- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The roles of RNA sequence/structure motifs, Packaging Signals (PSs), for regulating assembly of an HBV genome transcript have been investigated in an efficient in vitro assay
containing only core protein (Cp) and RNA. Variants of three conserved PSs, within the genome of a strain not used previously, preventing correct presentation of a Cp-recognition loop motif
are differentially deleterious for assembly of nucleocapsid-like particles (NCPs). Cryo-electron microscopy reconstruction of the _T_ = 4 NCPs formed with the wild-type gRNA transcript,
reveal that the interior of the Cp shell is in contact with lower resolution density, potentially encompassing the arginine-rich protein domains and gRNA. Symmetry relaxation followed by
asymmetric reconstruction reveal that such contacts are made at every symmetry axis. We infer from their regulation of assembly that some of these contacts would involve gRNA PSs, and
confirmed this by X-ray RNA footprinting. Mutation of the ε stem-loop in the gRNA, where polymerase binds in vivo, produces a poor RNA assembly substrate with Cp alone, largely due to
alterations in its conformation. The results show that RNA PSs regulate assembly of HBV genomic transcripts in vitro, and therefore may play similar roles in vivo, in concert with other
molecular factors. SIMILAR CONTENT BEING VIEWED BY OTHERS CRYOEM STRUCTURE AND ASSEMBLY MECHANISM OF A BACTERIAL VIRUS GENOME GATEKEEPER Article Open access 26 November 2022 MULTIPLE CAPSID
PROTEIN BINDING SITES MEDIATE SELECTIVE PACKAGING OF THE ALPHAVIRUS GENOMIC RNA Article Open access 17 September 2020 SHORT- AND LONG-RANGE INTERACTIONS IN THE HIV-1 5′ UTR REGULATE GENOME
DIMERIZATION AND PACKAGING Article Open access 28 March 2022 INTRODUCTION Hepatitis B Virus (HBV) has infected over 2 billion people worldwide1, ~240 million of whom are chronically infected
after failing to clear an acute primary infection. Within this cohort, failure to suppress the virus leads eventually to liver failure, cirrhosis and cancer, resulting in ~700,000 deaths
annually2. Despite an effective vaccine, over a million new infections occur every year3,4. For chronically infected patients therapy options are limited. Clinical therapy commonly uses
nucleos(t)ide analogue inhibitors of viral polymerase (Pol) but this rarely leads to a cure and elicits rapid resistance mutations5,6. HBV represents one of the largest health challenges of
any viral pathogen. A WHO Global Challenge has been established with the goal of making chronic infection treatable by 20307. Novel curative therapies require improved mechanistic
understanding of the HBV lifecycle. The basis of the chronic infection is a covalently-closed, circular DNA (cccDNA) copy of the viral genome which persists in the nucleus as a chromatinized
episome8,9. HBV is a para-retrovirus, i.e. a DNA virus that initially packages a positive-sense, single-stranded (ss) pre-genome, pgRNA10,11, into a nucleocapsid (NC) composed of multiple
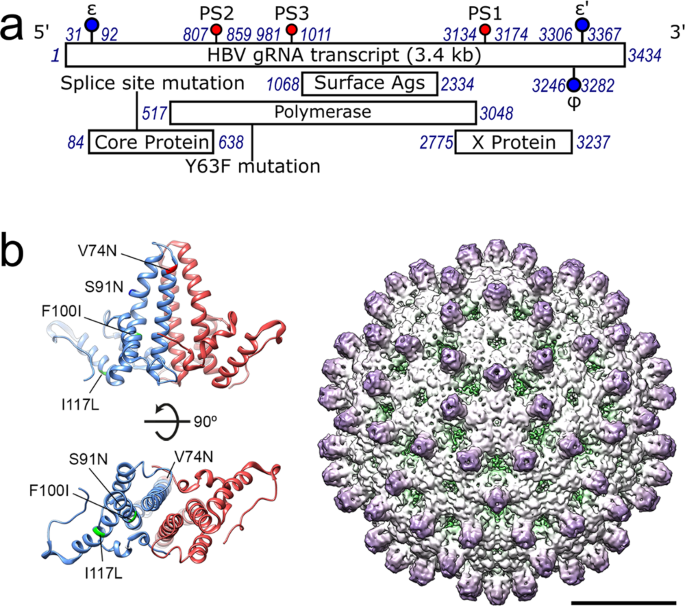
Cp dimers (Fig. 1). These form _T_ = 3 or _T_ = 4 surface lattices, the latter being dominant. The 3200 bp long genome encodes four overlapping reading frames for polymerase (Pol); surface
proteins (HBsAg); the cell regulatory factor protein X; and the core and pre-core proteins (HBcAg and HBeAg, respectively). Pol and Cp proteins are translated from the pgRNA, which also
serves as the template for reverse transcription within the NC shell. The pgRNA is a 5′-capped, terminally redundant, poly-A tailed, mRNA transcript ~3500 nts long. Previously, RNA
SELEX12,13 against recombinant full-length HBV Cp (183 aa long) dimers from strain NC_003977.1 was used to isolate aptamers whose sequences align with genomic sequences in the cognate pgRNA.
Analysis of these genomic matching sites identifies their common features. Each site has the potential to form a stem-loop with a defined loop sequence motif, -RGAG-. Such sites are highly
conserved across the pgRNAs of many strain variants. These HBV sites, as isolated RNA oligonucleotides, trigger in vitro sequence-specific NCP formation at nanomolar concentrations12. We
propose that they act as Packaging Signals (PSs) helping to ensure faithful encapsidation of the pgRNA, by analogy with the mechanism regulating assembly of many bona fide ssRNA
viruses14,15,16,17,18,19,20. Here we show that homologous PSs occur in similar genome locations in a commercially-available strain variant (JQ707375.1), which was not included in the
previous analysis. These sites regulate in vitro assembly of _T_ = 3 and _T_ = 4 nucleocapsid-like particles (NCPs) in the context of a long genomic gRNA fragment lacking a 5′ cap and a
poly-A tail. Regulation occurs at least in part through Cp recognition of the conserved loop -RGAG- motif of the PSs. Some of these contacts remain in the assembled particle. Symmetry
expansion21,22,23,24 of an icosahedrally-averaged ~3.2 Å resolution cryo-electron microscopy (cryo-EM) reconstruction of the reassembled _T_ = 4 NCPs reveals multiple contacts at all
symmetry axes between encapsidated density (the C-terminal arginine-rich domains (ARDs) of Cp’s & gRNA) and the globular Cp shell. X-ray RNA footprinting (XRF)25,26,27 confirms that the
most conserved PS makes one such contact via its -RGAG- motif. Assembly in vitro occurs in the absence of viral polymerase which binds a stem-loop (ε) on the pgRNA28, and post-translational
modifications of the Cp. Both features assist NCP formation in vivo28,29,30,31,32. Mutation to prevent ε interacting with the distal ϕ site creates a genomic fragment that is considerably
larger than both wild-type and PS mutant RNAs, as well as being a poor in vitro assembly substrate. These data highlight that PS-mediated assembly via the formation of multiple PS-Cp
contacts promotes NCP-like formation, in the absence of other mechanisms regulating assembly. RESULTS IDENTIFICATION OF PS SITES IN HBV STRAIN JQ707375.1 Evolutionarily conserved PS sites in
the pgRNA of NC_003977.1 (subtype _ayw_) were identified by aligning anti-Cp RNA aptamers against the sequences of 16 strain variant HBV pgRNAs, chosen at random from the ~750 sequences
then available in GenBank. Mfold33 suggests that each of the matched sites is potentially able to fold into a stem-loop with an over-represented sequence, 5′-RGAG-3′, in the loop12. The
three most highly conserved NC_003977.1 sites, PSs1–3, as oligonucleotides, trigger sequence/structure-specific in vitro assembly of Cp, mostly into _T_ = 4 NCPs. Sequence alignment and
Mfold33 readily identifies putative PS homologues of these sites in the JQ707375.1 RNA sequence (Fig. 2; ‘Methods’). Each of the JQ707375.134 PS homologues is predicted to fold into a
stem-loop, the latter presenting a purine-rich tetra-nucleotide motif. PSs 1 & 3, have ideal consensus Cp-recognition motifs, -GGAG- & -AGAG-, respectively, whilst the homologous
PS2, has a slight variation (-GAAG-). Their secondary structures and stabilities vary, as expected12. To investigate the PS-mediated NCP assembly hypothesis, and determine the roles of PSs
in assembly in the context of the genome, we used a transcript assembled from the commercially-available strain JQ707375.134. This was isolated from a patient with a lamivudine-resistant
infection and is therefore thought to be replication competent. The ~3400 nt assembled RNA is closer to the size of full-length pgRNA but lacks the 5′ cap and 3′ polyadenylation. It shares
90.7% nucleotide sequence identity with the same region of NC_003977.1, from which the recombinant Cp used in assembly assays was expressed12. There are just 4 amino acid sequence changes
between the Cp’s of these strains: (NC_003977.1 vs. JQ707375.1, respectively) V74N; S91N; F100I & I117L, all of which lie within the first 149 amino acids of core (Fig. 1b) which form
the outer portion of the NCP shell. None would be expected to interact with the pgRNA. NC_003977.1 Cp was therefore used in the in vitro assembly assays with the JQ707375.1 transcript. IN
VITRO ASSEMBLY WITH THE JQ707375.1 RNA TRANSCRIPT PS-mediated assembly of ssRNA viruses at nanomolar concentrations12,16,35 mimics the genome packaging specificity outcomes of natural
infections36. These conditions differ from most in vitro reassembly studies which are usually carried out at much higher concentrations37,38. Previously, PS oligonucleotide-induced HBV NCP
assembly was monitored using single-molecule (sm)fluorescence correlation spectroscopy (smFCS) with dye-labelled RNA oligonucleotides12,16,39. These assays monitor real-time changes to the
hydrodynamic radius (R_h_) of the labelled RNA, and end-products were analysed by negative stain EM (nsEM). However, the small sample sizes of such experiments prevent more detailed
analysis. To overcome that limitation here, NCPs were reassembled in 96-well plates under sm conditions using a liquid-handling robot (Fig. 3a). These samples were pooled and concentrated
(~10 fold) to allow subsequent fractionation and further analysis. Well over 90% of the input RNA was recovered from this step, presumably with the rest lost in the concentrator. The
subsequent analysis assumed that the material remaining soluble reflects the endpoint of each titration. Titrations of Cp dimer, prepared from recombinant NCPs using 1.5 M guanidine
hydrochloride (GuHCl), comprising 10 × 2 μL aliquots from one of six different stock concentrations, were made into an initial (180 μL) solution containing gRNA (1.1 nM). These dilute the
gRNA to a final concentration of 1 nM, whilst spanning sub-stoichiometric to a 4-fold molar excess for formation of a _T_ = 4 NCP around each gRNA. Each titration point was equilibrated for
10 min at room temperature (~20 °C) before addition of the next aliquot. This titration allows assembly initiation to occur at the highest affinity PSs on each gRNA molecule and then proceed
to completion37, and yields are >80% as judged by input gRNA. The final molar excess of Cp dimer ensures that any assembly defects detected are not a consequence of non-functional Cp
dimers. Assembled NCPs appear as separated, negative-stain excluding particles in TEM, with A260/280 ratios consistent with each particle containing a single, full-length copy of the
transcript (see below). The products from the titration reactions were analysed by absorption spectroscopy and the relative efficiency of complete NCP formation probed by treatment of one of
half of the sample with RNase A. Both aliquots were then fractionated by size-exclusion chromatography with the eluting material detected via multi-angle, laser light-scattering
(SEC-MALLS). Light-scattering peaks were collected, re-concentrated to ~1 mL, and their absorbance values re-measured. The SEC-MALLS signals were used to estimate assembly efficiency and the
hydrodynamic radii (R_h_) of eluting species (Fig. 3a, Table 1). For JQ707375.1 RNA transcripts the reassembly products prior to RNase treatment (Fig. 3b) elute as a single, symmetrical
peak, ~8.5 mL after application to the column. Unassembled Cp dimer, present from the titration, is much smaller than NCPs and invisible on these plots. RNase-treated aliquots elute
essentially identically with a very similar yield, although their peak has a slight low-side shoulder. Prior to chromatography, both aliquots contain identical stain-excluding particles in
nsEM, consistent with _T_ = 4 NCPs. Their apparent R_h_ values (~18–20 nm) match those for NCPs formed by Cp recombinant expression in _E. coli_ (~19 nm, Supplementary Fig. 1), and are
similar to values of ~25–32 nm determined by previous cryo-EM and smFCS measurements, respectively. The results show that NC_003977.1 Cp dimer successfully reassembles nuclease-resistant _T_
= 4 NCPs around JQ707375.1 gRNA (Table 1), validating the assumption about the common functionality of Cp’s between HBV strains. PROBING THE ROLE(S) OF PSS1-3 IN IN VITRO ASSEMBLY OF GRNA
Sequence variants were designed to investigate the role(s) of PSs in regulating assembly with JQ707375.1 transcripts in the in vitro assembly assay. Variants remove loop recognition motifs
and/or destabilize the secondary structures of the stems presenting the motif, whilst retaining wherever possible the global gRNA fold, as predicted by Mfold33. PS1 lies within an especially
G-rich (50%) region and consequently encompasses several potential alternative -RGAG- motifs. This made subtle motif ablation impossible. Instead, six G to U mutations were introduced into
the 5′ leg of the PS stem destabilising it (predicted free energy of folding going from −3.7 to −1.3 kcal mol−1), and increasing the bulge size, whilst moving the -RGAG- motif to a more
central position within the loop (Fig. 2). For PS2 and PS3, purine to U mutations can be introduced directly into the loops, replacing their recognition motifs -GAAG- and -AGAG-, with -UUAU-
and -UUUU-, respectively. It was also possible to destabilize the base-pairing in their stems via the following substitutions: in the 3′ leg of the PS2 stem-loop (-UUAAAAUUA- to -UAGCUUUG-
(nts 841–848)), and either side of the loop in PS3 (-AUAUAUUUUGGGAA- to -UUUUAUUUUGGCUU- (nts 992–1005)). These substitutions prevent formation of alternate -RGAG- motifs in either PS.
Disrupted PS2 is predicted to fold as a very stable (−8.9 kcal mol−1) stem-loop, with a multiply interrupted stem. The mutations within PS3 ablate all possible secondary structures using the
default Mfold parameters (Fig. 2). The global folding effects of these changes were estimated by folding a 300 nt region centred on each PS using a sliding 60 nt window noting the frequency
of defined structures. This procedure identified no long-range base-pairing issues and confirmed the greatly reduced occurrence of a Cp-recognition motif in a loop in the PS1 variant,
together with the complete ablation of PS2 and PS3 recognition signals and secondary structures. The effects of the variant PSs on NCP assembly were assessed in transcripts containing all
three variant PSs (ΔPS), or with individual variants (ΔPS1, ΔPS2 and ΔPS3) (Fig. 3c, Table 1). ΔPS gRNA does not assemble _T_ = 4 NCPs significantly under these conditions. Its pre-RNase
assembly product elutes later than a bona fide NCP (10 mLs after being applied to the column) and has a pronounced low-side tail. In nsEM, it appears to produce small quantities of malformed
and aggregated particles which are readily penetrated by stain. Absorbance measurements are consistent with this, the peak containing only ~10% of the wild-type amount of RNA. Particle
formation, however, remains RNA-dependent since RNase treatment eliminates all light-scattering material, revealing only a small number of apparently correctly assembled particles. This
result confirms the assumption that these sites act as PSs within JQ707375.1 gRNA. Alteration of only 24/3200 nucleotides in this RNA transcript completely prevents assembly. Since it is the
same length as the wild-type sequence, assembly cannot rely purely on favourable electrostatic interactions, as has been proposed for many single-stranded (ss) RNA viruses40,41,42,43,44,45,
i.e. it implies assembly regulation by the RNA. The sequence variations in ΔPS gRNA do not alter its hydrodynamic radius significantly (Supplementary Fig. 2) confirming that this regulation
is based on sequence-specific recognition of PS sites. ΔPS1 pgRNA also assembles poorly compared to the wild-type (Fig. 3c, Table 1), eluting (~9.5 mL vs ~8.5 mL for WT) as a roughly
symmetrical peak. The material before RNase treatment consists mostly of separate but misshapen particles of widely differing radii, which are mostly freely penetrated by negative stain. As
with the ΔPS variant, assembly is dependent on the RNA, with RNase treatment eliminating all light-scattering material, although there are many more particles roughly the size of an NCP than
with the ΔPS variant transcript. They are all freely penetrated by negative stain. These results arise from variation of just 6 nucleotides across the gRNA, and an -RGAG- motif is still
present in the smaller loop of the variant. Since the ΔPS and ΔPS1 transcripts have distinct assembly properties, PSs 2 & 3 must also contribute differentially to assembly regulation.
Analysis of their individual reassembly reactions suggests, however, that these are less significant than for PS1 (Supplementary Fig. 3). Both variants produce products that co-elute with
bona fide NCPs assembled around wild-type gRNA. They elute as symmetrical peaks containing mostly stain-excluding, single particles. Their hydrodynamic radii are also very close to that of
wild-type (~17–18 nm vs ~18 nm), although their susceptibility to RNase (Table 1) implies that they fail to form completely closed shells. ΔPS3 appears more deleterious in terms of closed
shell formation than ΔPS2. These results are consistent with a PS-mediated in vitro assembly mechanism for the HBV JQ707375.1 gRNA. Multiple PSs within the genomes of ssRNA viruses15,20,46
vary around a consensus sequence defining a hierarchy of binding affinities for cognate capsid proteins. This preferred assembly pathway around cognate viral RNA prevents multiple assembly
initiation events occurring on the same RNA, ensuring efficient capsid assembly, and avoiding the formation of off-pathway kinetic traps37. It implies that PSs act cooperatively but
differentially during assembly, as seen here. This mechanism creates a fixed spatial relationship between the pgRNA and Cp shell, and for several RNA bacteriophages these interactions have
been revealed directly by asymmetric cryo-EM reconstruction47,48. Zlotnick and colleagues49 have proposed that a preferred pgRNA conformation within the NCP would facilitate its reverse
transcription. To investigate that possibility, we determined the structure of the NCP assembled around the wild-type RNA transcript. THE GRNA CONTAINING NCP HAS ORDERED INTERNAL DENSITY A
cryo-EM reconstruction was calculated using particles of purified NCPs containing the HBV JQ707375.1 RNA transcript (‘Methods’ & Table 2). After data collection, 127,410 _T_ = 4 (84%)
and 23,257 _T_ = 3 (16%) particles were chosen for further analysis. The ratio of _T_ = 4:_T_ = 3 particles in this dataset is similar to that obtained following reassembly around the
NC_003977.1 PS1 oligonucleotide12. 2D classification reveals that most _T_ = 3 particles and ~18% of the _T_ = 4 particles were heterogeneous, with many particles appearing empty. No further
image processing was performed on these. A homogenous dataset of the remaining _T_ = 4 particles was subjected to icosahedrally-averaged refinement yielding a ~3.2 Å resolution map. This
reconstruction reveals a Cp layer similar to those seen in previous EM and crystal structures. Density for Cp chains is complete up to the start of the ARD (Fig. 4, Supplementary Fig.
4)50,51,52,53. The four Cp monomers of the asymmetric unit were built into the map using a previous structure of the NC_003977.1 Cp53 (PDB 3J2V) as a starting model. At this resolution, the
four distinct Cp monomers of the quasi-equivalent capsomer, chains A-D, can be built using polypeptide chains 144, 143, 144 and 147 residues long, respectively (Supplementary Figs. 4 and 5).
The icosahedrally-ordered Cp surface lattice surrounds a second, less-ordered internal layer of density, which may well include density for the encapsidated gRNA transcript and the Cp ARD
domains. Reconstruction of the same dataset without the imposition of symmetry fails to reveal any further molecular detail at 4.2 Å resolution, the internal density appearing as a mostly
continuous layer underneath the Cp shell (Supplementary Fig. 6). To reduce the impacts of the Cp layer on the reconstruction, the signal from the globular region of the Cp was subtracted
from the particles in the same dataset, and a 3D classification by alignment of the remaining density calculated revealing five similar classes (not shown). The icosahedrally-averaged
reconstruction of the _T_ = 4 particle was then symmetry expanded21,22,23,24 following removal of much of the Cp shell using a spherical mask. The remaining density was subjected to further
3D classification without alignment into a further five roughly equally populated classes (Supplementary Fig. 7a). These were reconstructed without imposing symmetry21,54, yielding maps with
global resolutions of 3.5–3.6 Å (Supplementary Fig. 7). The maps were low-pass filtered to 5 Å resolution for clarity revealing an asymmetric cage-like density under the Cp shell at this
lower resolution, presumably corresponding to either the C-terminal ARDs or gRNA, or both, in close contact with the Cp shell (Fig. 4, Supplementary Fig. 7). Superposition of the internal
density obtained by asymmetric reconstruction of the NCP assembled around NC_003977.1 PS1 (EMD-3714), into this map shows that this feature, which we presume includes multiple PS RNA
oligonucleotides12, overlaps with the internal density seen with the NCP gRNA. Additional refinement failed to resolve these features further here, perhaps due to conformational
heterogeneity or flexibility55,56. Interestingly, these maps reveal multiple fingers of density at every particle vertex that transit the gap between the Cp layer and the internal density.
Such bridges, illustrated in Fig. 4 based on the Class 3 data (Supplementary Fig. 7d) could be the result of the formation of multiple Cp-gRNA contacts in the NCP, i.e. as expected for
PS-mediated assembly. These density bridges 5-fold and 2-fold positions (Fig. 4c, d and g, h, respectively) appear to be different at the different symmetry axes. For both, the internal
density fuses with that of the Cp subunits adjacent to residue Pro144, i.e. just before the start of the ARD domain, and could represent extensions of the polypeptide chain at these points.
The bridging contacts at the 3-fold (Fig. 4a, b) and quasi-3-fold vertices (Fig. 4e, f) are much more similar to each other but distinct from those at 2- and 5-fold axes. They fuse into the
density of the Cp around residues E40 and C48. Both these amino acids are in well-ordered sections of the globular fold and there is no unassigned Cp density in this part of the map. In
order to test the idea that there are RNA PS-Cp contacts in the reassembled NCP, we used XRF27,57,58. To see if PS1, the most important PS within the JQ707375.1 transcript for regulating
assembly in vitro (Fig. 3c) contacts Cp in the reassembled NCP, XRF nucleotide reactivities across a genomic region encompassing PS1 were determined for both NCP and free transcript
(‘Methods’). Both reassembled NCP and transcript were flash-frozen and exposed to the X-ray beam on the customised beam-line at the NSLS-II in Brookhaven for various times ranging from 0 to
100 milliseconds59. Samples were returned to the host laboratory in the frozen state and RNA extracted/prepared for primer extension with a dye-labelled DNA primer annealed at nucleotide
positions 3308–3328 (Fig. 5). The reverse transcription extension products were then analysed using capillary electrophoresis (CE), and a combination of published and in-house software27,58.
Reassembled and protein-free gRNA is predicted to adopt the same stem-loop structure at PS1. This fold places the -RGAG- recognition motif (boxed in Fig. 5c) in the 3′ half of the loop, as
expected (Fig. 2). The reactivities of the nucleotides in these two RNA stem-loops are shown colour coded (dark green to red corresponding to unreactive to highly reactive, respectively),
and as a difference map in Fig. 5d. Nucleotides within the loop show the most significant differences in reactivity between the two states. In the transcript, the nucleotides of the -RGAG-
motif have high reactivities (red), with the 5′ and 3′ neighbouring nucleotides being much less reactive. In the NCP, however, these reactivity patterns across the motif are largely
reversed. All four nucleotides of the recognition motif become largely unreactive, whereas their 5′ and 3′ neighbours become more reactive, an effect that extends into the uppermost base
pair of the stem (Fig. 5b, c). These data are consistent with a direct interaction between the NCP Cp layer and the PS1 recognition motif. There are also slight conformational rearrangements
in the loop in the NCP resulting in increased flexibility of the non-contacted nucleotides. In vivo NCP assembly is known to be affected by Cp phosphorylation and/or the binding of
replicase at the ε site28,29,30,31,32, both of which are missing in the in vitro assay. The JQ707375.1 transcript encompass both ε and ϕ sites (Fig. 1), and these sites are thought to form
long-range base pairs making the RNA topology important for assembly60,61. In order to test this idea, we carried out reassembly with a variant RNA transcript of its ε site (Supplementary
Fig. 2) preventing it from base-pairing with the ϕ site. Strikingly, this variant gRNA, unlike those carrying the PS mutations, has a significantly increased hydrodynamic radius (27 ± 0.4
vs. 17 ± 0.3 nm, respectively). It is also a poor in vitro assembly substrate, although not as poor as the ∆PS and ∆PS1 variants, implying that in vitro and presumably also in vivo, both RNA
topology and PS-Cp contacts contribute to assembly efficiency. It has been argued that ε is the most important pgRNA sequence since in vivo it is possible to encapsidate heterologous RNAs
apparently encompassing only this site from the HBV genome. However, in these experiments the ε site was flanked by additional HBV genomic sequences which sequence/S-fold analysis
(Supplementary Methods) suggests contain multiple unrecognised PS sites akin to those analysed here. In addition, in a fusion of this fragment to a 3′ _lacZ_, an assumed non-specific
heterologous RNA construct, an additional PS site is added. The combinatorial effects of such sites would explain their packaging, ε sites being packaged whilst _lacZ_ alone is not
(Supplementary Tables 1–3). DISCUSSION The assembly of infectious HBV is a complex process that many people have studied in vivo in suitable cell culture systems. Here we have used a
minimised molecular system to investigate whether the RNA stem-loops identified previously stimulate in vitro assembly of an RNA transcript encompassing most of the coding region of a strain
not previously part of our analysis (JQ707375.1). The results establish that in such an assay this gRNA transcript is efficiently encapsidated. Modification of the gRNA to prevent or alter
presentation of the Cp-recognition sequence (-RGAG-), at three putative PS homologues of sites studied previously as oligonucleotides, results in varying assembly deficits. The -RGAG- Cp
recognition motif of one such site, PS1, appears to be selectively protected from X-ray modification in an HBV NCP, compared to a free RNA transcript. These results are consistent with NCP
formation in vitro occurring via a PS-mediated assembly mechanism. Given that the same RNA and Cp sequences will participate in NCP formation in vivo, it seems reasonable to conclude that a
similar mechanism contributes to such assembly. Cryo-EM reconstruction of the JQ707375.1 RNA transcript assembled NCP, reveals extensive density below the globular folds of the Cp shell.
Symmetry expansion suggests that there are multiple contacts between this internal density and the outer shell at particle symmetry axes. It is not possible at the resolutions obtained to
assign this internal density to either the C-terminal Cp ARD domains, the gRNA transcript or a complex of both. Since this density is not present in NCPs lacking encapsidated RNA12, it is
clearly a consequence of Cp-gRNA interaction. Previously, we showed that individual oligonucleotides encompassing PS sites will trigger sequence-specific NCP formation in vitro. Asymmetric
reconstruction of the NCP revealed an inner density larger than could be accounted for by a single RNA stem-loop12, although we assumed that PS1, or multiple copies of it, were part of this
density. Superposition of that density on the inner layer seen with the gRNA here shows that they are coincident with respect to radial positions, suggesting that this layer does contain
RNA. XRF of PS1 confirms this assumption. PSs only need to function, i.e. contact the protein shell of a virion, during assembly. For bacteriophage MS247, many of its RNA PSs dissociate from
the CP post-assembly. The fact that PS1 remains in contact after NCP assembly may reflect the need in a para-retrovirus to create an ‘RNA track’ along which the polymerase must move within
the NCP as it reverse transcribes the gRNA12,49. The in vitro assembly data with the three variant PS sites suggest that they constitute a novel, evolutionarily stable aspect to NCP assembly
regulation. The experiment with the ε variant suggests that gRNA topology is also important in efficient assembly, i.e. that there may be multiple factors that assist the process in vivo.
The PS-mediated aspect of assembly is a direct anti-viral drug target that could be exploited in the global challenge to treat chronic HBV infections62. There are currently only a limited
set of clinical interventions, including polymerase inhibitors Lamivudine and Tenofovir, and interferon63,64,65. Recently novel ligands that target additional aspects of the viral lifecycle
have been identified66,67,68,69,70. Amongst these are the ‘capsid assembly modulators, CAMs’ that include the hetero-aryl, dihydropyrimidine (HAP) compounds characterised by Zlotnick and
colleagues71,72. They have shown that HAPs bind Cp dimers altering their assembly behaviour resulting in formation of ‘empty’ NCP particles. Mathematical modelling suggests that targeting
capsid assembly via distinct routes would be synergic73,74. The recent development of a mouse model in which to probe the HBV lifecycle75 will hopefully allow these ideas to be tested
rapidly allowing the Global Challenge presented by this virus to be met. METHODS EXPRESSION AND PURIFICATION OF HBV CP HBV Cp was expressed in _E. coli_ BL21(DE3) cells (T7 Expression
strain, New England Biolabs), expressed from a pET28b plasmid. Induction with 1 mM isopropyl-β-D-thiogalactoside (IPTG) at an optical density (OD600) of ~0.6 was followed by growth for 20 h
at 21 °C. Cells were lysed using a Soniprep 150 and clarified by spinning at 11,000 × _g_ for 1 h. NCPs were then pelleted by centrifugation at 120,000 × _g_ for 14 h. Pellets were
resuspended in 20 mM HEPES (pH 7.5), 250 mM NaCl, 5 mM dithiothreitol (DTT) and applied to a prepacked Captocore 700 column (GE Life Sciences). Fractions containing NCPs were pooled and
precipitated with 40% (w/v) ammonium sulfate. NCPs were dissociated into Cp dimers by dialysis into 1.5 M guanidinium chloride (GuHCl) as previously described12,38. All steps after
sonication were performed in the presence of complete protease inhibitor tablets (1 tablet per 500 mL buffer, Thermofisher Scientific). Cp dimer concentration was determined by the
absorbance at 280 nm (ε280 of Cp2 = 55,920 L mol−1 cm−1). Cp fractions with an A260/280 ratio of ≤0.65 were used in assembly assays. IDENTIFICATION OF HOMOLOGOUS PS SITES IN STRAIN
JQ707375.1 PS sites within the JQ707375.1, homologous to those found within NC_003977.1, were identified by analysis of stem-loop folds within a 300 nt region of the genome that aligns to
the equivalent regions encompassing each PS in the latter strain. A 60 nt reference frame was slid across that region and secondary structure of the corresponding RNA sequence sampled,
recording each unique secondary structure fold with negative free energy12. Previously such PS sites were numbered according to the most frequently matched sites in a Bernoulli Plot of the
aptamer sequences against the cognate pgRNA12. We retain those labels for the homologous JQ707375.1 strain sites, although this means their numbering does not reflect their relative order.
In JQ707375.1, the PS1 equivalent lies between nucleotides 3134–3172 (Fig. 2). Mfold suggests that it retains the bulge in the base-paired stem and the -RGAG- loop motif seen in NC_003977.1,
although the latter is at the 5′ side of the loop as opposed to being central. This PS1 lies in the gene encoding the X protein, and its nucleotide sequence creates two non-synonymous
codons encoding conservative amino acid changes. PS2 (nts 807–859) and PS3 (nts 981–1011) sit in regions of the polymerase gene with much lower sequence identities than the overall genome
(67 & 75%, respectively). PS2 has a slightly modified loop motif (-RAAG-), which is centrally located, as opposed to being at the 3′ side of the loop, as it is in the NC_003977.1 strain.
The loop sits on top of a three base-paired duplex leading to single-stranded bulges of differing sizes. The upper region of this PS stem-loop is homologous to the PS2 oligonucleotide fold
in NC_003977.1 (Fig. 2). The JQ707375.1 PS3 retains the 3′ positioning of the -RGAG- motif within the loop but has a more extensively base-paired stem than in the previous strain (Fig. 2).
Nucleotide changes in JQ707375.1 PS2 create one conservative (F to Y), and two semi-conservative (T to N & H to L) amino acid substitutions relative to the NC_003977.1 polymerase. The
sequence of the PS3 region yields only synonymous coding changes. PREPARATION OF PREGENOMIC RNA A wild-type gRNA transcript clone was assembled using pAM6 39630TM, strain acc. no JQ707375.1,
purchased from ATCC®. The RNA sequence was copied from the purchased cDNA in fragments using PCR, and the fragments cloned into the correct order between the _BspHI_ and _HindIII_ sites in
a pACYC184 vector, using a Gibson Assembly® Master Mix, according to the manufacturer’s protocol (New England Biolabs). The sequence of this construct was confirmed by Sanger sequencing
(Source Bioscience, Nottingham). HBV pgRNA constructs encompassing PS mutations were produced synthetically, using gene fragments purchased from IDT. Gene fragments were cloned between the
_BglII_ and _HindIII_ sites within an empty pET22b vector using a NEBuilder® HiFi Master Mix, according to the manufacturer’s protocol (New England Biolabs). All pgRNA constructs were
designed with a T7 promoter sequence at the 5′ end. Transcription of pgRNA constructs were carried out using a Hiscribe™ T7 High Yield RNA Synthesis Kit (New England Biolabs), after
linearization of the DNA plasmid using _HindIII_. RNA was annealed prior to each experiment by heating to 70 °C for 90 secs and cooling slowly to 4 °C in a buffer containing 50 mM NaCl, 10
mM HEPES and 1 mM DTT at pH 7. Products were assessed using a 1% (v/v) denaturing formaldehyde agarose RNA gel. RNA concentration was determined using the A260 value (ε260 of pgRNA =
32,249,500 L mol−1 cm−1). HBV NCP ASSEMBLY ASSAYS 180 μL of 1.1 nM annealed (as above) gRNA transcript in a buffer containing 20 mM HEPES (pH 7.5), 250 mM NaCl and 5 mM DTT, was incubated in
each well of a 96-well plate at room temperature for 30 min. Cp dimer in dissociation buffer (as above) was then titrated into the RNA using a Biomek 4000 liquid handling robot (Beckmann
Coulter), step-wise up to a ratio of 1200:1 Cp dimer: RNA. Ten 2 μL titrations were performed, utilising six different stock Cp dimer solutions (indicated in brackets), cumulatively adding 1
nM (100 nM), 10 nM (1 μM), 25 nM (2.5 μM), 75 nM (7.5 μM), 120 nM (12 μM), 240 nM (12 μM), 480 nM (24 μM), 720 nM (24 μM), 960 nM (24 μM) and 1200 nM (24 μM Cp dimer. This results in a
final well volume of 200 μL, a cumulative volume of 19.2 mL/plate, and final concentrations of RNA and Cp of 1 nM and 1.2 μM, respectively. Cp aliquots were calculated to always reach 10% of
the final reaction volume limiting the final concentration of GuHCl to 0.15 M in each aliquot. Following incubation at room temperature for 1 hr, the samples were pooled and concentrated to
a final volume of 2 mL. The A260/280 ratio was measured using a Nanodrop™ One (Thermofisher Scientific) and RNA concentration was calculated using absorbance at 260 nm. This sample was
split into 2, with one half treated with 1 µM RNase A, and incubated overnight at 4 °C. After incubation, 5 µL of each sample was visualised by nsEM to assess particle shape and intactness,
and the remaining sample analysed by application to a TSK G6000 PWXL column (Tosoh), in a buffer containing 20 mM HEPES (pH 7.5), 250 mM NaCl and 5 mM DTT, using a SEC-MALLS system (ÄKTA
Pure (GE Heathcare) connected to a Optilab T-REX refractometer and miniDAWN® multiple angle laser light-scatterer fitted with a Wyatt QELS DLS module (Wyatt Technology)). Light scattering
peaks were collected and concentrated to ~1 ml, where the RNA content of particles and yield considering the RNA input was once again determined using the absorbance at 260 nm. Absorbance
values at 260 and 280 were corrected for light-scattering throughout, using the absorbance values at 310 and 340 nm as previously described76. All assembly assays were performed in
triplicate. R_H_ DETERMINATION OF GRNA TRANSCRIPTS 500 µL of gRNA transcript at a concentration of 500 ng/µL was applied to a TSK G6000 PWXL column (Tosoh) using the buffer and SEC-MALLS
system described above. All measurements were performed in triplicate. The hydrodynamic radius of assembled NCPs and gRNA transcripts were determined at the apex of peaks eluted from a TSK
G6000 PWXL column. The light scattering at this point is quantified via the second order correlation function, (1) where _I(t)_ is the intensity of scattered light at time _t_.
$${g}^{2}\left(\tau \right)=\frac{(I\left(t\right)I\left(t+\tau \right))}{(I{\left(t\right)}^{2})}$$ (1) The measured correlation function is fit to Eq. (2) using a nonlinear least squares
fitting algorithm to calculate the decay rate _g_. $${g}^{2}(\tau )=B+\beta \exp (-2\varGamma \tau )$$ (2) _G_ is then converted to the diffusion constant _D_ using Eq. (3), where _q_ is the
magnitude of the scattering vector given by Eq. (4). $$D=\frac{\varGamma }{{q}^{2}}$$ (3) $$q=\frac{4\pi {n}_{0}}{\lambda }{\sin }\left(\frac{\theta }{2}\right)$$ (4) _D_ is then fit to the
Stokes-Einstein Eq. (5) to give the R_h_. $${R}_{h}=\frac{{kT}}{6\pi {\eta D}_{t}}$$ (5) X-RAY FOOTPRINTING OF GRNA-CONTAINING NCPS To detect Cp-PS1 interactions in the NCP we used XRF,
comparing the footprint on the encapsidated gRNA with the transcript used in the assembly reaction. Both samples were flash frozen and exposed to synchrotron X-rays at the 17-BM beamline
(XFP) of the NSLS-II at the Brookhaven National Laboratory (‘Methods’)26. X-ray photons are largely absorbed by the solvent water which photolyzes to create hydroxyl radicals that then
modify the ribose sugars of solvent accessible nucleotides in a flexibility-dependent manner25,26. Ribose sugars of single-stranded nucleotides are more reactive than those in base-pairs or
involved in other molecular contacts. Modification leads to cleavage of the phosphodiester bond57, and the frequency of cleavage at each nucleotide can be determined by reverse transcription
using fluorescently-tagged primers annealed 3′ to the site of interest. NCPs were re-assembled as above around the wild-type JQ707375.1 pgRNA in three 96-well plates. These samples were
concentrated by centrifugation, and purified over a 10–50% (_w/v_) sucrose density gradient using a SW 40 _Ti_ rotor at 190,000 × _g_ for 2 h. Purified NCP samples were diluted in 10 mM
sodium phosphate buffer (pH 7.4) to 200 ng/μL with respect to pgRNA using a Nanodrop™ One, and flash-frozen using LN2 as 5 μL aliquots in 8 tube PCR strips. Heat-annealed (see above) pgRNA
only samples at the same concentration were also flash-frozen as a control. The samples were footprinted on beamline 17-BM (XFP) at the National Synchrotron Light Source II (Brookhaven
National Laboratory, NY, USA). A calibration curve of the X-ray induced photo-bleaching of Alexa488 fluorophore, diluted in 10 mM sodium phosphate buffer (pH 7.4) was performed to ascertain
beam strength, allowing adjustment of sample exposure times between runs to keep levels of RNA cleavage events similar between experiments. Samples were mounted in the beamline in a
temperature-controlled (−30 °C, ensuring samples remained frozen), 96-well motorised-holder, which accommodates strips of 8–12 PCR tubes. Beam exposure was controlled using a Uniblitz XRS6
fast shutter (Vincent Associates), exposing samples for 10, 25 and 50 msec, with each time point performed in triplicate. Ideally, exposed samples contained no more than 1 cleavage event
across the region of interest, defined as the length of the reverse transcript. This assumes that cleavages elsewhere do not cause large-scale conformational changes in the frozen gRNA
within the time span of the exposure (max = 100 milliseconds). Similar data on bacteriophage MS2 gRNA yield predicted secondary structures similar to those seen at atomic resolution by
cryoEM47,58, confirming that this is a reasonable assumption. ANALYSIS OF X-RAY INDUCED RNA MODIFICATION RNA was extracted from exposed NCPs using the phenol-chloroform (Thermofisher
Scientific) extraction technique according to the manufacturers’ protocols, with the exception that the RNAs were precipitated overnight at −20 °C in the presence of 1 volume isopropanol,
0.3 M sodium actetate (pH 5.2) and RNA-grade glycogen (0.01×, Thermofisher Scientific) which gave us higher yields. Recovered RNA was washed 3× with 70%(v/v) ethanol and allowed to air dry
for 5 min before resuspension with 12 μL nuclease-free water. The beamline-exposed extracted and free pgRNA were reverse transcribed using Superscript IV (Thermofisher Scientific) and a
sequence specific 5′ 5(6)-carboxyfluorescein (FAM) labelled primer that attached 3′ of the region of interest. Sequencing ladders were synthesised from in vitro transcribed RNA using
Hexachloro-fluorescein (HEX) or 5-carboxytetramethylrhodamine (TAMRA) labelled primers and the addition of a 3:1 molar concentration of ddATP or ddCTP, respectively. Bound RNA was degraded
with 5 units of RNase H (New England Biolabs) and the cDNA purified by ethanol precipitation overnight at −20 °C (3× volumes of ethanol, 0.3 M sodium acetate 0.01× volume of glycogen).
Experimental and sequencing ladder cDNAs were then resuspended in 20 μL formamide, their concentration measured by A260 absorbance and 500 ng of each sequencing ladder was spiked into each
experimental sample. The samples were heated to 65 °C for 10 min then transferred to a 96-well plate and frozen for shipping to DNASeq (Dundee, UK) for capillary electrophoresis (CE). A
reverse primer complementary to pgRNA sites 3328–3308 (sequence 3′–5′: AATTTATAAGGGTCAATGTC), was designed to analyse the region encompassing the PS1 signal which was synthesised (IDT) with
the appropriate flourophores, (as detailed above) for the aforementioned production of ladders and analysis of beamline-exposed samples. CE ANALYSIS Normalised reactivity profiles were
produced from the raw capillary electrophoresis (CE) data using the QuShape Software package43 in combination with the BoXFP wrapper module58. For each replicate, pre-processing of the CE
data was performed, including signal smoothing, decay correction and alignment of peaks in the ddA sequencing trace to corresponding peaks in the XRF trace. Peaks were identified and peaks
in different replicates aligned using the molecular weight size marker. Average and standard deviation of peak intensities were calculated across the replicates to produce the intensity
profile. Background correction of samples was performed using the average intensity profile across the unexposed background samples computed. The reactivity profile was calculated and
normalised, and error propagation was performed to obtain the error on the normalised intensities. Finally, the ddA traces across all samples from the same primer read was used to generate a
consensus sequence of U nucleotide locations across the primer read, which was then aligned to the reference genome to determine the position of the primer read in the genome. Average
Pearson correlation coefficients (PCCs) were calculated across the replicates for each treatment (transcript gRNA = 0.8459278, encapsidated gRNA = 0.94918026), and the normalisation factors
used in the computation of the normalised reactivity profiles were recorded for each case (transcript gRNA = 2678.25523, NCP gRNA = 1652.53287). The protection ratio across each primer read
was calculated as the normalisation factor of the transcript, divided by the normalisation factor for the encapsidated state of the gRNA, calculated as 1.620697097. DATA ACQUISITION FOR
CRYO-EM Lacey carbon 400-mesh copper grids coated with a <3 nm continuous carbon film (Agar Scientific, UK) were glow-discharged in air (10 mA, 30 s) before applying one 3 μL aliquot of
pgRNA containing HBV, reassembled and purified as described above for X-Ray Footprinting. Grids were blotted and vitrified in liquid nitrogen-cooled liquid ethane using a LEICA EM GP plunge
freezing device (Leica Microsystems). Chamber conditions were set at 4 °C and 95% relative humidity. Grids were stored in liquid nitrogen prior to imaging with an FEI Titan Krios
transmission electron microscope (ABSL, University of Leeds) at 300 kV, at a magnification of ×75,000 and a calibrated object sampling of 1.065 Å/pixel. Images were recorded on a FEI Falcon
III detector operating in integrating mode. Each movie comprises 59 frames with an exposure rate of 0.9 e− Å−2 per frame, with a total exposure time of 1.5 s and an accumulated exposure of
53.1 e− Å−2. Data acquisition was performed with EPU Automated Data Acquisition Software for Single Particle Analysis (ThermoFisher) at −0.7 µm to −2.5 µm defocus. IMAGE PROCESSING The
established RELION-3.0 pipeline was used for image processing77,78. Drift correction was performed using MOTIONCOR279, and the contrast transfer function estimated using Gctf80. A subset of
1000 particles were picked and classified using reference-free 2D classification and used as templates for the RELION autopicking procedure. Picked particles were sorted using 2D
classification, with 127,410 _T_ = 4 and 23,257 _T_ = 3 particles subsequently extracted. The large majority (~15,000) of _T_ = 3 particles were discarded due to heterogeneity. 104,337 _T_ =
4 particles were selected and subjected to 3D classification and subsequent auto-refinement with icosahedral symmetry (I1) imposed and without (C1), using EMD-3715 low pass filtered to 60 Å
resolution as a reference model. This reconstruction was post-processed to mask and correct for the B-factor of the map. The CTF refinement routine implemented in RELION-3.077 was used to
refine the reconstruction further, yielding a map with an overall resolution at 3.2 Å, based on the gold-standard (FSC = 0.143) criterion. To investigate the density of the pgRNA, a focussed
3D classification approach was employed. Each particle contributing to the final icosahedral symmetry-imposed reconstruction was assigned 60 orientations corresponding to its
icosahedrally-related views using the relion_symmetry_expand tool. SPIDER47 was used to generate a spherical mask placed just beneath the Cp shell of the NC, and the symmetry expanded
particles were subjected to masked 3D classification, sorting them into 5 classes without alignment, using a regularisation parameter of 25. Particles from these classes were reconstructed
using the relion_reconstruct tool, without imposing symmetry, and postprocessed, yielding maps with a resolution of 3.5 Å. UCSF Chimera was used for visualisation and figure generation81.
MODEL BUILDING AND REFINEMENT The structure of HBV NC (PDB 3J2V)53 was first manually docked as a rigid body into the density and followed by real space fitting with the Fit in Map routine
in UCSF Chimera81. A first step of real space refinement was performed in Phenix82. The model was then manually rebuilt in Coot83 to optimize the fit to the density. After icosahedral
symmetrisation to generate the entire capsid, a second step of real space refinement was performed in Phenix. Refinement statistics are listed in Supplementary Table 2. MODEL VALIDATION AND
ANALYSIS The FSC curve between the final model and map after post-processing in RELION (Model vs Map), is shown in Supplementary Fig. 3b. To perform cross-validation against overfitting, the
atoms in the final atomic model were displaced by 0.5 Å in random directions using Phenix. The shifted coordinates were then refined against one of the half-maps (work set) in Phenix using
the same procedure as for the refinement of the final model. The other half-map (test set) was not used in refinement for cross-validation. FSC curves of the refined shifted model against
the work set (FSCwork) and against the test set (FSCtest), are shown in Supplementary Fig. 3b. The FSCwork and FSCtest curves are not significantly different, consistent with the absence of
overfitting in the final models. The quality of the atomic model, including basic protein geometry, Ramachandran plots, clash analysis, was assessed and validated with Coot, MolProbity84 as
implemented in Phenix, and with the Worldwide PDB (wwPDB) OneDep System (https://deposit-pdbe.wwpdb.org/deposition). Graphics were produced by UCSF Chimera81. REPORTING SUMMARY Further
information on research design is available in the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY The data that supports the findings of this study are available
from the corresponding authors upon request. Correspondence and requests for materials should be addressed to N.P or P.G.S. The atomic coordinates for HBV NC were deposited in the Protein
Data Bank with code 7ABL. The icosahedrally averaged, symmetry-expanded and focused classified on genome and asymmetric cryo-EM density maps were deposited in the EM Data Bank with codes
EMD-11700, EMD-11702 and EMD-11701, respectively. REFERENCES * World Health Organization. Weekly epidemiological record. _Hepat. B Vaccines._ 84, 405–420 (2009). Google Scholar * Tillmann,
H. L. Antiviral therapy and resistance with hepatitis B virus infection. _World J. Gastroenterol._ https://doi.org/10.3748/wjg.v13.i1.125 (2007). * Murray, K. et al. Protective immunisation
against hepatitis B with an internal antigen of the virus. _J. Med. Virol._ https://doi.org/10.1002/jmv.1890230202 (1987). * World Health Organization. Progress report on HIV, viral
hepatitis and sexually transmitted infections. 1, 1–39 (2019). * Nassal, M. Hepatitis B viruses: Reverse transcription a different way. _Virus Res_.
https://doi.org/10.1016/j.virusres.2007.12.024 (2008). * Lok, A. S. et al. Long-term safety of lamivudine treatment in patients with chronic hepatitis B. _Gastroenterology_ 125, 1714–1722
(2003). Article CAS Google Scholar * World Health Organization. Combating hepatitis B and C to reach elimination by 2030. 1–16 (2016). * Bock, C. T. et al. Structural organization of the
hepatitis B virus minichromosome. _J. Mol. Biol._ 307, 183–196 (2001). Article CAS Google Scholar * Guo, Y. H., Li, Y. N., Zhao, J. R., Zhang, J. & Yan, Z. HBc binds to the CpG
islands of HBV cccDNA and promotes an epigenetic permissive state. _Epigenetics_ 6, 720–726 (2011). Article Google Scholar * Selzer, L. & Zlotnick, A. Assembly and release of hepatitis
B virus. _Cold Spring Harb. Perspect. Med._ 5, a021394 (2015). * Seeger, C. & Mason, W. S. Hepatitis B virus biology. _Microbiol Mol. Biol. Rev._ 64, 51–68 (2000). Article CAS Google
Scholar * Patel, N. et al. HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. _Nat. Microbiol._ 2, 17098
(2017). Article CAS Google Scholar * Bunka, D. H. J. & Stockley, P. G. Aptamers come of age—at last. _Nat. Rev. Microbiol._ https://doi.org/10.1038/nrmicro1458 (2006). * Stockley, P.
G. et al. Packaging signals in single-stranded RNA viruses: nature’s alternative to a purely electrostatic assembly mechanism. _J. Biol. Phys._ 39, 277–287 (2013). Article CAS Google
Scholar * Dykeman, E. C., Stockley, P. G. & Twarock, R. Packaging signals in two single-stranded RNA viruses imply a conserved assembly mechanism and geometry of the packaged genome.
_J. Mol. Biol._ 425, 3235–3249 (2013). Article CAS Google Scholar * Borodavka, A., Tuma, R. & Stockley, P. G. A two-stage mechanism of viral RNA compaction revealed by single molecule
fluorescence. _RNA Biol._ 10, 481–489 (2013). Article CAS Google Scholar * Bunka, D. H. et al. Degenerate RNA packaging signals in the genome of Satellite Tobacco Necrosis Virus:
implications for the assembly of a T = 1 capsid. _J. Mol. Biol._ 413, 51–65 (2011). Article CAS Google Scholar * Rolfsson, O. et al. Direct Evidence for Packaging Signal-Mediated Assembly
of Bacteriophage MS2. _J. Mol. Biol._ https://doi.org/10.1016/j.jmb.2015.11.014 (2015). * Shakeel, S. et al. Genomic RNA folding mediates assembly of human parechovirus. _Nat. Commun._ 8, 5
(2017). Article CAS Google Scholar * Twarock, R. & Stockley, P. G. RNA-mediated virus assembly: mechanisms and consequences for viral evolution and therapy. _Annu. Rev. Biophys._ 48,
495–514 (2019). Article CAS Google Scholar * Conley, M. J. et al. Calicivirus VP2 forms a portal-like assembly following receptor engagement. _Nature_
https://doi.org/10.1038/s41586-018-0852-1 (2019). * Scheres, S. H. W. Processing of structurally heterogeneous Cryo-EM data in RELION. _Methods Enzymol._
https://doi.org/10.1016/bs.mie.2016.04.012 (2016). * McElwee, M., Vijayakrishnan, S., Rixon, F. & Bhella, D. Structure of the herpes simplex virus portal-vertex. _PLoS Biol._
https://doi.org/10.1371/journal.pbio.2006191 (2018). * Zhou, M. et al. Atomic structure of the apoptosome: Mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. _Genes Dev._
https://doi.org/10.1101/gad.272278.115 (2015). * Adilakshmi, T., Soper, S. F. C. & Woodson, S. A. _Biophysical, Chemical, and Functional Probes of RNA Structure, Interactions and
Folding: Part A_ (Elsevier, 2009). * Asuru, A. et al. The XFP (17-BM) beamline for X-ray footprinting at NSLS-II. _J. Synchrotron Radiat._ https://doi.org/10.1107/S1600577519003576 (2019). *
Tetter, S. et al. Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein. _Science_ 372, 1220–1224 (2021). Article CAS Google Scholar *
Bartenschlager, R. & Schaller, H. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. _EMBO J._ 11, 3413–3420 (1992). Article
CAS Google Scholar * Gazina, E. V., Fielding, J. E., Lin, B. & Anderson, D. A. Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and
duck hepatitis B viruses. _J. Virol._ https://doi.org/10.1128/jvi.74.10.4721-4728.2000 (2000). * Lan, Y. T., Li, J., Liao, W. & Ou, J. Roles of the three major phosphorylation sites of
hepatitis B virus core protein in viral replication. _Virology_ 259, 342–348 (1999). Article CAS Google Scholar * Junker-Niepmann, M., Bartenschlager, R. & Schaller, H. A short
cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. _EMBO J._ https://doi.org/10.1002/j.1460-2075.1990.tb07540.x
(1990). * Pollack, J. R. & Ganem, D. An RNA stem-loop structure directs hepatitis B virus genomic RNA encapsidation. _J. Virol._ https://doi.org/10.1128/jvi.67.6.3254-3263.1993 (1993). *
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. _Nucleic Acids Res._ 31, 3406–3415 (2003). * Thai, H. et al. Convergence and coevolution of hepatitis B
virus drug resistance. _Nat. Commun._ 3, 789 (2012). Article CAS Google Scholar * Patel, N. et al. Revealing the density of encoded functions in a viral RNA. _Proc. Natl Acad. Sci. USA_
112, 2227–2232 (2015). Article CAS Google Scholar * Routh, A., Domitrovic, T. & Johnson, J. E. Host RNAs, including transposons, are encapsidated by a eukaryotic single-stranded RNA
virus. _Proc. Natl Acad. Sci. USA_ 109, 1907–1912 (2012). Article Google Scholar * Ford, R. J. et al. Sequence-specific, RNA-protein interactions overcome electrostatic barriers preventing
assembly of satellite tobacco necrosis virus coat protein. _J. Mol. Biol._ 425, 1050–1064 (2013). Article CAS Google Scholar * Porterfield, J. Z. et al. Full-length hepatitis B virus
core protein packages viral and heterologous RNA with similarly high levels of cooperativity. _J. Virol._ 84, 7174–7184 (2010). Article CAS Google Scholar * Borodavka, A., Tuma, R. &
Stockley, P. G. Evidence that viral RNAs have evolved for efficient, two-stage packaging. _Proc. Natl Acad. Sci. USA_ 109, 15769–15774 (2012). Article Google Scholar * Garmann, R. F. et
al. Role of electrostatics in the assembly pathway of a single-stranded RNA virus. _J. Virol._ 88, 10472–10479 (2014). Article CAS Google Scholar * Rudnick, J. & Bruinsma, R.
Icosahedral packing of RNA viral genomes. _Phys. Rev. Lett._ https://doi.org/10.1103/PhysRevLett.94.038101 (2005). * van der Schoot, P. & Bruinsma, R. Electrostatics and the assembly of
an RNA virus. _Phys. Rev. E Stat. Nonlin Soft Matter Phys._ 71, 61928 (2005). Article CAS Google Scholar * Belyi, V. A. & Muthukumar, M. Electrostatic origin of the genome packing in
viruses. _Proc. Natl Acad. Sci. USA_ https://doi.org/10.1073/pnas.0608311103 (2006). * Balint, R. & Cohen, S. S. The incorporation of radiolabeled polyamines and methionine into turnip
yellow mosaic virus in protoplasts from infected plants. _Virology_ 144, 181–193 (1985). Article CAS Google Scholar * Bruinsma, R. F. Physics of RNA and viral assembly. _Eur. Phys. J. E
Soft Matter_ 19, 303–310 (2006). Article CAS Google Scholar * Dykeman, E. C., Stockley, P. G. & Twarock, R. Solving a Levinthal’s paradox for virus assembly identifies a unique
antiviral strategy. _Proc. Natl Acad. Sci. USA_ 111, 5361–5366 (2014). Article CAS Google Scholar * Dai, X. et al. In situ structures of the genome and genome-delivery apparatus in a
single-stranded RNA virus. _Nature_ 541, 112–116 (2017). Article CAS Google Scholar * Meng, R. et al. Structural basis for the adsorption of a single-stranded RNA bacteriophage. _Nat.
Commun._ 10, 1–8 (2019). Article CAS Google Scholar * Wang, J. C., Nickens, D. G., Lentz, T. B., Loeb, D. D. & Zlotnick, A. Encapsidated hepatitis B virus reverse transcriptase is
poised on an ordered RNA lattice. _Proc. Natl Acad. Sci. USA_ 111, 11329–11334 (2014). Article CAS Google Scholar * Wynne, S. A., Crowther, R. A. & Leslie, A. G. W. The crystal
structure of the human hepatitis B virus capsid. _Mol. Cell_ https://doi.org/10.1016/S1097-2765(01)80009-5 (1999). * Böttcher, B., Wynne, S. A. & Crowther, R. A. Determination of the
fold of the core protein of hepatitis B virus by electron cryomicroscopy. _Nature_ https://doi.org/10.1038/386088a0 (1997). * Conway, J. F. et al. Visualization of a 4-helix bundle in the
hepatitis B virus capsid by cryo-electron microscopy. _Nature_ https://doi.org/10.1038/386091a0 (1997). * Yu, X., Jin, L., Jih, J., Shih, C. & Hong Zhou, Z. 3.5 Å cryoEM structure of
hepatitis B virus core assembled from full-length core protein. _PLoS ONE_ https://doi.org/10.1371/journal.pone.0069729 (2013). * Conley, M. J. & Bhella, D. Asymmetric analysis reveals
novel virus capsid features. _Biophys. Rev._ https://doi.org/10.1007/s12551-019-00572-9 (2019). * Ruan, L., Hadden, J. A. & Zlotnick, A. Assembly properties of hepatitis B virus core
protein mutants correlate with their resistance to assembly-directed antivirals. _J. Virol._ https://doi.org/10.1128/jvi.01082-18 (2018). Article PubMed PubMed Central Google Scholar *
Zlotnick, A. et al. Core protein: a pleiotropic keystone in the HBV lifecycle. _Antiviral Res._ https://doi.org/10.1016/j.antiviral.2015.06.020 (2015). * Sclavi, B., Woodson, S., Sullivan,
M., Chance, M. R. & Brenowitz, M. Time-resolved synchrotron x-ray ‘footprinting’, a new approach to the study of nucleic acid structure and function: Application to Protein-DNA
interactions and RNA folding. _J. Mol. Biol._ https://doi.org/10.1006/jmbi.1996.0775 (1997). * Chandler-Bostock, R. et al. RNA X-ray footprinting reveals the consequences of an in vivo
acquired determinant of viral infectivity. _bioRxiv 2021.08.10.455819_. Preprint at https://doi.org/10.1101/2021.08.10.455819 (2021). * Jain, R. et al. New high-throughput endstation to
accelerate the experimental optimization pipeline for synchrotron X-ray footprinting. _J. Synchrotron Radiat._ 28, 28 (2021). Article Google Scholar * Oropeza, C. E. & McLachlan, A.
Complementarity between epsilon and phi sequences in pregenomic RNA influences hepatitis B virus replication efficiency. _Virology_ 359, 371–381 (2007). Article CAS Google Scholar * Tang,
H. & McLachlan, A. A pregenomic RNA sequence adjacent to DR1 and complementary to epsilon influences hepatitis B virus replication efficiency. _Virology_ 303, 199–210 (2002). Article
CAS Google Scholar * Patel, N. et al. Dysregulation of hepatitis B virus nucleocapsid assembly with RNA-directed small ligands. _bioRxiv 2021.08.10.455820_. Preprint at
https://doi.org/10.1101/2021.08.10.455820 (2021). * Tillmann, H. L. et al. Safety and efficacy of lamivudine in patients with severe acute or fulminant hepatitis B, a multicenter experience.
_J. Viral Hepat._ https://doi.org/10.1111/j.1365-2893.2005.00695.x (2006). * Ying, C., De Clercq, E. & Neyts, J. Lamivudine, adefovir and tenofovir exhibit long-lasting anti-hepatitis B
virus activity in cell culture. _J. Viral Hepat._ https://doi.org/10.1046/j.1365-2893.2000.00192.x (2000). * Greenberg, H. B. et al. Effect of human leukocyte interferon on hepatitis B
virus infection in patients with chronic active hepatitis. _N. Engl. J. Med._ 295, 517–522 (1976). Article CAS Google Scholar * Wu, S. et al. Discovery and mechanistic study of benzamide
derivatives that modulate hepatitis B virus capsid assembly. _J. Virol._ https://doi.org/10.1128/jvi.00519-17 (2017). * Venkatakrishnan, B. et al. Hepatitis B virus capsids have diverse
structural responses to small-molecule ligands bound to the Heteroaryldihydropyrimidine pocket. _J. Virol._ https://doi.org/10.1128/jvi.03058-15 (2016). * Zhang, X. et al. Discovery of novel
hepatitis B virus nucleocapsid assembly inhibitors. _ACS Infect. Dis._ 5, 759–768 (2019). Article CAS Google Scholar * Schlicksup, C. J. et al. Hepatitis B virus core protein allosteric
modulators can distort and disrupt intact capsids. _Elife_ https://doi.org/10.7554/eLife.31473 (2018). * Yang, L. et al. Effect of a hepatitis B virus inhibitor, NZ-4, on capsid formation.
_Antiviral Res._ https://doi.org/10.1016/j.antiviral.2015.11.004 (2016). * Stray, S. J. & Zlotnick, A. BAY 41-4109 has multiple effects on Hepatitis B virus capsid assembly. _J. Mol.
Recognit._ https://doi.org/10.1002/jmr.801 (2006). * Stray, S. J. et al. A heteroaryldihydropyrimidine activates and can misdirect hepatitis B virus capsid assembly. _Proc. Natl. Acad. Sci.
USA_ https://doi.org/10.1073/pnas.0409732102 (2005). * Fatehi, F. et al. An intracellular model of hepatitis b viral infection: an in silico platform for comparing therapeutic strategies.
_Viruses_ 13, 11 (2021). * Fatehi, F., Bingham, R. J., Dykeman, E. C., Stockley, P. G. & Twarock, R. An age-structured model of hepatitis B viral infection highlights the potential of
different therapeutic strategies. arXiv 2108.01982. Preprint at https://arxiv.org/abs/2108.01982 (2021). * Strick-Marchand, H. et al. A novel mouse model for stable engraftment of a human
immune system and human hepatocytes. _PLoS ONE_ https://doi.org/10.1371/journal.pone.0119820 (2015). * Porterfield, J. Z. & Zlotnick, A. A simple and general method for determining the
protein and nucleic acid content of viruses by UV absorbance. _Virology_ 407, 281–288 (2010). Article CAS Google Scholar * Scheres, S. H. W. RELION: Implementation of a Bayesian approach
to cryo-EM structure determination. _J. Struct. Biol._ https://doi.org/10.1016/j.jsb.2012.09.006 (2012). * Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure
determination in RELION-3. _Elife_ https://doi.org/10.7554/eLife.42166 (2018). * Zheng, S. Q. et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron
microscopy. _Nat. Methods_ https://doi.org/10.1038/nmeth.4193 (2017). * Zhang, K. Gctf: Real-time CTF determination and correction. _J. Struct. Biol._
https://doi.org/10.1016/j.jsb.2015.11.003 (2016). * Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. _J. Comput. Chem._
https://doi.org/10.1002/jcc.20084 (2004). * Adams, P. D. et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. _Acta Crystallogr. Sect. D Biol.
Crystallogr._ https://doi.org/10.1107/S0907444909052925 (2010). * Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. _Acta Crystallogr. Sect. D Biol.
Crystallogr._ https://doi.org/10.1107/S0907444904019158 (2004). * Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. _Acta Crystallogr. Sect. D
Biol. Crystallogr._ https://doi.org/10.1107/S0907444909042073 (2010). Download references ACKNOWLEDGEMENTS We thank Prof Adam Zlotnick, Indiana University, for the gift of his Cp expression
construct and advice on its purification, and Ms Leah Wells who assisted with reassembly experiments during her BSc (Honours) undergraduate research project. We thank the Medical Research
Foundation for the award of a career development grant to N.P., and the UK MRC for previous grant funding to study HBV assembly (MRF-044-0002-RG-PATEL & MR/N021517/1). R.T. and P.G.S.
thank The Wellcome Trust (Joint Investigator Award Nos. 110145 & 110146 to P.G.S. and R.T., respectively) for funding, and we acknowledge the financial support of The Trust of
infrastructure and equipment in the Astbury Centre, University of Leeds (089311/Z/09/Z; 090932/Z/09/Z & 106692), and for their additional support, together with The University of Leeds,
of the Astbury Biostructure Facility. R.T. acknowledges additional funding via an EPSRC Established Career Fellowship (EP/R023204/1) and a Royal Society Wolfson Fellowship (RSWF\R1\180009).
Portions of this work used the XFP (17-BM) beamline at NSLS-II. Development of XFP was made possible by the National Science Foundation, Division of Biological Infrastructure (grant No.
1228549), while operations support of XFP was provided by the National Institutes of Health (grant No. P30-EB-009998). NSLS-II, a US Department of Energy (DOE) Office of Science User
Facility operated for the DOE Office of Science by Brookhaven National Laboratory, was supported under Contract No. DE-SC0012704. We thank DNA Sequencing & Services (MRC I PPU, School of
Life Sciences, University of Dundee, Scotland, www.dnaseq.co.uk) for DNA sequencing. AUTHOR INFORMATION Author notes * Eva U. Weiß Present address: Institute of Molecular Infection Biology
(IMIB), University of Würzburg, Josef-Schneider-Str. 2/D15, D-97080, Würzburg, Germany * Carlos P. Mata Present address: Electron and Confocal Microscopy Unit (UCCTs), National Centre for
Microbiology (ISCIII). Majadahonda, Madrid, Spain * Jen Bohon Present address: Los Alamos National Laboratory, Los Alamos, NM, 87545, USA AUTHORS AND AFFILIATIONS * Astbury Centre for
Structural Molecular Biology, University of Leeds, Leeds, LS2 9JT, UK Nikesh Patel, Carlos P. Mata, Daniel P. Maskell, Neil A. Ranson & Peter G. Stockley * Departments of Biology and
Mathematics & York Centre for Complex Systems Analysis, University of York, York, YO10 5DD, UK Sam Clark, Eva U. Weiß & Reidun Twarock * CWRU Center for Synchrotron Biosciences,
NSLS-II, Brookhaven National Laboratory, Upton, NY, 11973, USA Jen Bohon & Erik R. Farquhar Authors * Nikesh Patel View author publications You can also search for this author inPubMed
Google Scholar * Sam Clark View author publications You can also search for this author inPubMed Google Scholar * Eva U. Weiß View author publications You can also search for this author
inPubMed Google Scholar * Carlos P. Mata View author publications You can also search for this author inPubMed Google Scholar * Jen Bohon View author publications You can also search for
this author inPubMed Google Scholar * Erik R. Farquhar View author publications You can also search for this author inPubMed Google Scholar * Daniel P. Maskell View author publications You
can also search for this author inPubMed Google Scholar * Neil A. Ranson View author publications You can also search for this author inPubMed Google Scholar * Reidun Twarock View author
publications You can also search for this author inPubMed Google Scholar * Peter G. Stockley View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS E.U.W. and S.C. analysed XRF data, C.P.M. helped analyse cryo-EM data and produced structural figures. E.F. and J.B. oversaw experiments at the Brookhaven National Laboratory.
N.P. performed experiments and analysed data. N.P. and P.G.S. wrote the paper with help from R.T., N.A.R. and C.P.M. N.P., P.G.S. and R.T. conceived the project. CORRESPONDING AUTHORS
Correspondence to Nikesh Patel or Peter G. Stockley. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW INFORMATION _Communications Biology_
thanks Ju-Tao Guo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Karli Montague-Cardoso. Peer reviewer reports are
available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION PEER REVIEW FILE SUPPLEMENTARY INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA S1 SUPPLEMENTARY DATA S2
SUPPLEMENTARY DATA S3 REPORTING SUMMARY DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0
International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the
source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative
Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by
statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Patel, N., Clark, S., Weiß, E.U. _et al._ In vitro functional analysis of gRNA
sites regulating assembly of hepatitis B virus. _Commun Biol_ 4, 1407 (2021). https://doi.org/10.1038/s42003-021-02897-2 Download citation * Received: 12 December 2020 * Accepted: 16
November 2021 * Published: 16 December 2021 * DOI: https://doi.org/10.1038/s42003-021-02897-2 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content:
Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative