and BAI at follow-up. The baseline basic PA score was also associated with decreased
follow-up BMI and BAI. The baseline MET score was not associated with BMI change, or BAI change. The basic PA score was nominally associated with decreased BMI change and BAI change over
follow-up. EFFECT OF SNPS/GRS ON PHYSICAL ACTIVITY We first investigated the association of 14 obesity predisposing SNPs and corresponding GRS on basic PA score adjusting for sex, age,
ethnicity, glycemic status and BMI (Table 3). We observed a nominal association between three of these SNPs and the basic PA score: _NTRK2_ rs1211166, _BDNF_ rs1401635 and _NPC1_ rs1805081.
The association between the obesity risk GRS and the basic PA score was not significant. When adjusting for BAI rather than BMI the same three SNPs remained nominally associated with the
basic PA score with a consistent direction of effect (Supplementary Table 5). None of the 14 SNPs displayed a significant association with the MET score after adjustment for sex, age,
ethnicity, glycemic status and BMI (Table 3). The association between the obesity risk GRS and the MET score was also non-significant. Similar results were found when adjusting for BAI
rather than BMI (Supplementary Table 5). Of the 14 SNPs analyzed, only one (_NTRK2_ rs1211166) showed a nominal association with change in the basic PA score (Supplementary Table 6). None of
the 14 SNPs displayed a significant association with a change in the MET score. The obesity risk GRS was not associated with change in the basic PA score or change in the MET score
(Supplementary Table 6). EFFECT OF SNPS/GRS ON BMI/BAI At baseline, the obesity risk alleles of four SNPs were associated increased with BMI and BAI. _FTO_ rs1421085 and _CDKAL1_ rs2206734
were significantly associated with greater BMI/BAI, while _TNNI3K_ rs1514176 and _GIPR_ rs11671664 were nominally associated with increased BMI/BAI (Table 4). At baseline, the GRS was
significantly associated with greater BMI and BAI. After follow-up, three SNPs (_FTO_ rs1421085, _TNNI3K_, rs1514176, _GIPR_ 11671664) and the GRS were associated with increased BMI and BAI
(Table 4). _CDKAL1_ rs2206734 displayed a nominal association with reduced BMI and BAI change. The GRS was not associated with BMI or BAI change. INTERACTION ANALYSES Interaction tests with
PA were restricted to the subset of SNPs/GRS displaying a nominal or significant association with BMI/BAI at baseline and/or at follow-up. At baseline, the MET score modified the effect of
the _FTO_ risk allele on BMI and BAI (Table 5). Each additional _FTO_ risk allele (C) was associated with a (1) BMI increase of 0.60 kg/m2 (P = 1.1 × 10−4), BAI increase of 0.45 (P = 5.7 ×
10−3) in the lowest MET score tertile and (2) BMI increase of 0.26 kg/m2 (P = 0.05), BAI increase of 0.20 (P = 0.15) in the highest MET score tertile. This indicates that the effect of _FTO_
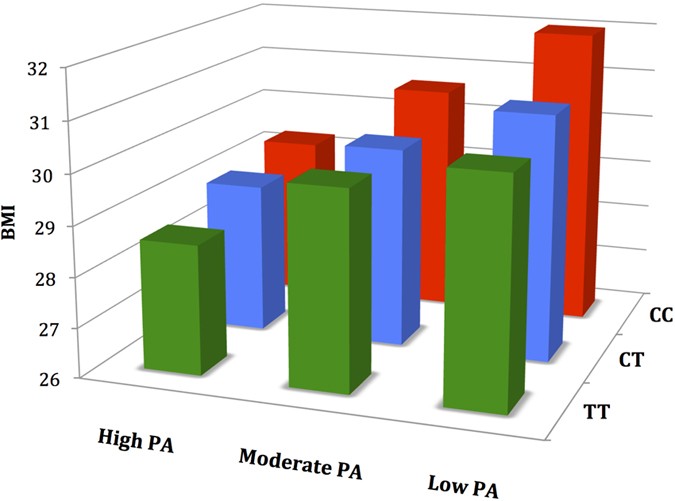
rs1421085 on BMI and BAI can be reduced by 57% and 56% respectively through PA. The basic physical score also modified the effect of the _FTO_ risk allele on BMI and BAI at baseline (Table
5, Fig. 1). Each additional obesity risk allele (C) was associated with a (1) BMI increase of 0.71 kg/m2 (P = 1.4 × 10−7), BAI increase of 0.62 (P = 2.1 × 10−5) in the inactive group and (2)
BMI increase of 0.35 kg/m2 (P = 0.03), BAI increase of 0.40 (P = 0.02) in the active group. This indicates that PA is associated with a 36–51% decrease in the effect of _FTO_ rs1421085 on
obesity measures at baseline. We also conducted a sensitivity analysis to analyze the BPA x _FTO_ rs1421085 interaction for baseline BMI in only non-diabetic patients and found similar
results (β = −0.40, 95% CI = −0.62 to −0.19, P = 2.4 × 10−4). The basic PA score interacted with _FTO_ rs1421085 in modulating BMI and BAI at follow-up. Each additional obesity risk allele
(C) was associated with a (1) BMI increase of 0.72 kg/m2 (P = 1.1 × 10−5), BAI increase of 0.75 (P = 3.4 × 10−4) in the inactive group and (2) BMI increase of 0.19 kg/m2 (P = 0.39), BAI
increase of 0.19 (P = 0.44) in the active group. This corresponds to a 74–75% decrease in the effect of _FTO_ rs1421085 on BMI/BAI at follow-up. No significant interactions were observed
between _TNNI3K_ rs1514176, _CDKAL1_ rs2206734, _GIPR_ rs11671664 or the obesity risk GRS and the basic PA/MET score on BMI/BAI at baseline, follow-up or change. Given that Ahmad _et al._11
reported the _FTO_ x PA interaction to be 10-fold larger in North American compared to European cohorts, we also analyzed a 3-way interaction (_FTO_ x PA x North American residence) among
the subgroup of European participants to follow-up this finding. Although the 3-way interaction was not significant (β = −0.01, 95% CI = −0.03 to 2.8 × 10−3, P = 0.10), we acknowledge that
our statistical power to detect 3-way interactions in this subgroup was limited and this association warrants further investigation. DISCUSSION We observed significant GEI between _FTO_
rs1421085 and PA at baseline and at follow-up in an international multiethnic population using both basic and quantitative assessments of PA. Although the interactions were nominally
significant, the Bonferroni correction we applied is overly conservative, particularly when testing highly correlated outcomes31 such as BMI/BAI, basic PA/MET score and we have confidence in
our results for several reasons. First, the power to detect GEI at the nominal level of association (P < 0.05) is adequate at baseline, although our power estimations for the Bonferroni
corrected threshold are moderate (see Supplementary Figures 3–6). Second, twin studies have shown that PA can substantially reduce the influence of genetic factors on BMI in adults7. Third,
the interaction between _FTO_ and PA has been demonstrated in several cross-sectional studies and currently represents the most robust example of GEI in the field of genetic
epidemiology8,9,10,11,18. Fourth, there is a plausible underlying biological process to substantiate this association. _FTO_ is a nucleic acid demethylase and _FTO_ intron 1 variation is
associated with different methylation profiles and BMI variance32,33,34. Since methylation of DNA is sensitive to environmental changes (e.g. PA and diet) there is a strong biological
rationale to identify GEI with _FTO_ as previously reported8,35. Two studies have shown that PA can change the methylation and mRNA expression pattern of genes, including _FTO_, in both
muscle and adipose tissue34,36. A more recent analysis demonstrated that variation at the _FTO_ locus represses mitochondrial thermogenesis in adipocyte precursor cells and causes a shift
from energy dissipating beige (brite) to energy-storing white adipocytes, which is accompanied by increased lipid storage and weight gain37. Exercise studies in humans and mouse models
indicate that exercise training increases the expression of the brown adipocyte marker uncoupling protein (UCP1) in both visceral and subcutaneous white adipose tissue (WAT)38. These changes
are associated with increases in brown-like adipocytes (browning or beiging), particularly in subcutaneous WAT38. The combined role of FTO and physical activity in obesity and adipocyte
browning, in conjunction with epigenetic mechanisms, strengthen the biological rationale and confidence in the statistical interaction39. Given the growing consensus that food intake may be
the main driver of the obesity epidemic40, it is important to note that both PA measures displayed significant associations with both adiposity measures at baseline and at follow-up. This
indicates that PA can influence obesity, despite the broad range of lifestyles among the participants. The value of PA for managing obesity has been recognized in a recent analysis of the
National Health and Nutrition Examination Survey (NHANES) cohort from 1988–2010, which found that PA had a larger impact on BMI and waist circumference trends than calorie intake41. Our
cross sectional analyses indicate that one hour of jogging or swimming (8.0 MET activities) per week was associated with approximately a 0.5 kg/m2 decrease in BMI. Together, these data
challenge the idea of attributing the obesity epidemic mainly to excessive caloric intake40 and support the universal value of PA to maintain a healthy body weight41. Although the MET score
provides a more comprehensive assessment of PA participation, only 11 015 (63%) participants completed the assessment of the MET score, compared to the 17 407 (99%) participants who
completed the basic PA score. The loss of power induced by the smaller sample size may have been compensated for by the added precision of the MET score. Simulations of GEI have shown that a
sample of about 2 000 participants with precisely measured environmental exposure and outcome data are needed to detect a GEI of large magnitude (a doubling of the genetic risk estimate in
the exposed group compared to the unexposed group) with reasonable power (95% power, P = 1 × 10−4)42. With less precise measurement of environmental exposure, the sample size requirement can
increase to 100 000 participants to detect the same interaction with comparable power42,43. Using a brief PA assessment may be the best compromise to balance the sample size requirements
with the need for sufficient precision, as recently suggested by Peters _et al._44. Measuring body fat content data is less feasible in large sample sizes for GEI studies. Only two small GEI
studies (N < 800) have used direct body fat content measures (DEXA and underwater weighing)45,46 and the meta-analysis of the PA x _FTO_ interaction which analyzed bioelectrical
impedance was over 95% Europeans16. To our knowledge, this is the first large-scale study to report an interaction between PA and _FTO_ rs1421085 using the BAI. Since assessing body fat
content is financially prohibitive in large-scale analyses, the BAI may be an acceptable method to complement the widely used BMI measure to assess adiposity in GEI studies. Assessing the
impact of the 14 SNPs on PA identified some interesting nominal associations. Despite being an obesity predisposing gene, _BDNF_ may be related to increased PA levels in our cohort. While
this appears contradictory, the loss of one functional copy of _BDNF_ gene has been associated with Mendelian obesity, cognitive impairment and hyperactivity in both humans47 and rodents48.
It has been proposed that increased PA may be a behavioral response to compensate for their proclivity to gain weight49. Alternatively, these SNPs have been hypothesized to be a remnant of
our hunter-gatherer past and may have been positively selected since they promote an “active-foraging” phenotype that induces a preference for energy-dense foods and the physical disposition
to attain those foods49. Together, this food seeking behaviour and overactive disposition represent a “Peppy-Thrifty Genotype.” In contrast, _NPC1_ variation appears to contribute to
sedentary behavior and may complement the thrifty genotype hypothesis50,51. Genetic variants increasing food intake and decreasing PA may have been positively selected based on their
parallel effects on energy balance51. The contrasting effects of these obesity predisposing SNPs on PA may account for the challenges in substantiating the lazy thrifty genotype
hypothesis52. Although there appears to be some shared genetic correlation between obesity and PA, additional studies are necessary to confirm our nominally significant associations and to
clarify the impact of additional obesity predisposing genes on PA. Limitations of this study include the multi-ethnic composition of the EpiDREAM cohort may have added significant
heterogeneity in the analyses, especially as important PA differences are observed in different ethnic backgrounds53. Although, the MET score was calculated with more objective criteria of
PA participation, recalling participation in 41 different activities introduces a source of error and or recall bias. Since most of the obesity predisposing SNPs selected in the study were
originally identified in European populations, they may not be ideal proxies for the causal SNP in other ethnic groups. We are aware that the 14 SNPs analyzed only represent a subset of the
current list of validated obesity SNPs. Lastly, the EpiDREAM population (participants identified for hyperglycemia risk) is not representative of the general population and the participants
missing from the follow-up analysis may have created a systematic bias in our sample. However, this bias may not have influenced our results since no significant differences in BMI (P =
0.25) or BAI (P = 0.21) were observed among those who completed follow-up and those that did not. Strengths of this analysis include the precise MET score, complementing the BMI with the
recently developed BAI, the prospective cohort design and the multi-ethnic sample. In summary, we identified an interaction between the _FTO_ SNP rs1421085 and PA in a prospective cohort of
six ethnic groups from 17 countries. While this has been demonstrated previously, this is the first study to analyze this interaction prospectively using a quantitative measure of PA while
comparing the recently developed BAI and BMI. Analyzing the impact of obesity predisposing SNPs on PA revealed novel associations, although further study is needed to confirm these effects.
These findings suggest that obesity prevention programs emphasizing vigorous PA for genetically at risk subgroups may be a valuable contribution to the global fight against obesity.
ADDITIONAL INFORMATION HOW TO CITE THIS ARTICLE: Reddon, H. _et al._ Physical activity and genetic predisposition to obesity in a multiethnic longitudinal study. _Sci. Rep._ 6, 18672; doi:
10.1038/srep18672 (2016). REFERENCES * Finucane, M. M. et al. National, regional and global trends in body-mass index since 1980: systematic analysis of health examination surveys and
epidemiological studies with 960 country-years and 9.1 million participants. Lancet 377, 557–567, 10.1016/S0140-6736(10)62037-5 (2011). Article PubMed PubMed Central Google Scholar *
Dixon, J. B. The effect of obesity on health outcomes. Mol Cell Endocrinol 316, 104–108, 10.1016/j.mce.2009.07.008 (2010). Article CAS PubMed Google Scholar * Fontaine, K. R., Redden, D.
T., Wang, C., Westfall, A. O. & Allison, D. B. Years of life lost due to obesity. Jama 289, 187–193 (2003). Article PubMed Google Scholar * McAllister, E. J. et al. Ten putative
contributors to the obesity epidemic. Crit Rev Food Sci Nutr 49, 868–913, 10.1080/10408390903372599 (2009). Article PubMed PubMed Central Google Scholar * Birch, L. L. & Ventura, A.
K. Preventing childhood obesity: what works? Int J Obes (Lond) 33 Suppl 1, S74–81, 10.1038/ijo.2009.22 (2009). Article Google Scholar * Wardle, J., Carnell, S., Haworth, C. M. &
Plomin, R. Evidence for a strong genetic influence on childhood adiposity despite the force of the obesogenic environment. Am J Clin Nutr 87, 398–404 (2008). Article CAS PubMed Google
Scholar * Mustelin, L., Silventoinen, K., Pietilainen, K., Rissanen, A. & Kaprio, J. Physical activity reduces the influence of genetic effects on BMI and waist circumference: a study
in young adult twins. Int J Obes (Lond) 33, 29–36, 10.1038/ijo.2008.258 (2009). Article CAS Google Scholar * Andreasen, C. H. et al. Low physical activity accentuates the effect of the
FTO rs9939609 polymorphism on body fat accumulation. Diabetes 57, 95–101 (2008). Article CAS PubMed Google Scholar * Xi, B. et al. [Impact on the risk of obesity due to interactions
between fat mass- and obesity-associated gene rs9939609 variants and behavioral factors, in the Chinese school-aged children]. Zhonghua Liu Xing Bing Xue Za Zhi 31, 737–741 (2010). CAS
PubMed Google Scholar * Demerath, E. W. et al. Interaction of FTO and physical activity level on adiposity in African-American and European-American adults: the ARIC study. Obesity (Silver
Spring) 19, 1866–1872, 10.1038/oby.2011.131 (2011). Article CAS Google Scholar * Ahmad, S. et al. Gene x physical activity interactions in obesity: combined analysis of 111,421
individuals of European ancestry. PLoS Genet 9, e1003607, 10.1371/journal.pgen.1003607 (2013). Article CAS PubMed PubMed Central Google Scholar * Mitchell, J. A. et al. FTO genotype and
the weight loss benefits of moderate intensity exercise. Obesity (Silver Spring) 18, 641–643, 10.1038/oby.2009.311 (2010). Article Google Scholar * Manolio, T. A., Bailey-Wilson, J. E.
& Collins, F. S. Genes, environment and the value of prospective cohort studies. Nat Rev Genet 7, 812–820, 10.1038/nrg1919 (2006). Article CAS PubMed Google Scholar * Grimes, D. A.
& Schulz, K. F. Cohort studies: marching towards outcomes. Lancet 359, 341–345, 10.1016/S0140-6736(02)07500-1 (2002). Article PubMed Google Scholar * Hein, R., Beckmann, L. &
Chang-Claude, J. Sample size requirements for indirect association studies of gene-environment interactions (G x E). Genet Epidemiol 32, 235–245 (2008). Article PubMed Google Scholar *
Ahmad, T. et al. Lifestyle interaction with fat mass and obesity-associated (FTO) genotype and risk of obesity in apparently healthy U.S. women. Diabetes Care 34, 675–680, 10.2337/dc10-0948
(2011). Article PubMed PubMed Central Google Scholar * Muller, M. J., Bosy-Westphal, A. & Krawczak, M. Genetic studies of common types of obesity: a critique of the current use of
phenotypes. Obesity reviews : an official journal of the International Association for the Study of Obesity 11, 612–618 (2010). Article CAS Google Scholar * Kilpelainen, T. O. et al.
Physical activity attenuates the influence of FTO variants on obesity risk: a meta-analysis of 218,166 adults and 19,268 children. PLoS Med 8, e1001116, 10.1371/journal.pmed.1001116 (2011).
Article PubMed PubMed Central Google Scholar * Li, S. et al. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective
population study. PLoS Med 7, 10.1371/journal.pmed.1000332 (2010). * Ainsworth, B. E. et al. 2011 Compendium of Physical Activities: a second update of codes and MET values. Med Sci Sports
Exerc 43, 1575–1581, 10.1249/MSS.0b013e31821ece12 (2011). Article PubMed Google Scholar * Bergman, R. N. et al. A better index of body adiposity. Obesity (Silver Spring) 19, 1083–1089,
10.1038/oby.2011.38 (2011). Article PubMed Central Google Scholar * Gerstein, H. C., Yusuf, S., Holman, R., Bosch, J. & Pogue, J. Rationale, design and recruitment characteristics of
a large, simple international trial of diabetes prevention: the DREAM trial. Diabetologia 47, 1519–1527, 10.1007/s00125-004-1485-5 (2004). Article CAS PubMed Google Scholar * Anand, S.
S. et al. Glucose levels are associated with cardiovascular disease and death in an international cohort of normal glycaemic and dysglycaemic men and women: the EpiDREAM cohort study. Eur J
Prev Cardiol 19, 755–764, 10.1177/1741826711409327 (2012). Article CAS PubMed Google Scholar * Keating, B. J. et al. Concept, design and implementation of a cardiovascular gene-centric
50 k SNP array for large-scale genomic association studies. PLoS One 3, e3583 (2008). Article ADS PubMed PubMed Central Google Scholar * Bergman, R. N. et al. A better index of body
adiposity. Obesity (Silver Spring) 19, 1083–1089, 10.1038/oby.2011.38 (2011). Article PubMed Central Google Scholar * Genuth, S. et al. Follow-up report on the diagnosis of diabetes
mellitus. Diabetes Care 26, 3160–3167 (2003). Article PubMed Google Scholar * Vanderweele, T. J., Ko, Y.-A. & Mukherjee, B. Environmental confounding in gene-environment interaction
studies. Am J Epidemiol 178, 144–152 (2013). Article PubMed PubMed Central Google Scholar * Thorgeirsson, T. E. et al. A variant associated with nicotine dependence, lung cancer and
peripheral arterial disease. Nature 452, 638–642, 10.1038/nature06846 (2008). Article CAS ADS PubMed PubMed Central Google Scholar * Janssens, A. C. et al. The impact of genotype
frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genet Med 9, 528–535, 10.1097GIM.0b013e31812eece0 (2007). Article PubMed Google Scholar *
Robiou-du-Pont, S. et al. Contribution of 24 obesity-associated genetic variants to insulin resistance, pancreatic beta-cell function and type 2 diabetes risk in the French population. Int
J Obes (Lond) 37, 980–985, 10.1038/ijo.2012.175 (2013). Article CAS Google Scholar * Shi, Q., Pavey, E. S. & Carter, R. E. Bonferroni-based correction factor for multiple, correlated
endpoints. Pharm Stat 11, 300–309, 10.1002/pst.1514 (2012). Article PubMed Google Scholar * Yang, J. et al. FTO genotype is associated with phenotypic variability of body mass index.
Nature 490, 267–272, 10.1038/nature11401 (2012). Article CAS ADS PubMed PubMed Central Google Scholar * Bell, C. G. et al. Integrated genetic and epigenetic analysis identifies
haplotype-specific methylation in the FTO type 2 diabetes and obesity susceptibility locus. PLoS One 5, e14040, 10.1371/journal.pone.0014040 (2010). Article CAS ADS PubMed PubMed Central
Google Scholar * Ronn, T. et al. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genet 9, e1003572,
10.1371/journal.pgen.1003572 (2013). Article CAS PubMed PubMed Central Google Scholar * Sonestedt, E. et al. Fat and carbohydrate intake modify the association between genetic variation
in the FTO genotype and obesity. Am J Clin Nutr 90, 1418–1425, 10.3945/ajcn.2009.27958 (2009). Article CAS PubMed Google Scholar * Nitert, M. D. et al. Impact of an exercise
intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes 61, 3322–3332, 10.2337/db11-1653 (2012). Article CAS PubMed
PubMed Central Google Scholar * Claussnitzer, M. et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med 373, 895–907, 10.1056/NEJMoa1502214 (2015). Article
CAS PubMed PubMed Central Google Scholar * Stanford, K. I., Middelbeek, R. J. & Goodyear, L. J. Exercise Effects on White Adipose Tissue: Beiging and Metabolic Adaptations. Diabetes
64, 2361–2368, 10.2337/db15-0227 (2015). Article CAS PubMed PubMed Central Google Scholar * Ioannidis, J. P. Why most published research findings are false. PLoS Med 2, e124,
10.1371/journal.pmed.0020124 (2005). Article PubMed PubMed Central Google Scholar * Speakman, J. R. & O’Rahilly, S. Fat: an evolving issue. Dis Model Mech 5, 569–573,
10.1242/dmm.010553 (2012). Article CAS PubMed PubMed Central Google Scholar * Ladabaum, U., Mannalithara, A., Myer, P. A. & Singh, G. Obesity, Abdominal Obesity, Physical Activity
and Caloric Intake in U.S. Adults: 1988-2010. Am J Med, 10.1016/j.amjmed.2014.02.026 (2014). * Wong, M. Y., Day, N. E., Luan, J. A., Chan, K. P. & Wareham, N. J. The detection of
gene-environment interaction for continuous traits: should we deal with measurement error by bigger studies or better measurement? Int J Epidemiol 32, 51–57 (2003). Article CAS PubMed
Google Scholar * Franks, P. W., Pearson, E. & Florez, J. C. Gene-environment and gene-treatment interactions in type 2 diabetes: progress, pitfalls and prospects. Diabetes Care 36,
1413–1421, 10.2337/dc12-2211 (2013). Article CAS PubMed PubMed Central Google Scholar * Peters, T. et al. Validity of a short questionnaire to assess physical activity in 10 European
countries. Eur J Epidemiol 27, 15–25, 10.1007/S10654-011-9625-Y (2012). Article PubMed Google Scholar * Rankinen, T., Rice, T., Teran-Garcia, M., Rao, D. C. & Bouchard, C. FTO
genotype is associated with exercise training-induced changes in body composition. Obesity (Silver Spring) 18, 322–326, 10.1038/oby.2009.205 (2010). Article Google Scholar * Rampersaud, E.
et al. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch Intern Med 168, 1791–1797, 10.1001/archinte.168.16.1791 (2008). Article
PubMed PubMed Central Google Scholar * Gray, J. et al. Hyperphagia, severe obesity, impaired cognitive function and hyperactivity associated with functional loss of one copy of the
brain-derived neurotrophic factor (BDNF) gene. Diabetes 55, 3366–3371, 10.2337/db06-0550 (2006). Article CAS PubMed Google Scholar * Kernie, S. G., Liebl, D. J. & Parada, L. F. BDNF
regulates eating behavior and locomotor activity in mice. Embo J 19, 1290–1300, 10.1093/emboj/19.6.1290 (2000). Article CAS PubMed PubMed Central Google Scholar * Jonsson A & F. P.
Obesity, FTO gene variant and energy intake in children. N Engl J Med 360, 1571–1572; author reply 1572 (2009). * Neel, J. V. Diabetes mellitus: a “thrifty” genotype rendered detrimental by
“progress”? Am J Hum Genet 14, 353–362 (1962). CAS PubMed PubMed Central Google Scholar * Pelleymounter, M. A. et al. Effects of the obese gene product on body weight regulation in ob/ob
mice. Science 269, 540–543 (1995). Article CAS ADS PubMed Google Scholar * Southam, L. et al. Is the thrifty genotype hypothesis supported by evidence based on confirmed type 2
diabetes- and obesity-susceptibility variants? Diabetologia 52, 1846–1851, 10.1007/s00125-009-1419-3 (2009). Article CAS PubMed PubMed Central Google Scholar * Belcher, B. R. et al.
Physical activity in US youth: effect of race/ethnicity, age, gender and weight status. Med Sci Sports Exerc 42, 2211–2221, 10.1249/MSS.0b013e3181e1fba9 (2010). Article PubMed PubMed
Central Google Scholar Download references ACKNOWLEDGEMENTS We would like to thank Sebastien Robiou du Pont, Aihua Li and Akram Alyass (Department of Clinical Epidemiology and
Biostatistics, McMaster University, Hamilton, Ontario, Canada) for their assistance in database management and statistical analyses. Sonia S. Anand holds the Heart and Stroke Foundation of
Ontario, the Michael G. DeGroote endowed Chair in Population Health and a Canada Research Chair in Ethnicity and Cardiovascular Disease, Hertzel C. Gerstein holds the Aventis PHRI Chair in
Diabetes, Salim Yusuf holds the Heart and Stroke Endowed Chair in Cardiovascular Research and David Meyre holds a Canada Research Chair in Genetics of Obesity. AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * Department of Clinical Epidemiology and Biostatistics, McMaster University, Hamilton, Ontario, Canada Hudson Reddon, Hertzel C. Gerstein, Dipika Desai, Salim Yusuf, Sonia S.
Anand & David Meyre * Population Health Research Institute, McMaster University and Hamilton Health Sciences, Hamilton General Hospital, Hamilton, Ontario, Canada Hertzel C. Gerstein,
Jackie Bosch, Dipika Desai, Salim Yusuf, Sonia S. Anand & David Meyre * Department of Medicine, McMaster University, Hamilton, Ontario, Canada Hertzel C. Gerstein, Salim Yusuf &
Sonia S. Anand * Departments of Medicine and Human Genetics, McGill University, Montreal, QC, Canada James C. Engert & Swneke D. Bailey * Madras Diabetes Research Foundation, Chennai,
India Viswanathan Mohan * ECLA—Academic Research Organization, Rosario, Argentina Rafael Diaz * Department of Pathology and Molecular Medicine, McMaster University, Hamilton, Ontario, Canada
David Meyre Authors * Hudson Reddon View author publications You can also search for this author inPubMed Google Scholar * Hertzel C. Gerstein View author publications You can also search
for this author inPubMed Google Scholar * James C. Engert View author publications You can also search for this author inPubMed Google Scholar * Viswanathan Mohan View author publications
You can also search for this author inPubMed Google Scholar * Jackie Bosch View author publications You can also search for this author inPubMed Google Scholar * Dipika Desai View author
publications You can also search for this author inPubMed Google Scholar * Swneke D. Bailey View author publications You can also search for this author inPubMed Google Scholar * Rafael Diaz
View author publications You can also search for this author inPubMed Google Scholar * Salim Yusuf View author publications You can also search for this author inPubMed Google Scholar *
Sonia S. Anand View author publications You can also search for this author inPubMed Google Scholar * David Meyre View author publications You can also search for this author inPubMed Google
Scholar CONTRIBUTIONS H.R., S.S.A., H.C.G. and D.M. designed research; H.C.G., J.C.E., V.M., J.B., D.D., R.D., S.Y. and S.S.A. conducted research; H.C.G., J.C.E., V.M., J.B., D.D., S.B.D.,
R.D., S.Y. and S.S.A. provided essential materials; H.R. and D.M. analyzed data; H.R. and D.M. wrote the manuscript; H.C.G., J.C.E., V.M., S.Y. and S.S.A. critically reviewed the manuscript
for important intellectual content; D.M. had primary responsibility for final content. All authors read and approved the final manuscript. ETHICS DECLARATIONS COMPETING INTERESTS The authors
declare no competing financial interests. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS This work is licensed under a Creative Commons Attribution 4.0
International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the
material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/ Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Reddon, H., Gerstein, H., Engert, J. _et al._ Physical activity and genetic
predisposition to obesity in a multiethnic longitudinal study. _Sci Rep_ 6, 18672 (2016). https://doi.org/10.1038/srep18672 Download citation * Received: 26 August 2015 * Accepted: 20
November 2015 * Published: 04 January 2016 * DOI: https://doi.org/10.1038/srep18672 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative







