- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Download PDF Article Open access Published: 05 May 2025 Molecular detection of SARS-CoV-2 and medically important respiratory and gastrointestinal virus pathogens on Thai currency Nattamon
Niyomdecha1, Chanakan Suttasit1, Attasit Boonyont1, Thanchira Saita2, Waratchaya Rodraksa2, Achiraya Phanitmas2, Nattapong Yamasamit2, Kantima Sangsiriwut3 & …Pirom Noisumdaeng2,4 Show
authors Scientific Reports volume 15, Article number: 15674 (2025) Cite this article
1674 Accesses
16 Altmetric
Metrics details
Subjects Applied microbiologyEnvironmental microbiologyInfectious diseasesVirology AbstractFomite-mediated viral transmission through using cash might be a potential risk to human health. Persistence of SARS-CoV-2, and other medically important viruses was investigated. A total of
300 samples (i.e., 150 banknotes and 150 coins) were randomly collected from nineteen fresh markets distributed across seventeen districts of Bangkok, Thailand. Every banknote or coin was
entirely swabbed and generated a total of 100 pool samples. Total viral nucleic acid was extracted and subjected for multiplex real-time qRT-PCR using Allplex™ SARS-CoV-2/FluA/FluB/RSV assay
and Allplex™ GI-virus assay. The results revealed detection rate of 4% (4/100), and they were only detected in banknote pooled samples. Two samples collected from fish shops tested positive
for SARS-CoV-2 (2%, 2/100); meanwhile, two samples (2%, 2/100) from pork and chicken shops tested positive for rotavirus A. None of pool samples were detected for influenza A and B viruses,
respiratory syncytial virus, norovirus genogroup I and II, adenovirus, astrovirus, and sapovirus. Phylogenetic analysis demonstrated that rotavirus A belonged to genotype G8; meanwhile,
SARS-CoV-2 resembled omicron GRA JN.1 sub variant. Our finding is the first report for demonstrating the presence of SARS-CoV-2 and rotavirus A in Thai banknotes on real-world situation,
implying the potential risk to human health and safety.
Similar content being viewed by others Surveillance of SARS-CoV-2 at the Huanan Seafood Market Article 05 April 2023 Detection ofSARS-CoV-2 in bioaerosols and surface samples from healthcare facilities in Klang Valley, Malaysia Article Open access 28 February 2025 SARS-CoV-2 delta variant infection in domestic dogs
and cats, Thailand Article Open access 19 May 2022 Introduction
Respiratory and gastrointestinal tract diseases caused by virus infections are a major significant challenge to medical and public health on a global scale. A variety of viruses are known to
be responsible for respiratory tract infections, including influenza A and B viruses, parainfluenza virus, respiratory syncytial virus (RSV), rhinovirus, and the recently discovered severe
acute respiratory syndrome coronavirus 2 (SARS-CoV-2)1,2,3. In the meantime, norovirus, rotavirus, astrovirus, sapovirus, enterovirus, and enteric adenovirus are primary causative agents of
gastrointestinal tract infections4,5,6. Multiple transmission pathways facilitated infection and dissemination, either directly or indirectly. These included airborne transmission via
inhalation or close contact through droplets from various secretions, such as mucus and saliva resulting from coughing and sneezing, among others7. The fecal-oral route is primarily
associated with the consumption of contaminated food and water8, as well as contact with contaminated environments. This is particularly evident in areas with high levels of contamination,
such as elevator buttons, public toilet flush buttons, fingerprint scanners, mobile phones, ATMs, and currency banknotes. These surfaces in public spaces like markets, hospitals, department
stores, and airports, where large groups of people congregate, have been significantly recognized as primary routes for infections9,10,11,12.
As above, one of the pathways for viral transmission occurs through contact (fomite-mediated transmission) with contaminated surfaces. It is evident that handling or coming into contact with
objects that serve as intermediaries between individuals may enhance the routes and possibilities for the transmission of bacteria and viruses. Currency in the form of banknotes and coins
serves as a crucial medium of exchange, significantly influencing daily human activities. Consequently, it is unavoidable that banknotes and coins will change hands, potentially heightening
the risk of contamination from bacteria or viruses present in the environment and through various human interactions. Multiple studies have indicated that banknotes and coins may serve as a
vehicle for the transmission of pathogens to individuals who come into contact with them through the use of cash within the circulating system11,13,14,15. Currency is frequently exchanged
among numerous individuals, and it is possible for droplets from coughing, sneezing, or bodily fluids to linger on the surface. This raises concerns about its potential role in the
transmission of infectious pathogens. Multiple bacterial species, including both opportunistic and pathogenic strains, were consistently identified on banknotes and coins across several
studied settings11,16,17. Nonetheless, there is limited data regarding the presence of viruses on currency banknotes and coins in real-world scenarios.
At present, individuals are increasingly attentive and cautious when handling cash from others, and cashless electronic payment methods have been implemented and widely adopted for currency
transactions. During the COVID-19 pandemic, there has been a notable increase in awareness and concern regarding the persistence of the infectious SARS-CoV-2, as well as other related viral
pathogens, including the influenza virus, rhinovirus, hepatitis A virus, rotavirus, norovirus, and poliovirus13,14,18,19,20,21. Viruses may be released from an infected individual through
respiratory secretion droplets, human excretions (such as feces and urine), and skin contact, leading to viral contamination of currency and an increased risk of fomite-based viral
transmission. Previous studies have demonstrated the stability and prevalence of detected SARS-CoV-2 in currency; however, there are limited studies addressing the real-world situation
regarding the presence of SARS-CoV-2 and other pathogenic viruses on banknotes and coins.
The currency in Thailand comprises banknotes and coins of various denominations, including banknotes of 1000 baht, 500 baht, 100 baht, 50 baht, and 20 baht, as well as coins of 10 baht, 5
baht, 2 baht, and 1 baht, with a substantial quantity of these notes and coins in circulation. Markets are the most well-known locations for individuals to conduct transactions with cash, as
they utilize substantial quantities of currency within the system. Therefore, the objective of this investigation has been to identify the presence of SARS-CoV-2 and other medically
significant respiratory and gastrointestinal pathogens on Thai banknotes and coins. Samples were taken from nineteen retail meat markets located in Bangkok, which reported the highest number
of cases of COVID-19, influenza, and gastroenteritis. Our findings offer valuable insights for public health initiatives, personal hygiene education, and the enhancement of epidemiological
surveillance for disease prevention and control.
Materials and methodsBiosafety approvalBiosafety issue was approved by the Institutional Biosafety Committee (TU-IBC), Thammasat University with the certificate of approval number 015/2567.
Study sites and sample collectionThis cross-sectional study was conducted between June and July 2024. Various values of Thai banknotes (500 baht, 100 baht, and 20 baht) and coins (10 baht, 5 baht, 2 baht and 1 baht) were
randomly collected from fresh meat shops at the markets in Bangkok, Thailand. Excellent fresh food markets certified by the Department of Internal Trade (DIT), Ministry of Commerce,
Thailand, located in various districts of Bangkok were chosen as study sites (Fig. 1). The banknote and coin samples were collected from three categories of shops: pork and chicken shops,
fish shops, and seafood shops. The samples received from each shop were kept in the sterile plastic bag under cold storage condition before testing.
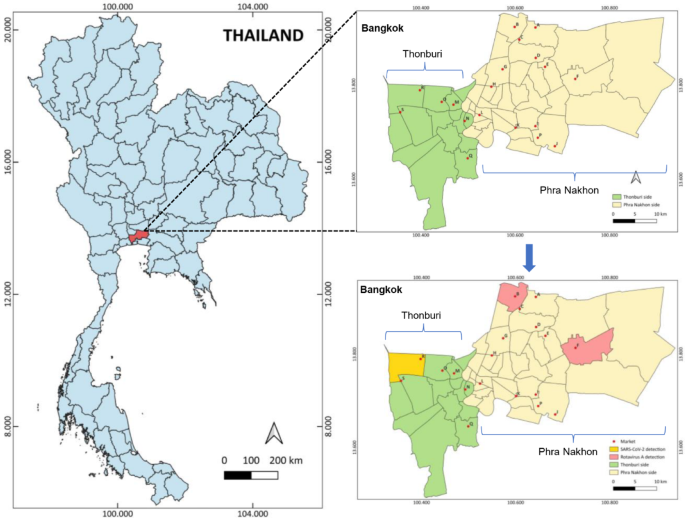
Fig. 1Study sites for currency banknote and coin collection in Bangkok, Thailand. A name of nineteen excellent fresh food markets was labeled as A to S, and the district areas showing the positive
detection results were highlighted with pink color for Rotavirus A and yellow color for SARS-CoV-2.
Full size imageSample processing and viral nucleic acid extractionA pooled sample was made by processing three banknotes, or three coins obtained from the same type of shop and the same market, as demonstrated in Fig. 2. Every banknote or coin was entirely
swabbed by synthetic nylon flocked swab, and the swab was then resuspended with 1.5 ml of ultrapure distilled water (UDW) and vigorously vortexed for 30 s. Next, the swab was discarded, and
the pooled sample was preserved at a temperature of -80 °C.
Fig. 2Schematic representation of sample collection and processing protocol.
Full size imageThe total viral genomic RNA or DNA was extracted on MagLEAD 12gC automated extraction platform (Precision System Science, Chiba, Japan) from 400 µl of a pooled sample. Extraction protocol
was performed according to the manufacturer’s instructions. Viral genomic RNA or DNA was eluted with 50 µl elution buffer and stored at -80 °C. The extracted viral genome was then subjected
for target nucleic amplification.
Medically important virus detection based on multiplex real-time PCR assaysThe extracted viral genome was used as a template to simultaneously detect the medically important respiratory (RT) viruses by using Allplex™ SARS-CoV-2/FluA/FluB/RSV Assay (abbreviated as
Allplex-SC2FabR) and the medically important gastrointestinal (GI) viruses by using Allplex™ GI-Virus Assay (abbreviated as Allplex-GI). These two commercial kits (Seegene, Seoul, Republic
of Korea) detected simultaneously specific viral gene targets based on one-step multiplex real-time reverse transcription PCR (qRT-PCR).
The Allplex-SC2FabR assay was used to detect three target genes (S gene, RdRP gene and N gene) of the SARS-CoV-2 RNA, M gene of influenza A (Flu A) RNA and NS2 gene of influenza B (Flu B)
RNA as well as M gene of human respiratory syncytial virus (RSV) RNA. The Allplex-GI assay is a recent one-step multiplex qRT-PCR kit that uses a multiple detection temperature technique
(MuDT)22. This kit is designed for simultaneously detecting rotavirus A (ROV), norovirus genogroup I (NVGI) and norovirus genogroup II (NVGII), adenovirus Type F (ADV-F) subtype 40/41,
astrovirus (ASV), and sapovirus (SV). All assays were done according to the manufacturer’s instructions. The amplification process was carried out using the CFX96 Touch Real-Time PCR
Detection System (Bio-Rad, California, USA). The PCR cycling conditions of the Allplex-SC2FabR assay were as followed: reverse transcription at 50 °C for 20 min, denaturation at 95 °C for 15
min, 3 cycles of 95 °C for 10 s, 60 °C for 40 s, and 72 °C for 20 s followed by 42 cycles of 95 °C for 10 s, 60 °C for 15 s, and 72 °C for 10 s with fluorescence detection at 60 °C and 72
°C. The PCR cycling conditions of the Allplex-GI assay were as followed: reverse transcription at 50℃ for 20 min, denaturation at 95℃ for 15 min, and 45 cycles of 95 °C for 10 s, 60 °C for 1
min, and 72 °C for 30 s.
The reliability of the tested assay was evaluated using a positive control, which produced valid results indicating positivity in all genes, and an RNase-free water negative control, which
yielded valid results showing negativity in all genes, both are included in each kit. The sample extraction and target amplification processes were validated through the use of the exogenous
internal control included in the Allplex-SC2FabR assay, designated as RP-V IC 2, and the Allplex-GI assay, referred to as GI-V IC. Additionally, the Allplex-SC2FabR assay utilized an
endogenous internal control to assess the quality of human sample collection. However, our sample was an environmental sample, not a clinical specimen. In contrast to the manufacturer’s
guideline, the interpretation did not account for the outcome of endogenous internal control. The positive detection criteria for the Allplex-SC2FabR assay in the real-world Thai currency
samples were as follows: a positive signal (quantification cycle, Cq value ≤ 40) in any gene of SARS-CoV-2, or a specific gene for other respiratory (RT) viruses with a positive signal of
exogenous internal control. For the Allplex-GI assay, the positive detection was defined as a positive signal (Cq value ≤ 40) in a specific gene for gastrointestinal (GI) viruses with a
positive signal of exogenous internal control.
Evaluation of protocol validityTo evaluate the validity of our protocol in virus detection from banknote and coin, an artificially spiked sample was created. A banknote or coin was cleaned with 70% alcohol, and any
remaining alcohol was eliminated using UDW cleaning. On a banknote or coin, twenty microliters (µl) of extracted SARS-CoV-2 RNA from 104 pfu/100 µl virus stock were randomly spotted. The
artificial samples were then dried at room temperature for 20 min. Using the sample processing protocol as indicated above, the extracted RNA was analyzed for the presence of SARS-CoV-2
target genes by the Allplex-SC2FabR. The experiment was done in duplicate.
Detection of SAR-CoV-2 and ROV by conventional PCR and DNA sequencingThe positive RNA samples were subsequently analyzed for SARS-CoV-2 strain or rotavirus A genotyping. The extracted RNA was used to generate cDNA by using SuperScript™ III First-Strand
Synthesis System (Invitrogen, Thermo Fisher Scientific, CA) according to the manufacturer’s instructions. The conventional PCR was conducted by using specific primer sets for target
amplification as followed: the nucleocapsid (N) primers for detecting SARS-CoV-2 (PPMU-28533F: 5’-TTGGCAATGTTGTTCCTTGA-3’ and PPMU-28778R: 5’- ACCGAAGAGCTACCAGACGA − 3’) kindly provided from
Prof. Pilaipan Puthavathana, Faculty of Medical Technology, Mahidol University, and the VP7 primers for detecting rotavirus A (sBeg9: 5’- GGCTTTAAAAGAGAGAATTTC-3’ and VP7-1’: 5’-
ACTGATCCTGTTGGCCATCCTTT-3’)23,24. The PCR products of 250 bp for SARS-CoV-2 and 395 bp for rotavirus A were visualized by electrophoresis on 2% agarose gel, purified by using QIAquick® gel
extraction kit (GIAGEN) and sent purified DNA for sequencing to Macrogen Inc., Seoul, Korea (https://dna.macrogen.com/main.do). All nucleotide sequences were deposited to the GenBank
database for the accession numbers PQ472486 for SARS-CoV-2 (strain: SARS-CoV-2/Environment/THA/Banknote-P55/2024) and PQ471577 for Rotavirus A (strain:
RVA/THA/Banknote-P41/2024).
Phylogenetic tree analysisSARS-CoV-2 isolate in the study was investigated for strain identification by comparing to reference SARS-CoV (Tor) and various SARS-CoV-2 variants as well as SARS-CoV-2 BLAST-hit retrieved
from GISAID (https://gisaid.org/) and NCBI databases (https://www.ncbi.nlm.nih.gov/) as shown in supplementary Table S1. The phylogenetic tree construction was generated based on N gene (250
bp).
The genotype of rotavirus A was investigated based on the classification systems previously described25. A total of 52 known RVA reference genotypes (G1 to G42) were retrieved from the
GenBank database and used in the construction of the phylogenetic trees. In addition, 10 RVA strains from BLAST-hit (https://blast.ncbi.nlm.nih.gov/Blast.cgi) were also included for
phylogenetic trees (supplementary Table S1).
The viral sequences were aligned using ClustalW multiple sequence alignment in the BioEdit version 7.0.5.226, and a phylogenetic tree was constructed using MEGA 7.0.26 software
(https://megasoftware.net/)27. The evolutionary distances were estimated using the neighbor-joining method and maximum composite likelihood algorithm. The reliability of the neighbor-joining
tree was estimated by bootstrapping analysis using 1000 replicate datasets.
Statistical analysisThis study employed descriptive statistical analysis. A distribution analysis was conducted using the frequency or percentage of data. The positive result of the multiplex real-time qRT-PCR
was repeated, and data are reported as mean ± SD.
ResultsValidity of the sample processing protocolPrior to collecting the real-world samples, an artificial spike of SARS-CoV-2 RNA was made on banknotes and coins. The swab sample was used in the Allplex-SC2FabR multiplex real-time PCR to
identify SARS-CoV-2 target genes. As shown in Table 1, our sample processing protocol was valid enough to detect 2 out of 3 SARS-CoV-2 target genes without yielding false positive results
from other viruses. Additionally, the Cq values derived from both sample types were comparable.
Table 1 SARS-CoV-2 detection by the Allplex-SC2FabR multiplex real-time PCR.Full sizetableSample collection and the multiplex real-time PCR detection
Banknotes and coins were collected from nineteen excellent fresh food markets (labeled A-S) located in different districts of Bangkok province across two zone areas of Phra Nakhon and
Thonburi (Fig. 1; Table 2). Any combination of three banknotes or three coins from the same type of food shop were collected as pooled swab samples (Fig. 2). A total of 100 pooled samples
was equally derived from the pooled samples from banknotes and coins, and the distribution data of sample collection was shown in Fig. 3. Most predominant pooled samples were collected from
the pork and chicken shops, seafood shops, and fish shops, respectively.
Table 2 Details of market locations and number of collected samples.Full size tableFig. 3The distribution of pooled sample collection in each shop type. Each bar represented the raw data of currency banknote and coin samples from each shop type.
Full size imageAs shown in Table 3, the positive detection rate for viruses was only 4% (4/100) and they were only detected in banknote pooled swab samples. Two samples collected from fish shops tested
positive for SARS-CoV-2 (2%, 2/100); meanwhile, two samples (2%, 2/100) from pork and chicken shops tested positive for rotavirus A. The positive results were found in different market
locations as indicated in Fig. 1. The correlation between the shop type and positive virus may indicate a potential risk source for viral infection. However, all Cq values of detection were
higher than 35 cycles.
Table 3 The positive samples detected by the multiplex real-time PCR assay targeted the panels of (A) respiratory viruses and (B) gastroenteritis viruses.Full sizetablePhylogenetic construction for viral genotyping
Among positive samples detected by conventional RT-PCR, the study conducted DNA sequencing and phylogenetic tree analysis. The rotavirus A phylogenetic analysis against the reference strains
revealed that our rotavirus A belonged to genotype G8. From nucleotide BLAST and phylogenetic analyzed, rotavirus A VP7 sequence was phylogenetically closely related to rotavirus A isolated
from clinically human stools reported in the United States in 2009, raw sewage in China in 2022 and bovine in China in 2023. (Fig. 4 and supplementary Table S1). Meanwhile, our SARS-CoV-2 N
gene sequence mostly resembled variant of interest (VOI) omicron GRA (JN.1 sub variant) isolated from humans in the United States and Canada in 2024 (Fig. 5 and supplementary Table
S2).
Fig. 4Phylogenetic construction for rotavirus A genotyping based on VP7 region (338 bp). The evolutionary distances were estimated using the neighbor-joining method and maximum composite
likelihood algorithm. The reliability of the neighbor-joining tree was estimated by bootstrapping analysis using 1000 replicate datasets.
Full size imageFig. 5Phylogenetic construction of SARS-CoV-2 N gene (248 bp) against SARS-CoV Tor strain and various variants of SARS-CoV-2. The evolutionary distances were estimated using the neighbor-joining
method and maximum composite likelihood algorithm. The reliability of the neighbor-joining tree was estimated by bootstrapping analysis using 1000 replicate datasets.
Full sizeimageDiscussion
The knowledge of possible potential transmission sources of clinically relevant pathogens is crucial for infectious disease prevention. Banknotes and coins have been discussed as fomite for
an indirect contact transmission because they are routinely exchanged between individuals. The handling of cash in routine businesses was hypothesized as a viral transmission vector in the
covid-19 era14, and the World Health Organization (WHO) and the Centers for Disease Control (CDC) have emphasized that people should be aware and using it with safety precautions, rather
than banning it.
The reliability of our sample processing protocol in an artificial setting experiment could promote the validity of true positive or true negative results in the real-world samples. The
recovery Cq data from the tested banknote and coin were similar, with the exception of the Cq values from the banknote being slightly lower than those found in the coin. This suggests that
there were no significant inhibitors from materials in the amplification reaction, and the lower Cq value in the tested banknote may correlate with its texture characteristics of a large and
flat surface, enhancing the efficacy of swab collection. However, only the S and RdRp genes were amplified. Three primary reasons account for the explanation: (1) the quantity of positive
genes is contingent upon the viral quantity in the sample and the integrity of the retrieved RNA. It implies that if the sample containing SARS-CoV-2 RNA had a very low concentration and was
not intact, the test would fail to detect any target genes, (2) the absence of N gene detection could be attributed to a mismatch of the primer and probe, and (3) the analytical sensitivity
of the N gene using the Allplex SARS-CoV-2/FluA/FluB/RSV kit was 95.51% (88.89–98.76), which is lower than that of the S gene, which is at 98.88% (93.90–99.97) and the RdRp gene which is at
97.75% (92.12–99.73) as demonstrated by Kim et al.28. That prior investigation identified 5 positive outcomes from 90 positive samples exhibiting amplified targets absent of the N gene28.
However, the results from the positive control showed the successful detection of all three target genes, demonstrating that the master mix used in this test was properly prepared and
capable of detecting the N gene, RdRp gene, and S gene. The positive control serves as a synthetic material used to validate the effectiveness of the reagent kit in amplifying each target
gene.
This study identified SARS-CoV-2 in 2% (2/100) of pooled samples collected from two distinct markets within the Thawi Wattana district, and rotavirus A in 2% (2/100) of samples from two
different markets located in the Bang Sue and Min Buri districts. Similarly, SARS-CoV-2 positive samples were identified in the absence of the N gene. From the beginning of week 24 to the
middle of week 29 in 2024 of our study periods, the Department of Disease Control (DDC) of the Ministry of Public Health (MoPH) in Thailand reported an average of 133–465 confirmed
SARS-CoV-2 cases per day, while the prevalence of confirmed rotavirus cases ranged from 26.32 to 27.12%, ranking second after norovirus GII cases, which were reported at 36.44–36.84%
(https://ddc.moph.go.th/). Nevertheless, the DDC reports did not provide the category data between infected cases and locations. Consequently, we were unable to elucidate the correlation
between the positive findings and the case incidence in the detected area.
Furthermore, another limitation of this investigation was the comparison of the positive viral detection rate with prior studies, as the data available was extremely few for both viruses.
The research conducted by Newey et al.29 found no SARS-CoV-2 RNA in any banknotes and coins collected from Utah, USA, using LAMP assay targeting ORF1ab and S genes. Conversely, the study by
Akter et al.11 reported a positive detection rate of 7.29% (31/425) for Bangladesh banknotes using real-time RT-PCR. In addition, to our knowledge, we have reported for the first time the
positive detection rate of currency contaminated with rotavirus. The factors associated with result variations may be influenced by the occurrence of cases at the study sites, hygienic
measures, material and surface of sample types, virus strain, various physical parameters, and sensitivity of detection methods30. Our chosen study sites are excellent fresh food markets
accredited by the Department of Internal Trade (DIT), Ministry of Commerce, Thailand. Stringent hygiene requirements are implemented in these markets to mitigate the risk of contamination.
Interestingly, only banknotes have tested positive detection. This may be explained by the difference in viral stability between on banknote and coin material and surfaces. Most Thai
banknotes are still made from 100% cotton fiber paper, except for the 20 baht note that has been recently changed from paper to polymer to improve the quality and make it more durable31.
Riddell et al.32 demonstrated that SARS-CoV-2 remains stable on paper and polymer banknotes for a minimum of 28 days at 20 °C. Kramer et al.33 have demonstrated that many gastrointestinal
viruses, including rotavirus, can persist on environmental inanimate objects for approximately 2 months. On the other hand, all Thai baht coins are made of various types of metal alloys
depending on each value, including nickel-clad iron, nickel-copper, and bimetallic between Cupronickel and Aluminium Bronze34. The numerous viruses can be effectively eradicated within 4 h
on copper and other alloy surfaces35,36,37. The antiviral effect may be due to the release of copper ions from metal surface, which results in the killing process38. Furthermore, coins
represent a lower risk than banknotes due to their significantly reduced exposed surface areas39.
Asymptomatic or mildly symptomatic individuals can act as a possible source of currency contamination by handling cash with contaminated hands that poses a risk of viral transmission. The
virions may infect another individual by being transferred via a finger from contact with contaminated currency to a face mucous membrane or through ingestion39. Conversely, the cause of
money contamination may arise not from interaction with infected individuals, whether vendors or buyers, but from food that is contaminated within the processing chain that is not entirely
eradicated. We presented the linked findings between the identified virus and the type of shop. The SARS-CoV-2 RNA samples have yielded positive results at the fish shop, whereas the
rotavirus A RNA samples have tested positive at the pork and poultry shop. Prior reports have documented the detection of SARS-CoV-2 on frozen aquatic food species and their products,
including packaging materials and storage environments40. Additionally, the rotavirus was previously identified in pork and chicken meat cuts available in retail markets41,42,43. Virus
contamination from contaminated foods to cash may occur through hand contact between them in the absence of proper hygienic precautions. The detection of viable virus is crucial for
demonstrating potential ability for viral transmission and infectivity. Unfortunately, our study faces a limitation with viral culture because it must be conducted in a Biosafety Level 3
(BSL3) facility, which is not generally available at our institute, and it requires expert technicians for the virus culture. Nonetheless, all positive Cq values obtained in this study were
above 35 cycles. It is possible that this is a viral remnant rather than infectious particles, which are at a reduced risk of causing viral transmission and infection. Schijven et al.39
demonstrated a minimal risk of banknotes serving as fomites for SARS-CoV-2 in cash transactions under real-world circumstances. Additionally, the successful infection requires high viral
loads, like those found in saliva and cough specimens of infected patient at levels of > 1010 copies/ml or > 106 PFU/ml44,45.
The isolated SARS-CoV-2 in the present investigation was classified as JN.1 sub-variants of the Omicron variant based on the genetic analysis and phylogenetic tree construction. We have also
analyzed nucleotide and amino acid sequence identities of our virus strain SARS-CoV-2/Environment/THA/Banknote-P55/2024 against multiple known SARS-CoV-2 variants by using Sequence Identity
Matrix application in BioEdit program. The result demonstrated that our identified virus was closely related to SARS-CoV-2 variant of Omicron GRA (JN.1) by showing 100.0% nucleotide
identity, and showed the 100% amino acid identity to all SARS-CoV-2 variants. In January 2024, Thailand experienced a significant increase in the number of hospitalizations and fatalities as
a result of the JN.1 sub-variant of the Omicron covid-19 variant46. The WHO has recently designated a novel strain, JN.1, as a variant of interest (VOI), emphasizing its potential
importance in the ongoing virus control effort. Despite the fact that existing antibodies offer some protection, variant JN.1 is characterized by unique mutations and rapid transmission.
Nevertheless, current vaccines demonstrate substantial potential47. Furthermore, scientists are expressing worries regarding the potential for the JN.1 variant of SARS-CoV-2 to specifically
target the intestinal tracts of individuals in addition to the respiratory tract48.
Rotavirus A is the most prevalent species and primarily infects humans, particularly young infants, resulting in acute gastroenteritis49. Based on the phylogenetic tree data, we identified
rotavirus A genotype G8 from our positive sample. The circulating strains of rotavirus in Thailand have been changed over time, more than twelve years period from 2008 to 2020, and the G8
genotype was one of predominant strains found in pediatric patients with diarrhea, especially from 2018 to 2020. Therefore, the G8 genotype became the most prevalent genotype49. Jiang et
al.50 indicated that children infected with G9 and G8 genotypes exhibited more severe symptoms, whereas the occurrence of fever and electrolyte imbalance did not significantly vary among the
other genotypes. Moreover, the rotavirus A genotype G8 has been identified in swine globally, although it is not the predominant strain49. Due to the prolonged persistence of rotavirus on
environmental surfaces33, there will be an increased risk of viral transmission and infection.
In conclusion, although banknotes and coins can act as carriers for virus transmission, the danger of causing illness appears to be minimal due to the low contamination rate. Nevertheless,
it is imperative to practice caution when handling cash and coins. Advancements in cashless and contactless transactions through online banking and digital wallets should be actively sought
wherever possible.
Data availabilityData is provided within the manuscript or supplementary information files. All nucleotide sequences generated in the study are available in the NLM-NCBI repository under GenBank accession
numbers PQ472486 and PQ471577.
References Calderaro, A. et al. Respiratory tract infections and laboratory diagnostic methods: a review with a focus on syndromic panel-based assays. Microorganisms 10 (9), 1856 (2022).
Article CAS PubMed PubMed Central Google Scholar
Dawre, S. & Maru, S. Human respiratory viral infections: current status and future prospects of nanotechnology-based approaches for prophylaxis and treatment. Life Sci. 278, 119561 (2021).
Article CAS PubMed PubMed Central Google Scholar
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579 (7798), 265–269 (2020).
Article ADS CAS PubMed PubMed Central Google Scholar
Orenstein, R. Gastroenteritis, viral. Encyclopedia Gastroenterol. 2nd Edition, 652–657. https://doi.org/10.1016/B978-0-12-801238-3.65973-1 (2020).
Article Google Scholar
Bishop, R. F., Kirkwood, C. D., Enteric & Viruses Encyclopedia Virol. 116–123. https://doi.org/10.1016/B978-012374410-4.00386-1 (2008).
Jagirdhar, G. S. K., Pulakurthi, Y. S., Chigurupati, H. D. & Surani, S. Gastrointestinal tract and viral pathogens. World J. Virol. 12 (3), 136–150 (2023).
Article PubMed PubMed Central Google Scholar
Wang, C. C. et al. Airborne transmission of respiratory viruses. Science 373 (6558), eabd9149 (2021).
Article ADS CAS PubMed PubMed Central Google Scholar
de Graaf, M. et al. Sustained fecal-oral human-to-human transmission following a zoonotic event. Curr. Opin. Virol. 22, 1–6 (2017).
Article PubMed Google Scholar
Castaño, N. et al. Fomite transmission, physicochemical origin of virus-surface interactions, and disinfection strategies for enveloped viruses with applications to SARS-CoV-2. ACS Omega. 6
(10), 6509–6527 (2021).
Article PubMed PubMed Central Google Scholar
Zhuang, L. et al. Fomite transmission in airports based on real human touch behaviors. Buildings 13 (10), 2582 (2023).
Article Google Scholar
Akter, S. et al. Prevalence and stability of SARS-CoV-2 RNA on Bangladeshi banknotes. Sci. Total Environ. 779, 146133 (2021).
Article CAS PubMed PubMed Central Google Scholar
Niyomdecha, N. et al. Detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and bacteria on highly common contaminated surfaces at urban hospital and public areas. Nat. Life
Sci. Commun. 22(1), e2023010 (2023).
Meister, T. L. et al. Stability of pathogens on banknotes and coins: A narrative review. J. Med. Virol. 95 (12), e29312 (2023).
Article CAS PubMed Google Scholar
Angelakis, E. et al. Paper money and coins as potential vectors of transmissible disease. Future Microbiol. 9 (2), 249–261 (2014).
Article MathSciNet CAS PubMed Google Scholar
Barbora, T. et al. Occasional paper series catch me (if you can): assessing the risk of SARS-CoV-2 transmission via Euro cash. (2021).
https://www.ecb.europa.eu/pub/pdf/scpops/ecb.op259~33b180d450.en.pdf
Saakshi, J. et al. Screening currency notes for microbial pathogens and antibiotic resistance genes using a shotgun metagenomic approach. PLoS One 10(6), e0128711 (2015).
Ofoedu, C. E. et al. Bacterial contamination of Nigerian currency notes: A comparative analysis of different denominations recovered from local food vendors. PeerJ 9, e10795 (2021).
Article PubMed PubMed Central Google Scholar
Thomas, Y. et al. Survival of influenza virus on banknotes. Appl. Environ. Microbiol. 74 (10), 3002–3007 (2008).
Article ADS CAS PubMed PubMed Central Google Scholar
Kraay, A. N. M. et al. Fomite-mediated transmission as a sufficient pathway: a comparative analysis across three viral pathogens. BMC Infect. Dis. 18 (1), 540 (2018).
Article PubMed PubMed Central Google Scholar
Sattar, S. A., Lloyd-Evans, N., Springthorpe, V. S. & Nair, R. C. Institutional outbreaks of rotavirus diarrhoea: potential role of fomites and environmental surfaces as vehicles for virus
transmission. J. Hyg. (Lond). 96 (2), 277–289 (1986).
Article CAS PubMed Google Scholar
Lopez, G. U. et al. Transfer efficiency of bacteria and viruses from porous and nonporous fomites to fingers under different relative humidity conditions. Appl. Environ. Microbiol. 79 (18),
5728–5734 (2013).
Article ADS CAS PubMed PubMed Central Google Scholar
Lee, Y. J., Kim, D., Lee, K. & Chun, J. Y. Single-channel multiplexing without melting curve analysis in real-time PCR. Sci. Rep. 4, 7439 (2014).
Article CAS PubMed PubMed Central Google Scholar
Kumthip, K., Khamrin, P., Ushijimam, H. & Maneekarn, N. Detection of six different human enteric viruses contaminating environmental water in Chiang Mai, Thailand. Microbiol. Spectr. 11(1),
e0351222 (2023).
Thongprachum, A. et al. Detection of nineteen enteric viruses in Raw sewage in Japan. Infect. Genet. Evol. 63, 17–23 (2018).
Article PubMed Google Scholar
Matthijnssens, J. et al. Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification working group (RCWG). Arch. Virol. 156 (8), 1397–1413 (2011).
Article CAS PubMed PubMed Central Google Scholar
Hall, T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 (1999).
CAS Google Scholar
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33 (7), 1870–1874 (2016).
Article CAS PubMed PubMed Central Google Scholar
Kim, H. N., Yoon, S. Y., Lim, C. S. & Yoon, J. Comparison of three molecular diagnostic assays for SARS-CoV-2 detection: evaluation of analytical sensitivity and clinical performance. J.
Clin. Lab. Anal. 36 (2), e24242 (2022).
Article CAS PubMed PubMed Central Google Scholar
Newey, C. R. et al. Presence and stability of SARS-CoV-2 on environmental currency and money cards in Utah reveals a lack of live virus. PLoS One 17(1), e0263025 (2022).
Chan, K. H. et al. The effects of temperature and relative humidity on the viability of the SARS coronavirus. Adv. Virol. 2011, 734690 (2011).
Bank of Thailand. (2024). Available from: https://www.bot.or.th/en/about-us.html (Accessed 14 October 2024).
Riddell, S., Goldie, S., Hill, A., Eagles, D. & Drew, T. W. The effect of temperature on persistence of SARS-CoV-2 on common surfaces. Virol. J. 17 (1), 145 (2020).
Article CAS PubMed PubMed Central Google Scholar
Kramer, A., Schwebke, I. & Kampf, G. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 6, 130 (2006).
Article PubMed PubMed Central Google Scholar
Royal Thai Mint. Available form: https://www.royalthaimint.net/ (Accessed 16 October 2024) (2024).
van Doremalen, N. et al. Aerosol and surface stability of SARS-CoV-2 as compared with SARS-CoV-1. N Engl. J. Med. 382 (16), 1564–1567 (2020).
Article PubMed Google Scholar
Suman, R. et al. Sustainability of coronavirus on different surfaces. J. Clin. Exp. Hepatol. 10 (4), 386–390 (2020).
Article PubMed PubMed Central Google Scholar
Abraham, J., Dowling, K. & Florentine, S. Can copper products and surfaces reduce the spread of infectious microorganisms and hospital-acquired infections? Mater. (Basel). 14 (13), 3444
(2021).
Article ADS CAS Google Scholar
Mathews, S., Hans, M., Mücklich, F. & Solioz, M. Contact killing of bacteria on copper is suppressed if bacterial-metal contact is prevented and is induced on iron by copper ions. Appl.
Environ. Microbiol. 79 (8), 2605–2611 (2013).
Article ADS CAS PubMed PubMed Central Google Scholar
Schijven, J. F. et al. Risk assessment of banknotes as a fomite of SARS-CoV-2 in cash payment transactions. Risk Anal. 43 (4), 700–708 (2023).
Article PubMed Google Scholar
Han, J., Zhang, X., He, S. & Jia, P. Can the coronavirus disease be transmitted from food? A review of evidence, risks, policies and knowledge gaps. Environ. Chem. Lett. 19 (1), 5–16 (2021).
Article CAS PubMed Google Scholar
Soares, V. M. et al. Detection of adenovirus, rotavirus, and hepatitis E virus in meat cuts marketed in Uruguaiana, Rio Grande do Sul, Brazil. One Health. 14, 100377 (2022).
Article CAS PubMed PubMed Central Google Scholar
Jones, T. H. & Muehlhauser, V. Frequency of hepatitis E virus, rotavirus and Porcine enteric calicivirus at various stages of pork carcass processing in two pork processing plants. Int. J.
Food Microbiol. 259, 29–34 (2017).
Article PubMed Google Scholar
Pereira, J. G. et al. Hepatitis A virus, hepatitis E virus, and rotavirus in foods of animal origin traded at the borders of Brazil, Argentina, and Uruguay. Food Environ. Virol. 10 (4),
365–372 (2018) .
Yang, Q. et al. Just 2% of SARS-CoV-2-positive individuals carry 90% of the virus circulating in communities. Proc. Natl. Acad. Sci. U S A 118(21), e2104547118 (2021).
Lin, Y. C. et al. Detection and quantification of infectious severe acute respiratory coronavirus-2 in diverse clinical and environmental samples. Sci. Rep. 12 (1), 5418 (2022).
Article ADS CAS PubMed PubMed Central Google Scholar
Krajangwit, J. Surge in JN.1 omicron cases triggers health alert. Available form








