- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT A grand challenge in terpene synthase (TS) enzymology is the ability to predict function from protein sequence. Given the limited number of characterized bacterial TSs and
significant sequence diversities between them and their eukaryotic counterparts, this is currently impossible. To contribute towards understanding the sequence-structure-function
relationships of type II bacterial TSs, we determined the structure of the terpentedienyl diphosphate synthase Tpn2 from _Kitasatospora_ sp. CB02891 by X-ray crystallography and made
structure-guided mutants to probe its mechanism. Substitution of a glycine into a basic residue changed the product preference from the clerodane skeleton to a _syn-_labdane skeleton,
resulting in the first _syn_-labdane identified from a bacterial TS. Understanding how a single residue can dictate the cyclization pattern in Tpn2, along with detailed bioinformatics
analysis of bacterial type II TSs, sets the stage for the investigation of the functional scope of bacterial type II TSs and the discovery of novel bacterial terpenoids. SIMILAR CONTENT
BEING VIEWED BY OTHERS MOLECULAR INSIGHTS INTO THE CATALYTIC PROMISCUITY OF A BACTERIAL DITERPENE SYNTHASE Article Open access 06 July 2023 FUNCTIONAL CHARACTERIZATION AND STRUCTURAL BASES
OF TWO CLASS I DITERPENE SYNTHASES IN PIMARANE-TYPE DITERPENE BIOSYNTHESIS Article Open access 30 September 2021 STRUCTURAL INSIGHT ON ASSEMBLY-LINE CATALYSIS IN TERPENE BIOSYNTHESIS Article
Open access 09 June 2021 INTRODUCTION Terpenoids are a large family of natural products biosynthesized from five-carbon building blocks. Acyclic precursors, such as the universal precursor
for the C20 diterpenes geranylgeranyl diphosphate (GGPP, 1), are often cyclized by terpene synthases (TSs) and the resulting mono- or polycyclic hydrocarbon skeletons are subsequently
modified by a variety of tailoring enzymes leading to the extraordinary diversity of structures and biological functions seen in this class of natural products1,2,3. Terpenoids are one of
the largest and most diverse families of natural products, with over 84,000 members currently characterized4. However, terpenoids of bacterial origin represent only a small fraction (<2%)
of this family of natural products1. For example, there are over 2300 clerodane natural products known in nature, but only four are from bacteria4,5,6,7. TSs employ carbocation chemistry to
convert acyclic prenyl diphosphates into diverse terpene skeletons. These reactions are quite complex, frequently involving changes in bonding, hybridization, or configuration for most the
carbons in the substrate through a variety of reactions including carbon-carbon bond formation, hydride and alkyl shifts, and eliminations8. The conformational flexibility of terpene
substrates paired with the inherent reactivity of each cationic intermediate allows TSs to provide a diverse array of molecular templates for folding the substrate into catalytically
relevant poses and stabilizing intermediates9,10,11. Canonical bacterial TSs are categorized into type I or type II depending on how they initiate catalysis. Type I TSs, which have conserved
metal ion-binding motifs DDxxD and NSE/DTE, abstract the diphosphate group from the substrate8,12,13,14. Type II TSs utilize a highly conserved DxDD motif, in which the central Asp residue
acts as a general acid, to protonate an alkene or epoxide8,15. Due to the limited number of known bacterial terpenoids, there is a lack of general sequence-structure-function knowledge
regarding the biosynthetic machinery for bacterial TSs. Only a handful of bacterial type II TSs have been functionally characterized and only three diterpene skeletons, labdane, clerodane,
and halimane, are currently known to be produced by these enzymes (Fig. 1a and Supplementary Fig. 1). Enzymes that form labdane skeletons include the _ent_-copalyl diphosphate (CPP) synthase
PtmT2 from platensimycin and platencin biosynthesis in _Streptomyces platensis_16,17, as well as other _ent_-CPP18,19,20 and _n_-CPP21,22 synthases. The clerodane synthase Cyc1, found in
the biosynthetic gene cluster (BGC) of the antitumor antibiotic terpentecin, is proposed to bicyclize GGPP into _syn_-CPP+ before a cationic cascade that ends with deprotonation at C3 to
form terpentedienyl diphosphate (TPP, 2), or _ent_-_neo_-_trans_-_trans_-clerodienyl diphosphate (CLPP, Supplementary Fig. 1)23. Haur_2145 forms (+)-kolavenyl diphosphate, the
_ent_-_neo_-_trans-cis_-CLPP diastereomer from the _n_-CPP+ intermediate24. Rv3377c, from _Mycobacterium tuberculosis_, also forms _n_-CPP+ but quenches the cationic cascade by deprotonation
at C6 on a halimane skeleton to yield tuberculosinyl diphosphate (TbPP)25. Only two of these enzymes, PtmT2 and Rv3377c, have been structurally characterized16,26. In this study, we aimed
to understand what controls labdane, halimane, or clerodane skeletal formation in bacterial TSs. To achieve this, we structurally and biochemically investigated Tpn2, a homolog of Cyc1. We
experimentally verified Tpn2 as a TPP synthase and determined its structure by X-ray crystallography at a resolution of 2.57 Å. This is only the third structurally determined bacterial type
II TS and the first from any organism that produces a clerodane skeleton. Using the structural data, we performed a series of mutations to probe its mechanism and identified a single-residue
switch, G485D, that controls the selectivity between clerodane and labdane formation. Finally, we used bioinformatics to categorize four major families of type II TSs in bacteria, predict
labdane- vs clerodane-forming TSs in actinobacteria, and support that uncharacterized BGCs harbor novel terpenoids. This study illustrates the need for additional characterization of
bacterial TSs to expand the current scope of terpene enzymology and improve functional prediction of TSs, ultimately providing opportunities for genome mining and discovery of otherwise
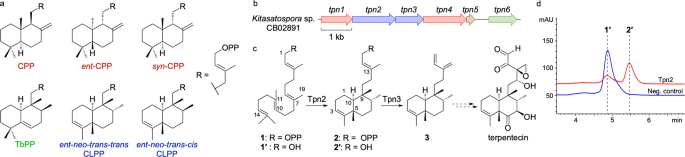
cryptic novel terpenoids. RESULTS AND DISCUSSION TPN2 IS A TERPENTEDIENYL DIPHOSPHATE SYNTHASE We identified a BGC (_tpn_) in _Kitasatospora_ sp. CB02891 identical in organization to the one
reported in _Kitasatospora griseola_ MF730-N6, the original terpentecin producer5,23,27. The _tpn_ BGC includes a GGPP synthase, two TSs, two cytochrome P450 enzymes, and a ferredoxin (Fig.
1b). Both TSs, the type II TS Tpn2 and the type I TS Tpn3, are homologous to Cyc1 and Cyc2 from _K. griseola_23,28 with 96 and 97% sequence identities, respectively, and are proposed to
form terpentetriene (3) from GGPP via TPP (2) (Fig. 1c). To confirm that Tpn2 is a TPP (2) synthase, we cloned and expressed _tpn2_ in _E. coli_ for protein production and purification
(Supplementary Fig. 2). Purified Tpn2 was incubated with GGPP and HPLC and GC-MS analysis of the enzyme reaction revealed one major product with an MS fragmentation pattern supporting a
clerodane diterpene (Supplementary Fig. 3). To simplify detection, reaction products were dephosphorylated and detected as their alcohol derivatives 1′ (geranylgeraniol, GGOH) and 2′
(_syn_-kolavenol) (Fig. 1d). To unambiguously confirm the function of Tpn2, 2 was isolated from large scale in vitro reactions and its structure was confirmed by 1H and 31P NMR analysis
(Supplementary Figs. S4 and S5 and Supplementary Table 4). FIRST CRYSTAL STRUCTURE OF A CLERODIENYL DIPHOSPHATE SYNTHASE We determined the crystal structure of Tpn2 at a resolution of 2.57 Å
(Fig. 2 and Supplementary Table 6). As seen in PtmT2 and Rv3377c16,26, Tpn2 adopts the characteristic double α-barrel βγ fold with the active site residing in the interfacial cavity. As is
typical for TSs from both eukaryotes and prokaryotes8, the active site cavity of Tpn2 is lined with both aromatic (H180, F250, W254, Y329, W384, W483, and Y489) and aliphatic (V116, A117,
T177, T181, G244, G287, L291, and G485) residues to form a hydrophobic pocket suitable for binding the long nonpolar tail of the substrate and preventing water or other nucleophiles from
prematurely quenching the reaction (Fig. 2b). At the bottom of the active site pocket lies the catalytically essential Asp-rich motif D294xDD. Docking experiments support that the central
Asp, D296, is properly positioned (3.2 Å from C14) for both the protonation of GGPP (Fig. 2c) and the deprotonation of the _syn_-cleroda-13_E_-en-4-yl+ intermediate (TPP+, Supplementary Fig.
1). D296 is also suitably positioned (2.8 Å) near H342 for activation. Tpn2 also includes other conserved residues found in type II TSs including K383, which was previously proposed to
interact with the negatively charged diphosphate group on the substrate in both plant and bacterial _ent_-CPPSs16,29. One structural feature of type II diterpene synthases that remains
ambiguous is the location of metal ions in type II TSs. Previous studies support that type II TSs are dependent on the presence of divalent ions with their absence either completely
inactivating or severely diminishing activity in both plant and bacterial TSs16,30,31. However, because divalent metal ions are not directly involved in catalysis, it is proposed that they
instead play an important role in binding the negatively charged diphosphate group of the substrate29. Tpn2, PtmT2, and Rv3377c each contain a D114xxxxE motif that resides near the opening
of the active site and is proposed to be responsible for metal-binding in bacterial type II TSs (Supplementary Fig. 6)16,26 This is supported by molecular modeling in both Tpn2 (Fig. 2c) and
Rv3377c26, as well as site-directed mutagenesis in PtmT216. As with PtmT2 and Rv3377c, no bound divalent metal ions can be inferred in the electron density of the crystal structure of Tpn2,
leaving the exact binding mechanism of divalent ions in bacterial type II TSs undetermined. Overall, Tpn2 is structurally homologous to both PtmT2 and Rv3377c with root-mean-square
deviations (rmsd) of 1.25 and 1.89 Å, respectively, for Cα atoms (Fig. 2a). Tpn2 shares 33% sequence identity and 45% similarity with PtmT2 and 26% identity and 38% similarity with Rv3377c
(Supplementary Fig. 6). A few key differences between these three enzymes give rise to changes in product selectivity. Analysis of the active site cavity of Tpn2 revealed that Tpn2 and PtmT2
possess cavities of similar size and shape; however, its pocket is much narrower than the one seen in Rv3377c (Fig. 2c–e). Most notably, W384 and L291 in Tpn2 (W380 and L290 in Rv3377c)
protrude into the domain interface resulting in a narrower cavity that likely restricts the conformation of GGPP. This establishes a template for _syn_-CPP+ formation, positioning residues
that may facilitate deprotonation near the proposed location of the cationic intermediates. Previous structural, docking, and mutation studies support that Y479 is responsible for
deprotonation of C6 of the halima-13-en-5-yl+ intermediate (TbPP+) to form TbPP (Supplementary Fig. 1)26,32. While this residue is strictly conserved across type II TSs in bacteria, its
function is not. Despite the presence of an analogous Tyr residue in Tpn2 (Y489), no detectable halimadienyl diphosphate is formed in the Tpn2 reaction. Structurally, this is supported by
the docking pose of GGPP and a distance of 6.2 Å between Y489 and C9 of GGPP (C6 of the halimane scaffold); this distance is only 2.7 Å in Rv3377c (Fig. 2c)26. With no other basic residues
appropriately positioned for deprotonation, Tpn2 forces _syn_-CPP+ through a series of methyl and hydride shifts to access the recently deprotonated D296 (Fig. 3). This mechanism, where the
central Asp acts as both initiating acid and quenching base, is consistent with labeling studies of type II mutant TSs that produce CLPPs33,34. Given the high conservation in active site
cavity shape and residues between Tpn2 and PtmT2, we reasoned that the residue at 485 controls product determination. In PtmT2 and _At_CPS, deprotonation of the CPP+ at C17 is proposed to
occur by a water molecule that is hydrogen bonded to D50216,29. Lacking an analogous Asp on the side opposing the DxDD motif, Tpn2 instead has a G485 situated 3.6 Å from C19 of the substrate
(Fig. 2c). Given the similarities in active site cavity and residues, and the position of G485 in relation to C7 of GGPP (C8 of the labdane scaffold), we hypothesized that a basic residue
at 485 would result in deprotonation of C19 and formation of CPP. STRUCTURE-GUIDED MUTAGENESIS CONVERTS TPN2 INTO A _SYN_-COPALYL DIPHOSPHATE SYNTHASE To assess whether we could stimulate
labdane production by Tpn2, we created a G485D mutant. Preliminary in vitro reactions supported that the G485D mutant produced at least two new products compared to native Tpn2
(Supplementary Fig. 7). Due to its low activity, we were unable to isolate enough product for structure determination. To overcome this challenge, we employed a terpenoid overproduction
system in _E. coli_. First, while it has been previously shown that endogenous _E. coli_ phosphatases can remove the diphosphate moiety25,35, we envisaged that using Tpn3 to eliminate the
diphosphate group via its native type I TS activity may provide a more efficient route for the production and isolation of diterpenes. In addition, it was reported that the Tpn3 homolog Cyc2
accepts both labdane and clerodane scaffolds36, demonstrating the potential substrate promiscuity needed for the investigation of Tpn2 mutants. Second, we utilized our recently reported
MKI4 system, a GGPP overproduction system that leverages two promiscuous kinases to phosphorylate exogenously added isoprenol37,38, to further enhance diterpene production in vivo
(Supplementary Fig. 8). We built a series of plasmids with _tpn2_ and _tpn3_, including a genetically fused _tpn2_-_tpn3_ construct39, to test for terpentetriene (3) production in _E. coli_
(Supplementary Table 3). Bacterial TSs are excellent candidates for building multidomain enzymes as fusing a type II and type I TS mimics the structure of naturally occurring bifunctional
TSs from plants and fungi8,40,41. Our initial screen found that the _tpn2-tpn3_ fusion led to the highest production of 3, likely due to the increased proximity of the TS active sites
(Supplementary Fig. 9). As expected, the major product was 3, isolated at a yield of 9.0 mg L−1 and structurally confirmed via 1H and 13C NMR (Supplementary Table 4, Supplementary Figs. 3,
10, and 11). This confirmed the function of Tpn3 as a terpentetriene synthase. Two additional products, 6 and 7, were also isolated with GC-MS and NMR confirming them as α- and β-springene,
respectively (Supplementary Figs. 12–14). Consistent with our results, it was previously shown that Cyc2 eliminates the diphosphate moiety from GGPP, resulting in the formation of various
acyclic diterpenes36. With an efficient diterpene production strategy in place, we cloned the Tpn2G485D-Tpn3 fusion construct and expressed it in the MKI4 system. GC-MS analysis of the crude
extract showed the production of three new products in addition to 3, 6, and 7 (Supplementary Fig. 12). All three new products showed fragmentation patterns representative of labdane
diterpenes (Supplementary Fig. 15). The major product, 5, exhibited an _m/z_ of 272.15; the two minor products exhibited _m/_z values of 290.39 and 272.28, respectively. NMR and optical
rotation analysis of 5, which was isolated at a yield of 8.0 mg L−1, revealed it to be _syn_-sclarene (Fig. 3, Supplementary Figs. 15–20, and Supplementary Table 5, also referred to as
griseolaene)36,42. The formation of _syn_-sclarene by Tpn2G485D-Tpn3 indicates that Tpn2G485D preferentially forms _syn_-CPP (4). Thus, G485D acts as a single-residue switch, exhibiting sole
control over the formation of the clerodane TPP or labdane _syn_-CPP (Fig. 3). We also probed if mutation to a Glu residue would affect the formation of 5. Tpn2G485E-Tpn3 also produced both
3 and 5, but at a ratio of 2.3:1 (Supplementary Fig. 21); Tpn2G485D-Tpn3 gave a ratio of 1.6:1. It is interesting that Tpn2G485D and Tpn2G485E are still able to produce 2, albeit not
surprising given that they retain both the active site contour and D296. Previous quantum chemical calculations also suggested that there is not a large energetic barrier between the initial
bicyclization of GGPP into _syn_-CPP+ and the final methyl shift to form the _syn_-cleroda-13_E_-en-4-yl+ final intermediate43. Conversely, although Tpn2 has all the machinery necessary to
produce the halimane diterpene TbPP, i.e., D296 for initiation, a non-basic residue at G485 (the analogous residue in Rv3377c is A475), and the proposed base Y489 (Y479 in Rv3377c), Tpn2
does not produce any halimane compounds. In addition, the Tpn2Y489A mutant is completely inactive (Supplementary Fig. 7). This supports that Y489 is not involved as a general base in the
Tpn2 reaction, but instead may play an important role in forming the active site contour. Therefore, it is evident that the structural differences between Tpn2 and Rv3377c lead to
significant differences in substrate binding and therefore control product specificity in these two TSs. SITE-DIRECTED MUTAGENESIS SUPPORTS THE MECHANISM OF CLERODANE BIOSYNTHESIS IN
BACTERIA While the residue analogous to H180 in Tpn2 is conserved among labdane synthases from plants, clerodane synthases in plants generally include a Phe or Tyr residue in this
position44,45. Exchanging this His in some plant TSs switched product selectivity from CPP to a CLPP or halimane diphosphate (HPP). For example, AtCPSH263Y switched activity from _ent_-CPP
to (–)-kolavenyl diphosphate (_neo_-_trans-cis_-CLPP)34 and OsCPS4H501F from _syn_-CPP to _syn_-HPP43. Correspondingly, SdCPS2F255H from _Salvia divinorum_ switched its activity from
neo-_trans_-_cis_-TPP to _ent_-CPP46. It is therefore intriguing that Tpn2 possesses a His residue at this position despite being a clerodane synthase. We created the Tpn2H180F and Tpn2H180V
mutants to examine if it would affect product formation; however, these mutants were inactive (Supplementary Fig. 7). These results imply that while this residue may give insight into
product selectivity in plant TSs, the same logic is not applicable to bacterial TSs. We made additional mutations in the Tpn2 active site to probe the importance of various residues for
catalysis (Supplementary Fig. 7). As expected, mutation of the central Asp, D296A, results in complete inactivation of the enzyme. It has been proposed that the central Asp is activated by
water in cyclases such as SHC or by an adjacent His residue in PtmT216,47. Accordingly, mutation of H342 to Ala or Phe abolished any product formation. These results, along with the
structural data, support that D296 is responsible for initiation via protonation and that H342 is required to activate D296 for catalysis. Finally, mutation of the conserved K383 also
resulted in inactivation of the enzyme (Supplementary Fig. 7), consistent with previous studies that suggested this residue is involved in diphosphate binding16. BIOINFORMATICS ANALYSIS OF
TYPE II TSS HIGHLIGHTS TERPENOID BIOSYNTHETIC POTENTIAL IN BACTERIA With a single residue mutation, Tpn2G485D became the first _syn_-CPP synthase from bacteria. Such a simple change supports
the notion that bacteria possess uncharacterized type II TSs that possess new functions. To assess this hypothesis, we performed a comprehensive bioinformatics analysis of all canonical
type II TSs in bacteria. Using the Enzyme Function Initiative - Enzyme Similarity Tool48, a total of 964 type II TSs were identified from a BLAST search of Tpn2 given a minimum e-value
threshold of 5; 876 of these were unique sequences. These TSs separated into four major subfamilies in a sequence similarity network (SSN) at an e-value of 10−60 (Fig. 4a). Tpn2 clustered
with actinobacterial CPP synthases (384 members), gibberellin-associated _ent_-CPP synthases clustered with Haur_2145 (336 members), Rv3377c was found in its own subfamily consisting of
terrabacterial TSs (126 members), and one family, of mainly actinobacterial TSs, has no characterized enzymes (108 members) but is highly similar to squalene-hopene cyclases (SHCs) from
other bacteria49. At an e-value of 10−88, Tpn2, the _n-_CPP synthases, and Haur_2145 were each found within distinct clusters (Fig. 4b). Sequence analysis of these four major subfamilies
revealed both conservation and variation of key motifs and residues (Fig. 4). As expected for type II TSs, the central Asp is almost 100% conserved in all families. In the Tpn2/CPP synthase
family, H180 (95.6%) and Y489 (96.9%) are all highly conserved; this is consistent with mutations at these positions rendering Tpn2 inactive. The residue at position 485 is either Asp
(84.1%) or Gly (13.3%) implicating that most of these TSs form labdanes. In the Rv3377c family, the basic Y479 (92.1%) is conserved; A475, the structurally analogous position to G485 in
Tpn2, shows more variation with Gly (58.7%), Ala (25.4%), Asp (6.3%), and Ser (3.2%). Overall, this is consistent with most members of this family forming a halimane skeleton. Contradictory
to the actinobacterial CPP synthases, the cluster containing the gibberellin-associated _ent_-CPP synthases and Haur_2145 mostly possess Gly (91.1%) at the position analogous to G485. While
this makes sense for the clerodane-forming Haur_2145, this suggests that the active site pocket of the gibberellin-associated TSs may be altered from the actinobacterial orthologues.
Finally, the putative SHCs have significantly different sequences (≅30% identity with ≅30% coverage) than the characterized TSs although they have strictly conserved (>99%) Glu and Tyr in
a similar C-terminal motif. To assess the diversity of BGCs containing the 964 type II TSs, we constructed a genome neighborhood network (GNN) from the SSN at an e-value of 10−88
(Supplementary Fig. 22)48. The subfamily with the most genetic diversity is the actinobacterial _ent_-CPPS cluster. Of the 342 BGCs, 67% encode a type I TS and 14% encode a likely UbiA
cyclase; this implies that the remaining 67 BGCs do not use a second cyclase, unless a noncanonical TS is utilized14. Many (44%) of the BGCs that contain homologs of Rv3377c have equivalents
of Rv3378c, the adenosine prenylation enzyme50, although 33% of these contain type I TSs. The family containing Haur_2145 is similar with 21% containing Rv3378c homologs and 36% containing
type I TSs. The BGCs containing Tpn2/Cyc1, _n_-CPP synthases, gibberellin-associated _ent_-CPP synthases, and SHCs are all extremely conserved in gene presence and organization supporting
production of tepentecin, cyslabdan/labdanmycins, gibberellins and hopanoids, respectively23,51,52,53,54. These findings suggest that there is more biosynthetic diversity than what is
currently known for bacterial diterpenoids, e.g., _ent_-CPP may not always be transformed into the polycyclic kauranes, atisanes, and pimaranes of currently known bacterial diterpenoids. Add
this to the recent discoveries of a monodomain type II TS from cyanobacteria55 and a bifunctional sesqui-TS that belongs to the haloacid dehalogenase-like hydrolase superfamily56 and the
terpenoid biosynthetic potential of bacteria is likely severely underestimated. In summary, we determined the structure of the TPP synthase Tpn2, a clerodane-forming TS, and mapped out
residues that are essential for catalysis and those that control the selectivity of product formation. We identified a single-residue switch, G485D, that converted a clerodane-specific TS
into a labdane-selective TS. This residue is different from those previously seen in plant TSs that controlled clerodane vs labdane formation, highlighting the sequence-function differences
between eukaryotic and prokaryotic TSs and illustrating the need to continue characterizing known and new bacterial TSs. Notably, TpnG485D is the first _syn-_CPP synthase from bacteria. This
strongly implies that the functional scope of bacterial TSs is much broader than currently known and supports continued discovery of novel TSs and terpenoids in bacteria. METHODS GENERAL
EXPERIMENTAL PROCEDURES 1H and 13C NMR experiments were performed in D2O or CDCl3 at 600 MHz for 1H and 151 MHz for 13C nuclei on a Bruker Avance III Ultrashield 600. 2D NOE experiments were
performed on a Bruker Avance-III-800 console equipped with a Bruker 18.8 T/54 mm Ascend Magnet at 25 °C. Preparative HPLC was conducted using an Agilent 1260 Infinity II LC equipped with an
Agilent Eclipse XDB-C18 column (250 mm × 21.2 mm, 7 μm). Analytical HPLC was performed on an Agilent 1260 Infinity II system equipped with an Agilent 5 HC-C18(2) column (150 × 4.6 mm, 5
μm). GC-MS experiments were run on a ThermoScientific Trace GC Ultra spectrometer equipped with an Rxi-5MS column (Restek Corp., 30 m × 0.25 mm i.d., 0.25 μm df). The injection temperature
was 250 °C, electron ionization was performed with 70 eV, ion source temperature was 250 °C, transfer line temperature was 280 °C, and mass scan range was from m/z 30–500 at 1500 μ s−1. The
program held at 50 °C for 3 min, increased the temperature at a rate of 20 °C min−1 up to 300 °C, and then was maintained at 300 °C for 3 min. Optical rotations were measured on a JASCO
P-1010 polarimeter. GENE CLONING Strains and plasmids used in this study are listed in Supplementary Tables 1 and 2, respectively. The _tpn2_ gene from _Kitasatospora_ sp. CB02891 (UniProt
accession A0A2M9LDX2, cloned starting at Met9) was amplified by PCR from genomic DNA using Q5 DNA polymerase (NEB) with primers pRSFtpn2_F and pRSFtpn2_R (Supplementary Table 3) following
the manufacturer’s protocols. The PCR product was purified and treated with T4 polymerase for cloning into pBS3080 (ref. 57) following ligation independent procedures58 to create pJR2001.
For site-directed mutagenesis, the _tpn2_ gene was amplified in two steps by primer extension59 using primers pRSFtpn2_F and pRSFtpn2_R, with internal primers containing the desired
mutation(s) as listed in Supplementary Table 3. The mutant genes were then cloned into pBS3080 as described previously to yield pJR2002–pJR2012. The _tpn2_ G485E mutant was amplified by
primer extension using primers Tpn2-3_F and Tpn2_HindIII_R and internal primers containing the mutation. The gene was then cloned into pCDF-Duet at the _Asc_I and _Hind_III sites using T4
DNA Ligase to afford pJR2019. For in vivo production, _tpn2_ (Tpn2 = A0A2M9LDX2), _tpn3_ (Tpn3 = A0A2M9LE16), and _tpn6_ (Tpn6 = A0A2M9LDW8) were amplified from _Kitasatospora_ sp. CB02891
genomic DNA using Q5 DNA polymerase and primers Tpn2-3_F and Tpn1-3_R for _tpn2_ and _tpn3_ and primers Tpn6_F and Tpn6_R for _tpn6_. _tpn2/tpn3_ and _tpn6_ were inserted into separate
multiple cloning sites in pET-Duet via restriction enzyme digestion using _Asc_I and _Hind_III for the former and _Nde_I and _Pac_I for the latter, followed by ligation via T4 DNA Ligase.
For the addition of a second ribosome-binding site and fusion protein construction, _tpn2_ was amplified from pJR2001 using Q5 DNA polymerase and primers NcoI_ala_Tpn2_F and either
Tpn2_RBSOP_R or Tpn2_GSlink_R, respectively. The full length _tpn3_ gene was similarly amplified using either primer RBSOP_Tpn3_F or GSlink_Tpn3_F and Tpn3_Hind_R for addition of a second
ribosome binding site or flexible linker upstream of Tpn3. Each construct was amplified by primer extension with primers NcoI_ala_Tpn2_F and Tpn3_Hind_R and inserted into pCDF-Duet
linearized with _Nco_I using T5 exonuclease-dependent assembly (TEDA)60, affording pJR2015. For antibiotic selection compatibility, wild-type (WT) _tpn2_ and the G485D mutant were subcloned
by digestion of pJR2001 and pJR2011 with _Bam_HI and _Pac_I and ligated into pCDF-Duet to form pJR2014 and pJR2015, respectively. For construction of the _tpn2_ G485D and G485E mutant fusion
proteins, mutant _tpn2_ was amplified from pJR2011 and pJR2019, respectively, using primers Tpn2-3_F and Tpn2_GSlink_R. The full length _tpn3_ gene was amplified from pJR2015 using primers
GSlink_Tpn3_F and Tpn1-3_R. The _tpn2_ mutants and _tpn3_ were amplified together by primer extension using primers Tpn2-3_F and Tpn1-3_R and similarly cloned into the _Asc_I and _Hind_III
sites affording pJR2018 and pJR2020. GENE EXPRESSION AND PROTEIN PRODUCTION For in vitro activity assays, pJR2001 was transformed into _E. coli_ NiCo cells (NEB) and grown in 500 mL of
lysogeny broth (LB) at 37 °C with shaking at 200 rpm until an OD600 of 0.6 was reached. The culture was cooled on ice before gene expression was induced by addition of 0.5 mM isopropyl
β-d-1-thiogalactopyranoside (IPTG). Flasks were grown overnight at 16 °C before the cells were harvested by centrifugation at 5000 × _g_ for 20 min at 4 °C. The cell pellet was prepared for
purification by resuspending in lysis buffer (50 mM Tris, 300 mM NaCl, and 10 mM imidazole, pH 8.0) and 1 mg mL−1 lysozyme before incubating on ice for 30 min. Cells were then sonicated and
centrifuged at 10,000 × _g_ for 30 min to separate soluble proteins before filtering with a 0.8 µM filter. The lysate was purified by nickel affinity chromatography and size exclusion
chromatography using an AKTA FPLC with a 5 mL HisTrap column and Superdex HiScale 16/40 80 mL column (GE Healthcare), respectively. Purified Tpn2 was concentrated using an Amicon Ultra-15
concentrator (Millipore) in SEC buffer (50 mM Tris, pH 8.0) before storing aliquots at −80 °C. Protein concentration was calculated at 280 nm using a molar absorptivity constant of 98,320
M−1 cm−1. Each of the Tpn2 site-directed mutants was produced and purified as described above using nickel affinity chromatography. CRYSTALLIZATION, DATA COLLECTION, AND STRUCTURE
DETERMINATION OF TPN2 Purified Tpn2 was concentrated to 30.0 mg mL−1 in 20 mM Tris buffer (pH 8.0) with 100 mM NaCl and crystallized using the hanging drop vapor-diffusion method in a screen
condition: 10 mM zinc sulfate heptahydrate, 25% v/v polyethylene glycol monomethyl ether 550, and 100 mM MES monohydrate, pH 6.5. The Tpn2 crystals were observed after two weeks. The
crystals were transferred to cryoprotectant solution containing 20% glycerol prior to the X-ray data collection. The diffraction data of Tpn2 were collected at National Synchrotron Radiation
Research Center (Taiwan) on the 15A1 beamline using a wavelength of 0.9732 Å with the ADSC QUANTUM 315r CCD detector. Diffraction data were indexed and scaled with HKL200061. The structure
of Tpn2 was solved by the molecular replacement using the structure of PtmT2 (PDB entry 5BP8) as a search model16. The model was refined with REFMAC62. The atomic coordinates and structure
factors of Tpn2 were deposited in the Protein Data Bank (PDB) with the accession code 7XKX (Supplementary Data 1). Data processing and refinement statistics are summarized in Supplementary
Table 6. ENZYMATIC ACTIVITY OF TPN2 GGPP was synthesized as previously described in the literature63,64. In vitro reactions were performed in 50 mM Tris pH 6.8, containing 1 mM MgCl2, 5 mM
β-mercaptoethanol (BME), 10% glycerol, 4 mM GGPP, and 20 µM Tpn2 in a total volume of 50 µL. Reactions were incubated for 16 h at 30 °C before undergoing dephosphorylation of reaction
products via addition of Quick-CIP (NEB) and incubation following the manufacturer’s protocols. The reaction mixture was then gently extracted with 100 µL acetonitrile and 50 µL saturated
NaCl before centrifugation for 1 min. The organic layer was then removed and injected on an HPLC system using an isocratic method of 95% acetonitrile in H2O. The substrate and product were
detected at 210 nm as the dephosphorylated analogs 1′ and 2′ and eluted at 5.4 min and 6.0 min, respectively. TPP (2) was isolated by scaling up in vitro reactions to yield approximately 0.5
mg of the product. Reactions were incubated for 16 h at 30 °C before extraction with equal volume of acetonitrile and placing on ice for 1 h. TPP (2) was purified via preparative HPLC using
a solvent gradient of 5–50% acetonitrile in NH4HCO3. Fractions containing 2 were combined and dried down via rotary evaporator (Supplementary Figs. 3, 4). IN VIVO PRODUCT ISOLATION AND
DETERMINATION For in vivo product isolation, pJR2014–pJR2018 were individually co-transformed along with pJR1064 into _E. coli_ BL21 Star (DE3) (Invitrogen) and grown in 9–12 L terrific
broth (TB) at 37 °C with shaking at 200 rpm until an OD600 of 1.0 was reached. The cultures were cooled on ice before gene expression was induced by the addition of 0.5 mM IPTG and 5 mM
isoprenol was supplemented in the culture. Flasks were shaken for 72 h at 28 °C before harvesting the cells by centrifugation. For the isolation 2′, 3, 5, 6, and 7, the cell pellets were
extracted with 1:1 methanol: acetone and the organic layer dried under air at room temperature. The extract was then redissolved in ethyl acetate and purified via silica chromatography with
a hexanes mobile phase before additional purification via preparative HPLC as previously described. Fractions containing the desired products were combined and dried down on a rotary
evaporator at 70 mbar at 20 °C. Compounds 3, 5, 6, and 7 were confirmed via NMR. The 1H and 13C spectra of 3 matched literature values (Supplementary Figs. 11, 12)28. The structure of
_syn_-sclarene (5) was confirmed via additional NMR experiments, including 1H and 13C, which matched literature values36, 1H-1H COSY, HSQC, and HMBC (Supplementary Figs. 13–16, Supplementary
Table 4). The absolute stereochemistry of 5 was confirmed by 2D NOESY and optical rotation experiments (Fig. 3 and Supplementary Fig. 17), the latter of which gave an \({[\alpha
]}_{{{{{{\rm{D}}}}}}}^{21}\) = +2.269 (c 0.005, CHCl3). Compound 2′ was confirmed via purification and comparison to in vitro reaction data via analysis by HPLC. Yields were calculated by
making a calibration curve using purified 3 and 5 and injecting on HPLC. Area under the curve and injection concentration was used to plot a standard curve. Tpn2-Tpn3 and Tpn2G485D-Tpn3
fusion proteins were fermented and extracted as previously described. Crude extracts were injected on HPLC and the area under the curve at λ = 210 nm was used to calculate yield.
COMPUTATIONAL MODELING The structure of the ligand, GGPP, was optimized for docking using the default parameters of MM2 energy minimization in ChemDraw 3D. This was saved in SDF format, then
converted to pdbqt in OpenBabel65. The receptor was prepared for docking in AutoDockTools 1.5.6 (ref. 66); hydrogen atoms were added and merged and Gasteiger charges calculated before
generating the final pdbqt file. The ligand and receptor were docked using AutoDock Vina67. Docking results were generated for a grid box of 30 × 25 × 20, default grid spacing of 0.375 Å,
and exhaustiveness = 20, using the coordinates of the central Asp side chain oxygen atom as the grid box center to ensure the entire active site was encompassed within the grid box.
BIOINFORMATICS The sequence alignment of plant and bacterial type II TSs obtained by aligning sequences from UniProt using ClustalW68 and visualized in ESPript 3.0 (ref. 69). Consensus
sequence logos of motifs were depicted with partial aligned sequences using WebLogo 3 (ref. 70). The collection of bacterial type II TSs for sequence similar network (SSN) analysis was
achieved using the Enzyme Function Initiative (EFI) Enzyme Similarity Tool (EST)48 with Tpn2 as the query sequence, taxonomy filter of “bacteria”, and an e-value of 5. The SSNs and genome
neighborhood networks (GNNs), created using the SSN Cluster Hub-Nodes function, were generated using EFI-EST and visualized in Cytoscape 3.9.1 (ref. 71). Taxonomic distribution of each
protein family was determined using the taxonomy function on EFI-EST. REPORTING SUMMARY Further information on research design is available in the Nature Research Reporting Summary linked to
this article. DATA AVAILABILITY All data generated during this study are available either in the main text, supplementary materials, or are deposited in online repositories. The atomic
coordinates and structure factors of Tpn2 (Supplementary Data 1) were deposited in the Protein Data Bank with the accession code 7XKX. REFERENCES * Rudolf, J. D., Alsup, T. A., Xu, B. &
Li, Z. Bacterial terpenome. _Nat. Prod. Rep._ 38, 905–980 (2021). Article CAS PubMed PubMed Central Google Scholar * Helfrich, E. J. N., Lin, G.-M., Voigt, C. A. & Clardy, J.
Bacterial terpene biosynthesis: Challenges and opportunities for pathway engineering. _Beilstein J. Org. Chem._ 15, 2889–2906 (2019). Article CAS PubMed PubMed Central Google Scholar *
Quin, M. B., Flynn, C. M. & Schmidt-Dannert, C. Traversing the fungal terpenome. _Nat. Prod. Rep._ 31, 1449–1473 (2014). Article CAS PubMed PubMed Central Google Scholar *
Dictionary of Natural Products, http://dnp.chemnetbase.com, Accessed (May 15, 2022). * Tamamura, T. et al. Isolation and characterization of terpentecin, a new antitumor antibiotic. _J.
Antibiot._ 38, 1664–1669 (1985). Article CAS Google Scholar * Kawada, S., Yamashita, Y., Fujii, N. & Nakano, H. Induction of a heat-stable topoisomerase II-DNA cleavable complex by
nonintercalative terpenoides, terpentecin and clerocidin. _Cancer Res._ 51, 2922–2925 (1991). CAS PubMed Google Scholar * Nakajima, M., Okazaki, T., Iwado, S., Kinoshita, T. &
Haneishi, T. New diterpenoid antibiotics, spirocardins A and B. _J. Antibiot._ 42, 1741–1748 (1989). Article CAS Google Scholar * Christianson, D. W. Structural and chemical biology of
terpenoid cyclases. _Chem. Rev._ 117, 11570–11648 (2017). Article CAS PubMed PubMed Central Google Scholar * Hare, S. R. & Tantillo, D. J. Dynamic behavior of rearranging
carbocations – implications for terpene biosynthesis. _Beilstein J. Org. Chem._ 12, 377–390 (2016). Article CAS PubMed PubMed Central Google Scholar * Shishova, E. Y., Di Costanzo, L.,
Cane, D. E. & Christianson, D. W. X-ray crystal structure of aristolochene synthase from _Aspergillus terreus_ and evolution of templates for the cyclization of farnesyl diphosphate.
_Biochemistry_ 46, 1941–1951 (2007). Article CAS PubMed Google Scholar * Baer, P. et al. Induced-fit mechanism in class I terpene cyclases. _Angew. Chem. - Int. Ed._ 53, 7652–7656
(2014). Article CAS Google Scholar * Aaron, J. A. & Christianson, D. W. Trinuclear metal clusters in catalysis by terpenoid synthases. _Pure Appl. Chem._ 82, 1585–1597 (2010). Article
CAS PubMed PubMed Central Google Scholar * Dickschat, J. S. Bacterial terpene cyclases. _Nat. Prod. Rep._ 33, 87–110 (2016). 2016. Article CAS PubMed Google Scholar * Rudolf, J. D.
& Chang, C. Y. Terpene synthases in disguise: Enzymology, structure, and opportunities of non-canonical terpene synthases. _Nat. Prod. Rep._ 37, 425–463 (2020). Article CAS PubMed
PubMed Central Google Scholar * Prisic, S., Xu, J., Coates, R. M. & Peters, R. J. Probing the role of the DXDD motif in class II diterpene cyclases. _ChemBioChem_ 8, 869–874 (2007).
Article CAS PubMed Google Scholar * Rudolf, J. D. et al. Structure of the ent-copalyl diphosphate synthase PtmT2 from _Streptomyces platensis_ CB00739, a bacterial type II diterpene
synthase. _J. Am. Chem. Soc._ 138, 10905–10915 (2016). Article CAS PubMed PubMed Central Google Scholar * Rudolf, J. D., Dong, L.-B. & Shen, B. Platensimycin and platencin:
Inspirations for chemistry, biology, enzymology, and medicine. _Biochem. Pharmacol._ 133, 139–151 (2017). Article CAS PubMed Google Scholar * Morrone, D. et al. Gibberellin biosynthesis
in bacteria: separate _ent_-copalyl diphosphate and _ent_-kaurene synthases in _Bradyrhizobium japonicum_. _FEBS Lett._ 583, 475–480 (2009). Article CAS PubMed Google Scholar * Hershey,
D. M., Lu, X., Zi, J. & Peters, R. J. Functional conservation of the capacity for _ent_-kaurene biosynthesis and an associated operon in certain rhizobia. _J. Bacteriol._ 196, 100–106
(2014). Article PubMed PubMed Central Google Scholar * Ikeda, C., Hayashi, Y., Itoh, N., Seto, H. & Dairi, T. Functional analysis of eubacterial _ent_-copalyl diphosphate synthase
and pimara-9(11),15-diene synthase with unique primary sequences. _J. Biochem._ 141, 37–45 (2007). Article CAS PubMed Google Scholar * Yamada, Y., Komatsu, M. & Ikeda, H. Chemical
diversity of labdane-type bicyclic diterpene biosynthesis in Actinomycetales microorganisms. _J. Antibiot._ 69, 515–523 (2016). Article CAS Google Scholar * Xu, M. et al. Characterization
of an orphan diterpenoid biosynthetic operon from _Salinispora arenicola_. _J. Nat. Prod._ 77, 2144–2147 (2014). Article CAS PubMed PubMed Central Google Scholar * Dairi, T. et al.
Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. _J. Bacteriol._ 183, 6085–6094 (2001). Article CAS PubMed PubMed Central Google
Scholar * Nakano, C., Oshima, M., Kurashima, N. & Hoshino, T. Identification of a new diterpene biosynthetic gene cluster that produces _O_-methylkolavelool in _Herpetosiphon
aurantiacus_. _ChemBioChem_ 16, 772–781 (2015). Article CAS PubMed Google Scholar * Nakano, C. & Hoshino, T. Characterization of the Rv3377c gene product, a type-B diterpene cyclase,
from the _Mycobacterium tuberculosis_ H37 genome. _ChemBioChem_ 10, 2060–2071 (2009). Article CAS PubMed Google Scholar * Zhang, Y. et al. Crystal structure and mechanistic molecular
modeling studies of _Mycobacterium tuberculosis_ Diterpene Cyclase Rv3377c. _Biochemistry_ 59, 4507–4515 (2020). Article CAS PubMed Google Scholar * Pan, G. et al. Discovery of the
leinamycin family of natural products by mining actinobacterial genomes. _Proc. Natl Acad. Sci. USA_ 114, E11131–E11140 (2017). Article CAS PubMed PubMed Central Google Scholar *
Hamano, Y. et al. Functional analysis of eubacterial diterpene cyclases responsible for biosynthesis of a diterpene antibiotic, terpentecin. _J. Biol. Chem._ 277, 37098–37104 (2002). Article
CAS PubMed Google Scholar * Köksal, M., Hu, H., Coates, R. M., Peters, R. J. & Christianson, D. W. Structure and mechanism of the diterpene cyclase _ent_-copalyl diphosphate
synthase. _Nat. Chem. Biol._ 7, 431 (2011). Article PubMed PubMed Central Google Scholar * Prisic, S. & Peters, R. J. Synergistic substrate inhibition of _ent_-copalyl diphosphate
synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. _Plant Physiol._ 144, 445–454 (2007). Article CAS PubMed PubMed Central Google Scholar * Peters,
R. J. & Croteau, R. B. Abietadiene synthase catalysis: Conserved residues involved in protonation-initiated cyclization of geranylgeranyl diphosphate to (+)-copalyl diphosphate.
_Biochemistry_ 41, 1836–1842 (2002). Article CAS PubMed Google Scholar * Lemke, C. et al. Investigation of acid–base catalysis in halimadienyl diphosphate synthase involved in
_Mycobacterium tuberculosis_ Virulence. _ACS Biol. Med. Chem. Au_. 2, 490–498 (2022). * Schulte, S., Potter, K. C., Lemke, C. & Peters, R. J. Catalytic bases and stereocontrol in
Lamiaceae class II diterpene cyclases. _Biochemistry_ 57, 3473–3479 (2018). Article CAS PubMed Google Scholar * Potter, K. C. et al. Blocking deprotonation with retention of aromaticity
in a plant _ent_-copalyl diphosphate synthase leads to product rearrangement. _Angew. Chem. - Int. Ed._ 55, 634–638 (2016). Article CAS Google Scholar * Nakano, C., Okamura, T., Sato, T.,
Dairi, T. & Hoshino, T. _Mycobacterium tuberculosis_ H37Rv3377c encodes the diterpene cyclase for producing the halimane skeleton. _Chem. Commun_. 1016–1018 (2005). * Nakano, C. et al.
Substrate specificity of the CYC2 enzyme from _Kitasatospora griseola_: production of sclarene, biformene, and novel bicyclic diterpenes by the enzymatic reactions of labdane- and
halimane-type diterpene diphosphates. _Tetrahedron Lett._ 51, 125–128 (2010). Article CAS Google Scholar * Xu, B., Ning, W., Wei, X. & Rudolf, J. D. Mutation of the eunicellane
synthase Bnd4 alters its product profile and expands its prenylation ability. _Org. Biomol. Chem._ https://doi.org/10.1039/D2OB01931K (2022). * Lund, S., Hall, R. & Williams, G. J. An
artificial pathway for isoprenoid biosynthesis decoupled from native hemiterpene metabolism. _ACS Synth. Biol._ 8, 232–238 (2019). Article CAS PubMed PubMed Central Google Scholar *
Chen, X., Zaro, J. L. & Shen, W. C. Fusion protein linkers: property, design, and functionality. _Adv. Drug Deliv. Rev._ 65, 1357 (2013). Article CAS PubMed Google Scholar * Cao, R.
et al. Diterpene cyclases and the nature of the isoprene fold. _Proteins_ 78, 2417 (2010). Article CAS PubMed PubMed Central Google Scholar * Zhou, K. et al. Insights into diterpene
cyclization from structure of bifunctional abietadiene synthase from _Abies grandis_. _J. Biol. Chem._ 287, 6840–6850 (2012). Article CAS PubMed PubMed Central Google Scholar * Jia, M.,
Potter, K. C. & Peters, R. J. Combinatorial biosynthesis and the basis for substrate promiscuity in class I diterpene synthases. _Metab. Eng._ 37, 24–34 (2016). Article CAS PubMed
PubMed Central Google Scholar * Potter, K. C., Jia, M., Hong, Y. J., Tantillo, D. J. & Peters, R. J. Product rearrangement from altering a single residue in the rice _syn_-copalyl
diphosphate synthase. _Org. Lett._ 18, 1060–1063 (2016). Article CAS PubMed PubMed Central Google Scholar * Köksal, M., Potter, K., Peters, R. J. & Christianson, D. W. 1.55
Å-resolution structure of _ent_-copalyl diphosphate synthase and exploration of general acid function by site-directed mutagenesis. _Biochim. Biophys. Acta_ 1840, 184–190 (2014). Article
PubMed Google Scholar * Pelot, K. A., Hagelthorn, D. M., Hong, Y. J., Tantillo, D. J. & Zerbe, P. Diterpene synthase-catalyzed biosynthesis of distinct clerodane stereoisomers.
_ChemBioChem_ 20, 111–117 (2019). Article CAS PubMed Google Scholar * Pelot, K. A. et al. Biosynthesis of the psychotropic plant diterpene salvinorin A: discovery and characterization of
the _Salvia divinorum_ clerodienyl diphosphate synthase. _Plant J._ 89, 885–897 (2017). Article CAS PubMed Google Scholar * Wendt, K. U., Lenhart, A. & Schulz, G. E. The structure
of the membrane protein squalene-hopene cyclase at 2.0 A resolution. _J. Mol. Biol_. 286, 175–187 (1999). * Zallot, R., Oberg, N. & Gerlt, J. A. The EFI web resource for genomic
enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. _Biochemistry_ 58, 4169–4182 (2019). Article CAS PubMed Google
Scholar * Lee, S. & Poulter, C. D. Cloning, solubilization, and characterization of squalene synthase from _Thermosynechococcus elongatus_ BP-1. _J. Bacteriol._ 190, 3808–3816 (2008).
Article CAS PubMed PubMed Central Google Scholar * Layre, E. et al. Molecular profiling of _Mycobacterium tuberculosis_ identifies tuberculosinyl nucleoside products of the
virulence-associated enzyme Rv3378c. _Proc. Natl Acad. Sci. USA_ 111, 2978–2983 (2014). Article CAS PubMed PubMed Central Google Scholar * Ikeda, H., Shin-ya, K., Nagamitsu, T. &
Tomoda, H. Biosynthesis of mercapturic acid derivative of the labdane‐type diterpene, cyslabdan that potentiates imipenem activity against methicillin-resistant _Staphylococcus aureus_:
Cyslabdan is generated by mycothiol-mediated xenobiotic detoxification. _J. Ind. Microbiol. Biotechnol._ 43, 325–342 (2016). Article CAS PubMed Google Scholar * Xiong, Z. J. et al.
Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. _Org. Chem. Front._ 5, 1272–1279 (2018). Article CAS Google Scholar * Nett, R. S. et al. Elucidation of
gibberellin biosynthesis in bacteria reveals convergent evolution. _Nat. Chem. Biol._ 13, 69–74 (2016). Article PubMed PubMed Central Google Scholar * Pan, J. J. et al. Biosynthesis of
squalene from farnesyl diphosphate in bacteria: Three steps catalyzed by three enzymes. _ACS Cent. Sci._ 1, 77–82 (2015). Article CAS PubMed PubMed Central Google Scholar * Moosmann, P.
et al. A monodomain class II terpene cyclase assembles complex isoprenoid scaffolds. _Nat. Chem._ 12, 968–972 (2020). Article CAS PubMed PubMed Central Google Scholar * Vo, N. N. Q.,
Nomura, Y., Kinugasa, K., Takagi, H. & Takahashi, S. Identification and characterization of bifunctional drimenol synthases of marine bacterial origin. _ACS Chem. Biol._ 5, 1226–1238
(2022). Article Google Scholar * Lohman, J. R., Bingman, C. A., Phillips, G. N. & Shen, B. Structure of the bifunctional acyltransferase/decarboxylase LnmK from the leinamycin
biosynthetic pathway revealing novel activity for a double-hot-dog fold. _Biochemistry_ 52, 902–911 (2013). Article CAS PubMed Google Scholar * Aslanidis, C. & de Jong, P. J.
Ligation-independent cloning of PCR products (LIC-PCR). _Nucleic Acids Res._ 18, 6069–6074 (1990). Article CAS PubMed PubMed Central Google Scholar * Ho, S. N., Hunt, H. D., Horton, R.
M., Pullen, J. K. & Pease, L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. _Gene_ 77, 51–59 (1989). Article CAS PubMed Google Scholar * Xia,
Y., Li, K., Li, J., Wang, T., Gu, L. & Xun, L. T5 exonuclease-dependent assembly offers a low-cost method for efficient cloning and site-directed mutagenesis. _Nucleic Acids Res._ 47,
e15 (2019). Article PubMed Google Scholar * Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. _Methods Enzymol._ 276, 307–326 (1997).
Article CAS PubMed Google Scholar * Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. _Acta Crystallogr. Sect.
D. Biol. Crystallogr._ 53, 240–255 (1997). Article CAS Google Scholar * Roe, S. J., Oldfield, M. F., Geach, N. & Baxter, A. A convergent stereocontrolled synthesis of [3-(14)
C]solanesol. _J. Label. Compd. Radiopharm._ 56, 485–491 (2013). Article CAS Google Scholar * Davisson, V. J. et al. Phosphorylation of isoprenoid alcohols. _J. Org. Chem._ 51, 4768–4779
(1986). Article CAS Google Scholar * O’Boyle, N. M. et al. Open Babel: An open chemical toolbox. _J. Cheminform._ 3, 1–14 (2011). Google Scholar * Morris, G. M. et al. AutoDock4 and
AutoDockTools4: Automated docking with selective receptor flexibility. _J. Comput. Chem._ 30, 2785–2791 (2009). Article CAS PubMed PubMed Central Google Scholar * Trott, O. & Olson,
A. J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. _J. Comput. Chem._ 31, 455–461 (2010). CAS PubMed
PubMed Central Google Scholar * Goujon, M. et al. A new bioinformatics anaylsis tools framework at EMBL-EBI. _Nucleic Acids Res._ 38, W695–W699 (2010). Article CAS PubMed PubMed
Central Google Scholar * Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. _Nucleic Acids Res._ 42, W320–W324 (2014). Article CAS
PubMed PubMed Central Google Scholar * Crooks, G. E., Hon, G., Chandonia, J.-M. & Brenner, S. E. WebLogo: A sequence logo generator. _Genome Res._ 14, 1188–1190 (2004). Article CAS
PubMed PubMed Central Google Scholar * Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. _Genome Res._ 13, 2498–2504 (2003).
Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS This work funded in part by NIH Grants R00 GM124461 and R35 GM142574 (JDR) and MOST Grant
110-2113-M-A49-026-MY3 (CYC). We wish to thank James Rocca and Ion Ghiviriga for excellent NMR support. We acknowledge the University of Florida’s Center for Nuclear Magnetic Resonance
Spectroscopy and the McKnight Brain Institute at the National High Magnetic Field Laboratory’s Advanced Magnetic Resonance Imaging and Spectroscopy (AMRIS) Facility, which is supported by
the US NSF Cooperative Agreement No. DMR-1644779 and the State of Florida. Some NMR spectra were acquired using a unique 1.5 mm High Temperature Superconducting Cryogenic Probe developed
with support from the NIH (R01 EB009772). We also thank Jodie Johnson and the University of Florida’s Mass Spectrometry Research and Education Center (MSREC), which is supported by the NIH
S10 OD021758-01A1, for GC-MS support. We thank the experimental facility and technical services provided by the National Synchrotron Radiation Research Center (NSRRC) in Taiwan. We thank Ben
Shen for bacterial strains. Finally, we acknowledge Alisha Das, Adriana LaVopa, and Santiago Velez for their assistance in cloning TpnG485E as part of the University of Florida’s
Course-Based Undergraduate Research Experience (CURE). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Chemistry, University of Florida, Gainesville, FL, 32611, USA Emma A.
Stowell, Michelle A. Ehrenberger & Jeffrey D. Rudolf * Department of Biological Science and Technology, National Yang Ming Chiao Tung University, Hsinchu, 30010, Taiwan, ROC Ya-Lin Lin
& Chin-Yuan Chang * Center for Intelligent Drug Systems and Smart Bio-devices, National Yang Ming Chiao Tung University, Hsinchu, 30010, Taiwan, ROC Chin-Yuan Chang Authors * Emma A.
Stowell View author publications You can also search for this author inPubMed Google Scholar * Michelle A. Ehrenberger View author publications You can also search for this author inPubMed
Google Scholar * Ya-Lin Lin View author publications You can also search for this author inPubMed Google Scholar * Chin-Yuan Chang View author publications You can also search for this
author inPubMed Google Scholar * Jeffrey D. Rudolf View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS J.D.R. conceived the project; E.A.S.,
M.A.E., C.-Y.C., and J.D.R. designed the experiments; E.A.S., M.A.E., Y.-L.L, C.-Y.C., and J.D.R. performed the experiments; E.A.S., M.A.E., Y.-L.L, C.-Y.C., and J.D.R. analyzed the results;
and E.A.S., C.-Y.C., and J.D.R. wrote the manuscript with inputs from all co-authors. CORRESPONDING AUTHORS Correspondence to Chin-Yuan Chang or Jeffrey D. Rudolf. ETHICS DECLARATIONS
COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Communications Chemistry_ thanks the anonymous reviewers for their contribution to the
peer review of this work. Peer reviewer reports are available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps
and institutional affiliations. SUPPLEMENTARY INFORMATION PEER REVIEW FILE SUPPLEMENTARY INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2
REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation,
distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and
indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to
the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will
need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE Stowell, E.A., Ehrenberger, M.A., Lin, YL. _et al._ Structure-guided product determination of the bacterial type II diterpene synthase Tpn2. _Commun Chem_ 5, 146 (2022).
https://doi.org/10.1038/s42004-022-00765-6 Download citation * Received: 19 September 2022 * Accepted: 21 October 2022 * Published: 08 November 2022 * DOI:
https://doi.org/10.1038/s42004-022-00765-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative






