- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
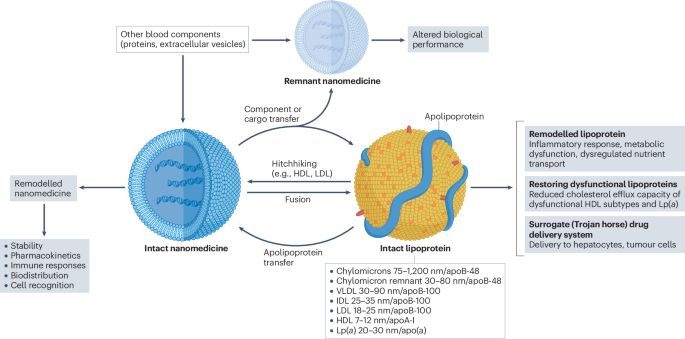
Nanomedicines interact with lipoproteins in the blood, which might result in alterations in both nanomedicine and lipoprotein structure and function. Such interactions could also be
exploited for site-specific delivery through a ‘lipoprotein Trojan Horse’ mechanism and to restore dysfunctional lipoproteins. Access through your institution Buy or subscribe This is a
preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 digital issues and online access to articles
$119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are
calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support REFERENCES * Shah, A. S., Tan, L., Long,
J. L. & Davidson, W. S. Proteomic diversity of high-density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. _J. Lipid Res._ 54, 2575–2585
(2013). Article MATH Google Scholar * Dilliard, S. A. & Siegwart, D. J. Passive, active and endogenous organ-targeted lipid and polymer nanoparticles for delivery of genetic drugs.
_Nat. Rev. Mater._ 8, 282–300 (2023). Article MATH Google Scholar * Williams, K. J., Phillips, M. C. & Rodrigueza, W. V Structural and metabolic consequences of liposome-lipoprotein
interactions. _Adv. Drug Deliv. Rev._ 32, 31–43 (1998). Article MATH Google Scholar * Courtney, H. S., Zhang, Y.-M., Frank, M. W. & Rock, C. O. Serum opacity factor, a streptococcal
virulence factor that bind to apolipoproteins A-I and A-II and disrupts high density lipoprotein structure. _J. Biol. Chem._ 281, 5515–5521 (2006). Article Google Scholar * Woodburn, K.
& Kessel, D. The alteration of plasma lipoproteins by Cremophor EL. _J. Photochem. Photobiol._ 22, 197–201 (1994). Article MATH Google Scholar * Hamad, I., Hunter, A. C. &
Moghimi, S. M. Complement monitoring of Pluronic 127 gel and micelles: Suppression of copolymer-mediated complement activation by elevated serum levels of HDL, LDL, and apolipoproteins AI
and B-100. _J. Control. Release_ 170, 167–174 (2013). Article Google Scholar * Wolfrum, C. et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. _Nat. Biotechnol._
25, 1149–1157 (2007). Article MATH Google Scholar * Sobot, D. et al. Circulating lipoproteins: A Trojan horse guiding squalenoylated drugs to LDL-accumulating cancer cells. _Mol. Ther._
25, 1596–1605 (2017). Article MATH Google Scholar * Michell, D. L. et al. Elucidation of physico-chemical principles of high-density lipoprotein-small RNA binding interactions. _J. Biol.
Chem._ 298, 101952 (2022). Article MATH Google Scholar * Guevara, J. G. Jr., Kang, D. & Moore, J. P. Nucleic acid-binding properties of low-density lipoproteins: LDL as a natural gene
vector. _J. Protein Chem._ 18, 845–857 (1999). Article MATH Google Scholar * Engelen, S. E. et al. Lipoproteins act as vehicles for lipid antigen delivery and activation of invariant
natural killer T cells. _JCI Insight_ 8, 158089 (2023). Article MATH Google Scholar * Szebeni, J. et al. Formation of complement-activating particles in aqueous solutions of Taxol:
possible role in hypersensitivity. _Int. Immunopharmacol._ 1, 721–735 (2001). Article Google Scholar * Wasan, K. M. et al. Poloxamer 407-mediated alterations in the activities of enzymes
regulating lipid metabolism in rats. _J. Pharm. Pharmaceut. Sci._ 6, 189–197 (2003). Google Scholar * Rosenson, R. S. et al. Dysfunctional and atherosclerotic cardiovascular disease. _Nat.
Rev. Cardiol._ 13, 48–60 (2016). Article Google Scholar * Kudinov, V. A. et al. High-density lipoprotein remodeling by phospholipid nanoparticles improves cholesterol efflux capacity and
protects from atherosclerosis. _Biomed. Pharmacother._ 141, 111900 (2021). Article MATH Google Scholar Download references ACKNOWLEDGEMENTS S.M.M. acknowledges support by the European
Union’s Horizon 2020 programme funded under H2020-EU.1.3. – Excellent Science – Marie Skłodowska-Curie Actions, grant agreement ID. 956544 (DIRNANO: Directing the immune response through
designed nanomaterials). D.S. acknowledges support by the National Institutes of Health grant R01AI154959. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * School of Pharmacy, Newcastle
University, Newcastle upon Tyne, UK S. Moein Moghimi * Translational and Clinical Research Institute, Faculty of Health and Medical Sciences, Newcastle University, Newcastle upon Tyne, UK S.
Moein Moghimi * Department of Pharmaceutical Sciences, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of Colorado Anschutz Medical Campus, Aurora, CO, USA S. Moein
Moghimi & Dmitri Simberg * Colorado Center for Nanomedicine and Nanosafety, University of Colorado Anschutz Medical Campus, Aurora, CO, USA S. Moein Moghimi & Dmitri Simberg *
Translational Bio-Nanosciences Laboratory, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of Colorado Anschutz Medical Campus, Aurora, CO, USA Dmitri Simberg Authors * S.
Moein Moghimi View author publications You can also search for this author inPubMed Google Scholar * Dmitri Simberg View author publications You can also search for this author inPubMed
Google Scholar CORRESPONDING AUTHOR Correspondence to S. Moein Moghimi. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW
INFORMATION _Nature Reviews Bioengineering_ thanks Jan Grimm and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Moghimi, S.M., Simberg, D. Nanomedicine–lipoprotein interactions. _Nat Rev Bioeng_ (2025). https://doi.org/10.1038/s44222-025-00291-9
Download citation * Published: 26 February 2025 * DOI: https://doi.org/10.1038/s44222-025-00291-9 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this
content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative