- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Emerging evidence suggests Jumonji domain-containing proteins are epigenetic regulators in diverse biological processes including cellular differentiation and proliferation. RNA
interference-based analyses combined with gene expression profiling can effectively characterize the cellular functions of these enzymes. We found that the depletion of Jumonji
domain-containing protein 6 (JMJD6) and its paralog protein Jumonji domain-containing protein 4 (JMJD4) individually by small hairpin RNAs (shRNAs) slowed cell proliferation of mouse NIH3T3
fibroblasts. We subsequently performed gene expression profiling on both JMJD6- and JMJD4-depleted mouse NIH3T3 fibroblasts using the Affymetrix GeneChip Mouse Exon 1.0 ST Array. Here we
report the gene profiling datasets along with the experimental procedures. The information can be used to further investigate how JMJD6 and JMJD4 affect gene expression and cellular
physiology. Design Type(s) parallel group design • RNA Interference Measurement Type(s) transcription profiling assay Technology Type(s) DNA microarray Factor Type(s) biological replicate
role • shRNA Sample Characteristic(s) Mus musculus • NIH-3T3 cell Machine-accessible metadata file describing the reported data (ISA-Tab format) SIMILAR CONTENT BEING VIEWED BY OTHERS
EFFICIENT COMBINATORIAL TARGETING OF RNA TRANSCRIPTS IN SINGLE CELLS WITH CAS13 RNA PERTURB-SEQ Article 22 December 2022 SCPCOR-SEQ ENABLES CO-PROFILING OF CHROMATIN OCCUPANCY AND RNAS IN
SINGLE CELLS Article Open access 08 July 2022 INTEGRATIVE TRANSCRIPTOMIC AND PROTEOMIC PROFILING OF THE EFFECTS OF CELL CONFLUENCY ON GENE EXPRESSION Article Open access 12 June 2024
BACKGROUND & SUMMARY Jumonji domain-containing proteins are iron- and 2-oxoglutarate-dependent oxygenases that act on diverse substrates including proteins, nucleic acids and small
molecules1,2. These enzymes either hydroxylate or demethylate their substrates in an oxygen-dependent manner. Many JmjC domain proteins play a key role in the epigenetic regulation of
mammalian development and of diseases such as cancer2,3. Phylogenetic analysis classifies the Jumonji domain containing proteins into several subgroups3. One subgroup comprises the
asparaginyl hydroxylase FIH4,5, the ribosomal hydroxylases MINA53 and NO66 (ref. 6) , the lysyl hydroxylase JMJD4 (ref. 7), the lysine demethylase/hydroxylase JMJD5 (refs 8,9) , the lysyl
hydroxylase/arginine demethylase JMJD6 (refs 10,11) , the tRNA hydroxylase TYW5 (ref. 12), and the enzymatically uncharacterized proteins JMJD8 and HSPBAP1. Although these enzymes share the
conserved JmjC domain with the histone lysine demethylases, they generally lack chromatin-binding domain, indicating potential functions other than modifying histones. Among the members of
this functionally diverse subgroup, JMJD6 was recently characterized as a crucial regulator for gene expression at the level of histone modification10,13, transcriptional elongation14 and
RNA splicing11,15,16. _Jmjd6_-deficient mice show various developmental defects and perinatal lethality17,18. Moreover, JMJD6 is required for angiogenesis15, adipocyte differentiation19 and
T cell proliferation20. Elevation of JMJD6 expression has been observed in breast21,22, lung23, colon24, and oral cancer25. Depletion of JMJD6 by RNA interference reduced the proliferation
of human cancer cell lines21,24. Therefore, JMJD6 is essential for cellular proliferation and differentiation. In contrast, the paralog protein JMJD4 that shares 34% sequence identity with
JMJD6 was shown to be involved in translational termination by hydroxylating the translational termination factor eRF1 (ref. 7). Other biological functions of JMJD4 have not been
demonstrated. We conducted a loss-of-function study using an RNA interference approach to reduce the levels of endogenous JMJD6 and JMJD4 in proliferating mouse NIH3T3 fibroblasts. We found
that depletion of either JMJD6 or JMJD4 alone significantly reduced cell proliferation. In order to determine whether and how these JmjdC domain proteins affect gene expression on a global
scale, we performed gene expression profiling on the JMJD6 and JMJD4 knockdown NIH3T3 cells using the Affymetrix Exon Arrays. In this report, we provide a detailed description of the gene
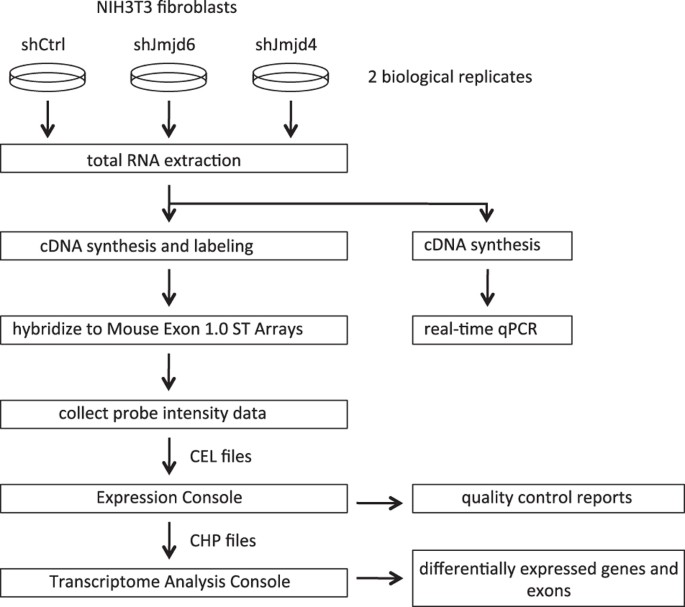
expression profiling datasets. METHODS The scheme of the experimental procedures is presented in Fig. 1. CELL CULTURE The mouse NIH3T3 fibroblast cell line was maintained in DMEM high
glucose medium (Invitrogen) containing 10% calf serum (Sigma) and 100 U ml−1 penicillin/streptomycin (Invitrogen). Human 293T cells were maintained in DMEM high glucose medium containing 10%
fetal calf serum (Sigma) and 100 U ml−1 penicillin/streptomycin (Invitrogen). To evaluate cell proliferation, 1×105 cells were seeded in 6-well plates, and the number of viable cells was
counted every other day with a hemocytometer. PLASMID CONSTRUCTION AND VIRUS TRANSDUCTION The preparation of small hairpin RNA (shRNA) lentiviral constructs was performed as previously
described (Campeau, 2009). The shJMJD6 target sequence is 5′-GCTGACACCCAGAGAACAA-3′. The shJMJD4 target sequence is GAAGAATTCTGGAGAGCAT. The control shRNA sequence is
TGGCGGCGAGTGAAGTACGTGATAA. The shRNA lentiviral constructs were co-transfected with pLP1, pLP2 and pVSVG packaging vectors into 293T cells with Lipofectamine 2000 reagent (Invitrogen). The
viral supernatant was harvested after 48 h incubation and filtered through a 0.45 μm syringe filter (Millipore). To infect NIH3T3 cells, one million cells were incubated with 2 ml of the
filtered viral supernatant supplemented with 4 μg ml−1 of polybrene (Sigma) for 48 h. The infected cells were subsequently selected in media containing 2.5 μg ml−1 puromycin (Invitrogen) for
7 days. RNA ISOLATION AND REAL-TIME QPCR ANALYSIS One million cells were plated overnight in 10 cm culture dishes. The cells were washed with 5 ml cold PBS and then lysed directly in the
culture dish by adding 1 ml of TRIzol reagent (Invitrogen). Total RNA was isolated from TRIzol lysates using RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. An
on-column DNAse digestion step is included in this protocol. cDNA was prepared from 1 μg of total RNA by Superscript III reverse transcriptase kit (Invitrogen). Quantitative PCR was
performed on a StepOne Plus real-time PCR machine with Fast SYBR Green Master mix (Applied Biosystems). The relative gene expression levels were calculated as 2^ (Ct_Eef1a1_−Ct_gene_) and
were normalized to the scrambled shRNA control as indicated. The primers are listed below: _Eef1a1_ (forward): 5′-AGCTTCTCTGACTACCCTCCACTT-3′ _Eef1a1_ (reverse): 5′-GACCGTTCTTCCACCACTGATT-3′
_Jmjd6_ (forward): 5′-GTTCCAGCTCGTCAGACTCG-3′ _Jmjd6_ (reverse): 5′-TGCCCCTAAGACATGACCAC-3′ _Jmjd4_ (forward): 5′-TCCTGCTGGAATGTCGCACCTGT-3′ _Jmjd4_ (reverse): 5′-ACCCCAAATAGGGACCGGAGGC-3′
MICROARRAY ANALYSIS The RNA samples were processed at the Genomics Core Facility of UMass Medical School. RNA quantity and quality were assessed using an Agilent Bioanalyzer 2100. cDNA was
synthesized from 500 ng RNA using the Ambion WT kit (Life Technologies) and the single stranded cDNA was prepared using the Affymetrix GeneChip WT Terminal Labeling Kit (Affymetrix)
according to the manufacturer’s instructions. The labeled mix was hybridized to GeneChip Mouse Exon 1.0 ST Array (Affymetrix) in the Gene Chip Hybridization Oven 640 overnight. Probe
intensities were measured using the Affymetrix GeneChip Scanner 3000 7G. The probe cell intensity data (CEL) from GeneChip Mouse Exon 1.0 ST Arrays was analyzed in the Affymetrix Expression
Console software to generate CHP files using the Robust Multichip Analysis (RMA)-sketch algorithm workflow. The transcript structure confidence levels for both gene and exon level analyses
were set as Core, which limits analysis to exon-level probe sets that map to BLAT alignments of mRNA with annotated full-length coding sequence (CDS) regions. Differentially expressed genes
and exons were identified by Transcriptome Analysis Console (TAC) 3.0 software (Affymetrix). DATA RECORDS Gene expression profiling on RNA samples collected from the control shRNA-treated
(shCtrl_R1 and shCtrl_R2), JMJD6 knockdown (shJMJD6_R1 and shJMJD6_R2), JMJD4 knockdown (shJMJD4_R1 and shJMJD4_R2) cells was performed using GeneChip Mouse Exon 1.0 ST Arrays. Two
biological replicates were performed. Both gene level and exon level expression were analyzed. All samples and datasets are described in Table 1. The primary data are available at the NCBI
Gene Expression Omnibus (GEO) under the accession numbers(Data Citation 1). TECHNICAL VALIDATION CONFIRMATION OF THE SHRNA-MEDIATED KNOCKDOWN The shRNA-mediated knockdown reduced cell
proliferation is shown in Fig. 2a. The signal intensity values of expression from the _Jmjd6_ and _Jmjd4_ genes in the microarray datasets are shown in Fig. 2b. The expression levels from
the endogenous _Jmjd6_ and _Jmjd4_ genes were confirmed by real-time qPCR and are shown in Fig. 2c. QUALITY CONTROL OF MICROARRAY DATA The probe cell intensity values in each individual
array were generated from the Affymetrix Expression Console software and are presented in the box plot (Fig. 3a). The data indicate that the probe cell intensities from the individual arrays
were similar. The reproducibility of the microarray results is shown by the correlation analyses on the biological replicates. The correlation between the samples at the gene level (Fig.
3b) and at the exon level (Fig. 3c) are presented in the scatter plots with the squared Pearson correlation coefficient (R2). All pairs of biological replicates have very high correlation
(R2≥0.98 for gene level analysis; R2≥0.96 for exon level analysis). Knockdown of JMJD4 has more profound impact on gene expression than knockdown of JMJD6 (R2=0.97 versus R2=0.99 for gene
level analysis; R2=0.96 versus R2=0.98 for exon level analysis). USAGE NOTES The full quality control report can be accessed using the Affymetrix Expression Console software. The
differentially expressed genes and exons can be identified using the Affymetrix Transcriptome Analysis Console software. ADDITIONAL INFORMATION HOW TO CITE THIS ARTICLE: Hu, Y.-J. &
Imbalzano, A. N. Global gene expression profiling of JMJD6- and JMJD4-depleted mouse NIH3T3 fibroblasts. _Sci. Data_ 3:160022 doi: 10.1038/sdata.2016.22 (2016). REFERENCES REFERENCES *
Klose, R. J., Kallin, E. M. & Zhang, Y. JmjC-domain-containing proteins and histone demethylation. _Nat. Rev. Genet._ 7, 715–727 (2006). Article CAS Google Scholar * Loenarz, C. &
Schofield, C. J. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. _Trends. Biochem. Sci._ 36, 7–18 (2011). Article
CAS Google Scholar * Johansson, C. et al. The roles of Jumonji-type oxygenases in human disease. _Epigenomics_ 6, 89–120 (2014). Article CAS Google Scholar * Hewitson, K. S. et al.
Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. _J. Biol. Chem._ 277, 26351–26355 (2002).
Article CAS Google Scholar * Lando, D. et al. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. _Genes. Dev._ 16,
1466–1471 (2002). Article CAS Google Scholar * Ge, W. et al. Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. _Nat. Chem. Biol._ 8, 960–962 (2012). Article
CAS Google Scholar * Feng, T. et al. Optimal translational termination requires C4 lysyl hydroxylation of eRF1. _Mol. Cell_ 53, 645–654 (2014). Article CAS Google Scholar * Hsia, D. A.
et al. KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation. _Proc. Natl Acad Sci. USA_ 107, 9671–9676 (2010). Article ADS
CAS Google Scholar * Youn, M. Y. et al. JMJD5, a Jumonji C (JmjC) domain-containing protein, negatively regulates osteoclastogenesis by facilitating NFATc1 protein degradation. _J. Biol.
Chem._ 287, 12994–13004 (2012). Article CAS Google Scholar * Chang, B., Chen, Y., Zhao, Y. & Bruick, R. K. JMJD6 is a histone arginine demethylase. _Science_ 318, 444–447 (2007).
Article ADS CAS Google Scholar * Webby, C. J. et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. _Science_ 325, 90–93 (2009). Article ADS CAS
Google Scholar * Noma, A. et al. Expanding role of the jumonji C domain as an RNA hydroxylase. _J. Biol. Chem._ 285, 34503–34507 (2010). Article CAS Google Scholar * Unoki, M. et al.
Lysyl 5-hydroxylation, a novel histone modification, by Jumonji domain containing 6 (JMJD6). _J. Biol. Chem._ 288, 6053–6062 (2013). Article CAS Google Scholar * Liu, W. et al. Brd4 and
JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. _Cell_ 155, 1581–1595 (2013). Article CAS Google Scholar * Boeckel, J. N. et al. Jumonji
domain-containing protein 6 (Jmjd6) is required for angiogenic sprouting and regulates splicing of VEGF-receptor 1. _Proc. Natl Acad Sci. USA_ 108, 3276–3281 (2011). Article ADS CAS
Google Scholar * Heim, A. et al. Jumonji domain containing protein 6 (Jmjd6) modulates splicing and specifically interacts with arginine-serine-rich (RS) domains of SR- and SR-like
proteins. _Nucleic Acids Res._ 42, 7833–7850 (2014). Article CAS Google Scholar * Kunisaki, Y. et al. Defective fetal liver erythropoiesis and T lymphopoiesis in mice lacking the
phosphatidylserine receptor. _Blood_ 103, 3362–3364 (2004). Article CAS Google Scholar * Bose, J. et al. The phosphatidylserine receptor has essential functions during embryogenesis but
not in apoptotic cell removal. _J. Biol._ 3, 15 (2004). Article Google Scholar * Hu, Y. J. et al. Transcriptional and post-transcriptional control of adipocyte differentiation by Jumonji
domain-containing protein 6. _Nucleic Acids Res._ 43, 7790–7804 (2015). Article CAS Google Scholar * Chen, C. F. et al. Regulation of T cell proliferation by JMJD6 and PDGF-BB during
chronic hepatitis B infection. _Sci. Rep._ 4, 6359 (2014). Article CAS Google Scholar * Lee, Y. F. et al. JMJD6 is a driver of cellular proliferation and motility and a marker of poor
prognosis in breast cancer. _Breast Cancer Res._ 14, R85 (2012). Article CAS Google Scholar * Poulard, C. et al. Role of JMJD6 in Breast Tumourigenesis. _PLoS One_ 10, e0126181 (2015).
Article Google Scholar * Zhang, J., Ni, S. S., Zhao, W. L., Dong, X. C. & Wang, J. L. High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. _Tumour Biol._ 34,
2397–2401 (2013). Article CAS Google Scholar * Wang, F. et al. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. _PLoS Biol._ 12, e1001819 (2014).
Article Google Scholar * Lee, C. R. et al. Elevated expression of JMJD6 is associated with oral carcinogenesis and maintains cancer stemness properties. _Carcinogenesis_ 37, 119–128
(2015). Article Google Scholar DATA CITATIONS * Hu, Y. J., & Imbalzano, A. N. _Gene Expression Omnibus_ GSE76758 (2016) Download references ACKNOWLEDGEMENTS We thank P. Spatrick and R.
Barutcu for technical assistance. This work was supported by funding from NIH grant GM56244 to A.I. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Cell and Developmental
Biology, University of Massachusetts Medical School, Worcester, 01655, Massachusetts, USA Yu-Jie Hu & Anthony N. Imbalzano Authors * Yu-Jie Hu View author publications You can also
search for this author inPubMed Google Scholar * Anthony N. Imbalzano View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Y.H. and A.I.
designed the experiments and wrote the manuscript. Y.H. generated RNA samples and performed data analyses. CORRESPONDING AUTHOR Correspondence to Anthony N. Imbalzano. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing financial interests. ISA-TAB METADATA ISA-TAB METADATA RIGHTS AND PERMISSIONS This work is licensed under a Creative Commons Attribution
4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if
the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0 Metadata associated with this Data Descriptor is available at http://www.nature.com/sdata/ and is released under the CC0 waiver to maximize reuse.
Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Hu, YJ., Imbalzano, A. Global gene expression profiling of JMJD6- and JMJD4-depleted mouse NIH3T3 fibroblasts. _Sci Data_ 3,
160022 (2016). https://doi.org/10.1038/sdata.2016.22 Download citation * Received: 13 January 2016 * Accepted: 08 March 2016 * Published: 12 April 2016 * DOI:
https://doi.org/10.1038/sdata.2016.22 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative