- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Normal veins could develop to varicose vein (VV) by some risk factors, and might further progress to shallow vein thrombosis (SVT). However, the molecular mechanism of key genes
associated with the progression and regression of VV are still not thorough enough. In this study, the healthy control (HC), VV, and SVT vascular samples were collected for transcriptome
sequencing. The differentially expressed genes (DEGs) were screened by “DESeq2”, including DEGs1 (HC vs. VV), DEGs2 (HC vs. SVT) and DEGs3 (VV vs. SVT). And their functional enrichment
analyses were conducted by “ClusterProfiler”. The receiver operating characteristic (ROC) curve was used to obtain the key genes (KGs) of the pathogenesis of VV and SVT. The qRT-PCR assay
was performed to validate the expressions of KGs. Immune cell infiltration analyses were conducted based on ssGSEA method. The competitive endogenous RNAs (ceRNAs) regulatory network was
constructed. The target drugs of KGs were predicted using DrugBank database. The biofunctions of DACT3 were further investigated through a series of experiments in vitro. All of these DEGs
were associated with inflammation and immunity related functions. Immune cell infiltration was significantly different between VV and SVT. Six key genes including PLP2, DACT3, LRRC25, PILRA,
MSX1 and APOD that were associated with the progression and regression of VV were screened. The expression of LRRC25 and PILRA was significantly negatively associated with central memory T
cell, and significantly positively associated with B cell. Besides, XIST was the critical regulator of multiple KGs. Cimetidine was potential drug for VV and SVT therapy. Overexpression of
DACT3 significantly inhibited the proliferation and migration of vascular smooth muscle cells (VSMCs), and affected their cell cycle and phenotypic transition. This study identified six key
genes associated with the progression and regression of VV. Among them, DACT3 was proved to hinder VV progression. These findings may help to deepen understanding its underlying mechanisms.
SIMILAR CONTENT BEING VIEWED BY OTHERS IDENTIFICATION OF IMPORTANT GENES RELATED TO HVSMC PROLIFERATION AND MIGRATION IN GRAFT RESTENOSIS BASED ON WGCNA Article Open access 12 January 2024
CAUSAL ROLE OF IMMUNE CELLS IN VARICOSE VEINS: INSIGHTS FROM A MENDELIAN RANDOMIZATION STUDY Article Open access 04 June 2025 A VEIN WALL CELL ATLAS OF MURINE VENOUS THROMBOSIS DETERMINED BY
SINGLE-CELL RNA SEQUENCING Article Open access 31 January 2023 INTRODUCTION Varicose vein (VV) of the lower extremities, is a common manifestation in the field of vascular surgery1. If
there is a valve malfunction or blockage in the great saphenous vein, blood from the femoral vein will flow back into the great saphenous vein2,3. When people are standing, because the
venous valve is not fully closed, the blood flow in the deep vein will backflow into the superficial vein of the lower limb under the gravity of gravity4. The venous pressure in the lower
leg will be greatly increased by about 20mmHg5. The elevated pressure often results in several classical symptoms, such as tortuous dilation of superficial veins and impairment of the skin’s
microvascular system, which can manifest as skin pigmentation, lipid sclerosis, and ulcer formation6. The incidence of varicose veins is as high as 20–40%, and more than half of people over
the age of 40 will have varying degrees of varicose veins in their lower limbs7. Venous blood builds up in the branches or main trunk of the great saphenous vein, eventually leading to
fatigue and swelling in the legs. In addition, the abnormal supply of oxygen and nutrients can cause skin symptoms such as atrophy, desquamation, itching, and ulcers8,9. Superficial venous
thrombosis of the lower extremities is a common disorder reported to affect 3–11% of the general population10,11. In patients with varicose veins, the incidence of shallow vein thrombosis
(SVT) ranges from 4 to 59%, and the most commonly affected site is the tributary of varicose veins12. Patients with VV have multiple complications, the most common of which is SVT13, and the
vast majority of SVT cases are diagnosed in patients with VV14. In conclusion, normal veins first develop into VV under the induction of some risk factors, and then may further develop into
SVT15. However, the key genes and underlying mechanisms that influence disease development are not yet known, and in recent years, microarray technology, together with comprehensive
bioinformatics analysis, has been used to identify novel genes associated with various diseases that may serve as disease diagnostic and prognostic biological markers16,17. Therefore, this
study conducted mRNA sequencing by collecting samples of normal veins, varicose veins, and varicose veins with thrombus to explore the key genes involved in the progression of varicose vein
disease and to further explore the functions of key genes and their relationship with immune cells18. This study also predicted its regulatory mechanisms and targeted drugs, while clarifying
the function of these genes in the development and progression of varicose veins19. MATERIALS AND METHODS DATA COLLECTION, RNA EXTRACTION AND LIBRARY CONSTRUCTION In this study, totals of 5
healthy control (HC), 10 VV and 10 SVT vascular samples were acquired for total RNA extraction and sequencing. This study was approved by the Shaanxi Provincial People’s Hospital ethical
review committee (No.SP20230672). All patients provided written informed consent before sequencing. All methods were carried out in accordance with relevant guidelines and regulations. Total
RNA was isolated and purified using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and the RNA amount and purity of each sample were quantified using NanoDrop ND-1000 (NanoDrop,
Wilmington, DE, USA). The poly(A) RNA was fragmented into small pieces and reverse-transcribed to create the cDNA. Next, second-stranded DNAs were synthesized with E.coli DNA polymerase I,
RNase H and dUTP Solution. Single- or dual-index adapters were ligated to the fragments, and size selection was performed with AMPureXP beads. The U-labeled second-stranded DNAs were treated
by the heat-labile UDG enzyme (NEB, cat.m0280, USA), and the ligated products were amplified with polymerase chain reaction (PCR). The average insert size for the final cDNA library was 300
± 50 bp. At last, we performed the 2 × 150 bp paired-end sequencing (PE150) on an Illumina NovaSeqTM 6000 (LC-Bio Technology CO, Ltd, Hangzhou, China)20 following the vendor’s recommended
protocol. Finally, the mRNA sequencing data of 5 HC, 10 VV and 10 SVT samples were obtained. The quality assessment of the sequencing data was analyzed by “FastQC” (version 0.11.9) on
[2022.11.20]21,22,23. The low quality data were filtered to remove contamination and adaptor sequences, and the clean data were finally obtained by “Trimmomatic” (version 0.39) on
[2022.11.20]24. Besides, the clean data was aligned to the reference genome (hg19) by “hisat2” (version 2.2.1) on [2022.11.20]25. IDENTIFICATION AND FUNCTIONAL ENRICHMENT ANALYSES OF THE
DIFFERENTIALLY EXPRESSED GENES (DEGS) The DEGs1 between HC and VV samples, the DEGs2 between HC and SVT samples, and the DEGs3 between VV and SVT samples were compared by “DESeq2” R package
(version 1.30.1) on [2022.12.23] (|log2FC| > 0.5, adj._p_.value < 0.05), respectively26. The functional enrichment analyses of DEGs1, DEGs2 and DEGs3 were conducted by
“clusterprofiler” R package (version 3.18.1) and “org.Hs.eg.db” R package (version 3.12.0) on [2022.12.24], respectively (adj._p_.value < 0.05)27. THE IMMUNE MICRO-ENVIRONMENT ANALYSES
AMONG DIFFERENT GROUPS In this study, the proportions of 24 immune cells in HC, VV and SVT samples were calculated by “ssGSEA” algorithm and compared by “rank-sum test” on [2022.12.24].
Moreover, the expression of marker genes that belong to differential immune cells was compared by “rank-sum test”28. IDENTIFICATION AND FUNCTIONAL ANALYSIS OF TARGETED GENES ASSOCIATED WITH
THE PROGRESSION AND REGRESSION OF VV The targeted genes (TGs) of each group were obtained by intersecting the DEGs1, DEGs2 and DEGs3 using “venn” R package (version 1.11) on [2022.12.24].
Among these, TGs1, which were associated with the occurrence of VV, were unique to the VV groups. TGs2, associated with the development of VV (inducing VV to SVT), were unique genes in the
SVT groups. TGs3, associated with the progression and regression of VV, were the common genes of DEGs1, DEGs2, and DEGs3. Moreover, the functional enrichment analyses of TGs1, TGs2 and TGs3
were conducted by “Metascape” on [2022.12.25], respectively (https://metascape.org/gp/index.html#/main/step1). CORRELATION ANALYSES BETWEEN KEY GENES AND DIFFERENTIAL IMMUNE CELLS Firstly,
the receiver operating characteristic (ROC) curves of each target gene were drawn by “pROC” R package (version 1.17.0.1) on [2022.12.25]. TGs1 and TGs2, with a top 10 area under the ROC
curve (AUC) value, and TGs3, with an AUC value greater than 0.8, were screened and defined as the key genes (KGs1, KGs2, and KGs3)29. Based on it, the correlations between key genes and
differential immune cells were further studied by “Spearman”, respectively. CONSTRUCTION OF COMPETITIVE ENDOGENOUS RNAS (CERNAS) REGULATORY NETWORK OF KGS3 The potential miRNA targets for
KGs3 were predicted using the miRwalk database (with a binding score > 0.95)30,31 (http://mirwalk.umm.uni-heidelberg.de/), and the lncRNA targets of these predicted miRNAs were identified
using the Starbase database32 (with clipExpNum > 1, degraExpNum > 0, and geneType = lncRNA) (http://starbase.sysu.edu.cn/index.php). Then, the ceRNAs network of KGs3 was constructed
by “Cytoscape” (version 3.7.2) on [2022.12.25]33. DRUG PREDICTION In this study, the targeted drugs of KGs1, KGs2, and KGs3 were predicted in DrugBank online database34,35 (DGIdb,
http://www.dgidb.org/), and the gene-drug networks were constructed by “Cytoscape” (version 3.7.2) on [2022.12.25], respectively. VALIDATION OF THE EXPRESSION OF KGS3 We conducted
quantitative real-time PCR (qRT-PCR) to confirm the expression of KGs3 in vascular tissue samples from HC, VV, and SVT. Total RNA was extracted using TRIZol (Thermo Fisher, Shanghai, CN) and
then reverse transcribed into cDNA to analyze mRNA expression. Subsequently, qRT-PCR reactions were carried out using the SureScript First-strand cDNA synthesis kit (Servicebio, Wuhan, CN).
CELL CULTURE AND TRANSFECTION Human vascular smooth muscle cells (VSMCs) were used to explore the biofunctions of DACT3 via the experiments in vitro. All cells were purchased from JingFeng
biology science and technology company (Shanghai, China). HVSMCs cells were cultured in DMEM medium with 10% FBS (Fetal bovine serum). Specific short-hairpin RNA targeting DACT3 (sh-DACT3)
and its amplification plasmids (OE-DACT3) were purchased from HanHeng Biotechnology (Shanghai, China). The cells were transfected by Lentiviruses according to manufacturer’s protocols
(HanHeng Biotechnology, Shanghai, China). 10 WESTERN BLOT ASSAY Five pairs of VV and healthy clinical samples were collected from department of Vascular Surgery, Shaanxi Provincial People’s
Hospital. Western blot detection on these clinical samples was approved by the Shaanxi Provincial People’s Hospital ethical review committee (No.SP20230672). All patients provided written
informed consent before detections. All methods were carried out in accordance with relevant guidelines and regulations. Western blot assay was conducted as described previously36. Briefly,
transfected cells were lysed on ice by RIPA buffer (Beyotime, China). After centrifugation at 12,000 rpm for 4 min, the supernatant was collected and moved into an EP tube. Protein
concentration was measured using BCA kit (Beyotime, China). Proteins were separated by 10% SDS-PAGE and were transferred to PVDF membranes through electrophoresis. The PVDF membranes were
blocked by 5% skim milk at 37 °C for 2 h and were incubated with the primary and secondary antibodies in turn. All antibodies were both purchased from Abcam company (Shanghai, China), as
follows: anti-DACT3 rabbit polyclonal antibody (ab79047), anti-OPN rabbit polyclonal antibody (ab75285), anti-SM22α rabbit polyclonal antibody (ab10135), anti-GAPDH rabbit polyclonal
antibody (ab9485) and goat anti-rabbit IgG H&L (HRP) (ab6721). 11 CCK8 ASSAY HVSMCs were seeded into 96-well plate at a concentration of 5 × 104 cells per well. At each time point, CCK8
reagent (Beyotime, China) was added into each well and were incubated with cells for 4 h. Using a microplate reader, optical density (OD) at 490 nm was measured, by which cell viability was
assessed. 12 TRANSWELL MIGRATION ASSAY Transfected cells (1 × 105 per well) were seeded in 24-well transwell chambers (Corning, NY, USA). Serum-free medium was added into upper chambers,
while complete medium with 10% FBS was added into lower ones. Next, migrated cells were fixed by paraformaldehyde and stained by 0.1% crystal violet. Cells were counted in five randomly
selected visual fields of the chamber. 13 FLOW CYTOMETRIC ANALYSIS Cells with logarithmic phase were trypsinized and collected. After PBS washing, cells were fixed in 70% ethanol pre-cooled
by ice. Then, the cells were resuspended by 0.5 ml PBS and 100 µl RNase A (50 µl/ml) was added for digestion at 37˚C for 30 min. 100 µl PI dye solution (C1052, Beyotime, China) was added for
staining at 4˚C in the dark for 30 min. The red fluorescence using flow cytometry was detected at the excitation wavelength of 488 nm. RESULTS DEGS WERE ASSOCIATED WITH INFLAMMATION AND
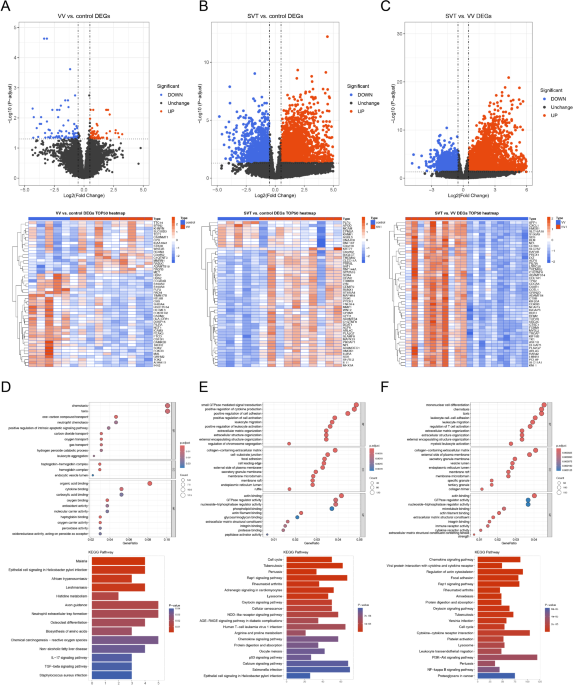
IMMUNITY RELATED FUNCTIONS There were 142 DEGs1 (59 up-regulated and 83 down-regulated) between 5 HC and 10 VV samples, 3,563 DEGs2 (2,287 up-regulated and 1,276 down-regulated) between 5 HC
and 10 SVT samples, and 4,712 DEGs3 (3,012 up-regulated and 1,700 down-regulated) between 10 VV and 10 SVT samples (Fig. 1A–C). Functionally, it was shown that the 142 DEGs1 were mainly
enriched to neutrophil chemotaxis, positive regulation of intrinsic apoptotic signaling pathway, oxygen transport, organic acid binding, antioxidant activity and etc. In addition, DEGs1 were
associated with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways such as histidine metabolism, IL-17 signaling pathway, TGF-beta signaling pathway and etc. (Fig. 1D). It was found
that the 3,563 DEGs2 were enriched for gene ontology (GO) functions such as positive regulation of cell adhesion and cytokine production, regulation of small GTPase mediated signal
transduction and etc. In addition, DEGs2 were associated with KEGG pathways such as cellular senescence, chemokine signaling pathway, NOD-like receptor signaling pathway and etc. (Fig. 1E).
The 4,712 DEGs3 were enriched for GO functions such as regulation of T cell activation, leukocyte cell-cell adhesion, leukocyte migration and etc. And DEGs3 were associated with KEGG
pathways such as focal adhesion, protein digestion and absorption, PI3K-Akt signaling pathway and etc. (Fig. 1F). It is worth noting that both DEGs2 and DEGs3 were associated with the rap1
signaling pathway, and all of these DEGs were associated with the functions of inflammation and immunity. IMMUNE CELL INFILTRATION WAS SIGNIFICANTLY DIFFERENT BETWEEN VV AND SVT In this
study, the proportion of mast cells was significantly different between HC and SVT groups, and the proportions of B cells, cytotoxic cells, central memory T cells (Tcm), follicular helper T
cells (Tfh) were significantly different between VV and SVT groups (Fig. 2A and B). Moreover, the expressions of marker genes were consistent with above results (Fig. 2C and D). TOTALS OF 12
GENES WERE ASSOCIATED WITH THE PROGRESSION AND REGRESSION OF VV Totals of 46 TGs1 that were associated with occurrence of VV, 2,257 TGs2 that were associated with development of VV, and 12
TGs3 were that associated with the progression and regression of VV were screened (Fig. 3A). Moreover, the TGs1 and TGs3 were associated with the functions of metabolic process, biological
regulation and etc. The TGs2 were associated with the functions of immune system process, homeostatic process, regulation of the biological processes and etc. (Fig. 3B–D). B CELL AND TCM
WERE THE KEY IMMUNE CELLS _PHF21B_, _IRS2_, _DUSP15_, _NUPR1_, _AC138969.1_, _OLFML3_, _POM121_, _CCDC124_, _SOX2_ and _CADM2_ were defined as the KGs1, and _KIF20A_, _SELL_, _MEFV_,
_POU2F2_, _METTL7B_, _PTCRA_, _CTRC_, _AURKB_, _BIRC7_ and _SKA3_ were defined as the KGs2 (Fig. 4A and B). Among them, the expressions of _CCDC124_ and _PHF21B_ were positively correlated
with B cell, the expression of _SOX2_ was positively correlated with Tcm, and the expression of _POM121_ was negatively correlated with Tcm (_p_ < 0.05, |cor| > 0.3). All KGs2 were
positively correlated with B cell, cytotoxic cell, mast cell, Tfh, and negatively correlated with Tcm (Fig. 4C and D). Six genes, including _PLP2_, _DACT3_, _LRRC25_, _PILRA_, _MSX1_ and
_APOD_ were screened with AUC value > 0.8 and defined as the KGs3 (Fig. 4E). The correlation analysis results showed that the expressions of _LRRC25_ and _PILRA_ were significantly
negatively associated with Tcm (_p_ < 0.05, |cor| > 0.3), and significantly positively associated with B cell (_p_ < 0.01, |cor| > 0.3) (Fig. 4F and G). THE CONSTRUCTION OF CERNA
REGULATORY NETWORK The ceRNA regulatory network was constructed with 6 KGs3, 92 miRNAs, and 37 lncRNAs. The hsa-miR-107 was capable of simultaneously regulating _LRRC25_, _PILRA_, and
_APOD_, while hsa-miR-25-3p could simultaneously regulate _PLP2_, _DACT3_, and _PILRA_, and hsa-miR-5010-5p could simultaneously regulate _DACT3_, _APOD_, and _PLP2_. There were four common
miRNAs (hsa-miR-125a-5p, hsa-miR-3126-5p, hsa-miR-514a-5p, and hsa-miR-6875-5p) between _LRRC25_ and _MSX1_. There were three common miRNAs between _LRRC25_ and _PILRA_ (hsa-let-7e-5p,
hsa-miR-345-3p and hsa-miR-6763-5p), three common miRNAs between _DACT3_ and _MSX1_ (hsa-miR-129-1-3p, hsa-miR-129-2-3p, and hsa-miR-6866-3p), and three common miRNAs between _DACT3_ and
_LRRC25_ (hsa-miR-378b, hsa-miR-513a-5p and hsa-miR-516b-5p). There were two common miRNAs between _APOD_ and _LRRC25_ (hsa-let-7d-5p and hsa-miR-103a-3p), two common miRNAs between _LRRC25_
and _PLP2_ (hsa-miR-1343-3p and hsa-miR-4525), and two common miRNAs between _PLP2_ and _DACT3_ (hsa-miR-378a-3p and hsa-miR-378i). Besides, there was only one miRNA (hsa-miR-1296-5p)
between _PLP2_ and _PILRA_. It was worth noting that XIST could regulate 24 miRNAs at the same time, NEAT1 could regulate 15 miRNAs at the same time, and MALAT1 could regulate 15 miRNAs at
the same time (Fig. 5). DRUG PREDICTION In this study, the targeted drugs of KGs1, KGs2 and KGs3 were predicted. The results indicated that the targeted drugs of KGs1 included Betamethasone,
Phosphate, Cimetidine, Samarium (153Sm) lexidronam, Ethylhexyl methoxycrylene, Sodium acetate, Tetradecyl hydrogen sulfate (ester), Girentuximab I-124, and etc. (Fig. 6A). Degarelix,
Diamorphine, Cimetidine, Ursodeoxycholic acid, Fluvastatin, Inositol, Flecainide, Caffeine, Leuprolide and others were the targeted drugs of KGs2 (Fig. 6B). Nitrazepam, Cimetidine,
Dactinomycin, Calcitriol, Pilocarpine, Ethylhexyl methoxycrylene and others were the targeted drugs of KGs3 (Fig. 6C; Table 1). EXPRESSION VERIFICATION The results of qRT-PCR showed that the
expression of PLP2, APOD and DACT3 were significantly decreased in the process of occurrence and development of VV (_p_ < 0.05) (Fig. 7). Given that the pivotal functions of DACT3 in
cell growth, DACT3 was selected for further investigation. DACT3 MEDIATES THE ONSET AND PROGRESSION OF VV Similar to PCR results, the protein expressions of DACT3 in VV were significantly
downregulated compared to normal samples, as four pairs of clinical samples determined (Fig. 8A). sh-DACT3 and OE-DACT3 can effectively manipulate DACT3 expressions in HVSMCs (Fig. 8BC).
CCK8 assays revealed that overexpression of DACT3 inhibited HVSMCs proliferation, whereas DACT3 deletion suppressed this process (Fig. 8D). As expected, overexpression of DACT3 blocked the
transition from the G0/G1 phase to the S phase (Fig. 8E). Quantitative data confirmed above observations (Fig. 8F). The proportion of cell at G0/G1 phase in overexpression group was
significantly higher than that in control group, whereas that in silencing group was markedly lower (Fig. 8F). Moreover, DACT3 also mediated the vascular migration and phenotypic transition.
Transwell assay indicated that overexpression of DACT3 significantly inhibited VSMCs migration, but silencing DACT3 promoted the migrative process (Fig. 8G). The similar trends were
observed in cell counting (Fig. 8H). Meanwhile, the expression of OPN, a common marker of the extracellular matrix (ECM) synthesis, was significantly increased in overexpression group
compared to other groups (Fig. 8I). However, the expression of SM22α, a classical marker of the VSMCs contraction, was significantly increased in overexpression group (Fig. 8I). Clearly,
overexpression of DACT3 can promote the transition from synthetic to contractile phenotype. Collectively, DACT3 was closely involved in the proliferation, cell cycle, migration and phenotype
transition of VSMCs, in turn regulating the progression of VV. DISCUSSION Initially, it was noted that the number of differentially expressed genes in the control versus VV comparison was
relatively low (142). Conversely, the number of differentially expressed genes in the control versus SVT (3563) and VV versus SVT (4712) comparisons was quite high. This suggests that in the
early stages of VV formation, only a small number of genes related to metabolism exhibited differential expression, triggering metabolic abnormalities that ultimately lead to the
development of VV. In both the control versus SVT and VV versus SVT comparisons, there was an abundance of differentially expressed genes, primarily concentrated in signaling pathways
associated with inflammatory responses and immune function. The PI3K/Akt signaling pathway has been reported to play a vital role in the development of variosevein of the great saphenous
vein37. This indicated that the generation of SVT was closely related to the immune environment. Immunological analysis showed significant differences in Mast cells between the control vs.
SVT; and the other four cells (Tcm, Tfh, B cells, and Cytotoxic cells) showed significant differences between VV vs. SVT, once again proving this point. Previous research had shown that
thrombi in varicose veins could induced infiltration of mast cells, T cells, and B cells, which might be involved in the remodeling of venous walls38. Increased infiltration of activated
mast cells has also been recently implicated in the pathophysiology of varicose veins39. Another study proved that immunological memory might be involved in the chronic venous disorders
development and Tcm cell played a role in it40. Secondly, there were 46 TGs1 associated with the occurrence of VV in region A, 2,257 TGs2 linked to the development of VV in region G, and 12
TGs3 associated with the progression and regression of VV in region D. We could find that the number of genes in the A region (46) was much less than that in the G region (2257), which was
consistent with the trend of the numbers of differentially expressed genes between different groups. Most of the six key genes identified in the D region were related to immunity. For
example, _LRRC25_ played a role in the inhibition of NF-kappa-B signaling pathway and inflammatory response41. _DACT3_ was a negative regulator of Wnt/beta-catenin signaling that was
transcriptionally repressed in colorectal cancer42. Overexpression of _PLP2_ increased tumour metastasis and knockdown of _PLP2_ inhibited the growth and metastasis of melanoma cells43.
PILRA was an immune inhibitor receptor, that was widely involved in the regulation of the immune system44. It could be seen that immune related cells and pathways were closely related to the
occurrence and development of VV. Thirdly, in the ceRNA regulatory network we constructed, a large number of miRNA (92) related to VV occurrence were found, among which the role of
miR-125a-5p in VV occurrence has been reported in Wei et al.‘s paper37, indicating that our results were consistent with previous studies and had further in-depth analysis value. There were
37 lncRNAs in the network, among which, XIST, as an important regulator of cell growth and development, was one of the central lncRNAs in the network45, indicating that XIST had a regulatory
role in the inflammatory response, which indirectly regulated the development of VV. Fourthly, SVT was usually treated with anti-inflammatory medicine or anticoagulant46. In this study, we
found that Cimetidine was identified in all three groups KGs1, KGs2, and KGs3, indicating that it was the key targeted drug for VV and SVT. Previous research reported that pituitrin combined
with Cimetidine could significantly reduced portal vein pressure, improved liver blood circulation, and effectively controlled esophageal variceal rupture bleeding in cirrhosis47. Meanwhile
our genetic drug network presented other interesting results. We found that caffeine was associated with three key genes (DACT3, PILRA, and MSX1). As we all known, caffeine could increase
vasoconstriction and blood pressure, and aggravate VV. Therefore, caffeine or similar drugs may play an important role in the development of VV. In addition, we observed that APOD only
interacted with two sedative drugs, anileridine and nitrazepam, which diverged from the network. So sedative drugs may also have an effect on the treatment of VV. Lastly, we focused the
biofunctions of DACT3 in VV progression due to its critical roles in cell growth. It has been established that DACT3 involved in negative regulation of canonical Wnt signaling pathway and
negative regulation of cell growth42. Similarly, as our experimentations determined, overexpression of DACT3 significantly inhibited the proliferation of VSMCs and retarded their transition
from G0/G1 to S phase. Moreover, the phenotypic change of VSMCs profoundly affects the onset and progression of VV. For instance, FOXC2-AS1 can aggravate VV through regulating the phenotypic
transition of VSMCs48. Herein, overexpression of DACT3 also promoted the transition from synthetic to contractile phenotype of VSMCs, the latter was the dynamics basis of vascular
operation. Clearly, DACT3 was confirmed as a critical regulator in the pathogenesis of VV, and its agonists hold great potentials for VV therapy. However, our study has some limitations. The
study was mainly based on transcriptomic data, and other histological methods had not yet been applied. In addition, the study mainly relies on bioinformatics analyses of relevant samples,
which have not yet been experimentally validated. In order to enhance the reliability of the conclusions, animal experiments will be conducted in the next step. At the same time, we plan to
combine proteomic data for integrated multi-omics analyses in order to draw more accurate and valuable conclusions. DATA AVAILABILITY The datasets used and/or analyzed in the current study
are available from the corresponding author upon reasonable request. REFERENCES * De Maeseneer, M. G. et al. (eds) Choice - European Society for Vascular Surgery (ESVS) 2022 Clinical
Practice Guidelines on the Management of Chronic Venous Disease of the Lower Limbs. Eur J Vasc Endovasc Surg. 63(2): 184–267. (2022). * Lurie, F. et al. The 2020 update of the CEAP
classification system and reporting standards. _J. Vasc Surg. Venous Lymphat Disord. _8(3), 342–352 (2020). Article PubMed Google Scholar * Rajeeva Pandian, N. K. & Jain, A. In silico
analyses of blood flow and oxygen transport in human micro-veins and valves. _Clin. Hemorheol. Microcirc. _81(1), 81–96 (2022). Article CAS PubMed Google Scholar * Gianesini, S. et al.
Comparison between Duplex Ultrasound and Multigate Quality Doppler Profile Software in the Assessment of Lower Limb Perforating Vein Direction. _Eur. J. Vasc Endovasc Surg. _55 (5), 688–693
(2018). Article PubMed Google Scholar * Vincent, J. R., Jones, G. T., Hill, G. B. & van Rij, A. M. Failure of microvenous valves in small superficial veins is a key to the skin
changes of venous insufficiency. _J. Vasc Surg. _54(6 Suppl): 62S-9S.e1-3 (2011). * Perrin, M. & Ramelet, A. A. Pharmacological treatment of primary chronic venous disease: rationale,
results and unanswered questions. _Eur. J. Vasc Endovasc. Surg. _41(1), 117–125 (2011). Article CAS PubMed Google Scholar * Gordon, P., Widener, J. M. & Heffline, M. Venous leg
ulcers: impact and dysfunction of the venous system. _J. Vasc. Nurs. _33(2), 54–59 (2015). Article PubMed Google Scholar * Farah, M. H. et al. A systematic review supporting the Society
for Vascular Surgery, the American venous forum, and the American Vein and Lymphatic Society guidelines on the management of varicose veins. _J. Vasc Surg. Venous Lymphat. Disord. _10(5),
1155–1171 (2022). Article PubMed Google Scholar * Passman, M. A. et al. Validation of venous clinical severity score (VCSS) with other venous severity assessment tools from the American
venous forum, national venous screening program. _J. Vasc Surg. _54(6 Suppl), 2S–9S (2011). Article PubMed Google Scholar * Stücker, M. [Superficial venous thrombosis, varicose veins, and
chronic venous insufficiency: an update for clinical practice. _Inn. Med. (Heidelb.) _63(6), 612–618 (2022). PubMed Google Scholar * Litzendorf, M. E. & Satiani, B. Superficial venous
thrombosis: disease progression and evolving treatment approaches. _Vasc. Health Risk Manag. _7, 569–575 (2011). Article PubMed PubMed Central Google Scholar * Poredos, P., Kozak, M.,
Antignani, P. L. & Jezovnik, M. K. From varicose veins to venous thromboembolic events. _Int. Angiol. _42(3), 254–259 (2023). Article PubMed Google Scholar * KakkosSK et al. Editor
(eds) ‘s Choice European Society for Vascular Surgery (ESVS) 2021 clinical practice guidelines on the management of venous thrombosis. _Eur. J. Vasc Endovasc Surg. _61(1), 9–82 (2021).
Article PubMed Google Scholar * Geersing, G. J., Cazemier, S., Rutten, F., Fitzmaurice, D. A. & Hoes, A. W. Incidence of superficial venous thrombosis in primary care and risk of
subsequent venous thromboembolic sequelae: a retrospective cohort study performed with routine healthcare data from the Netherlands. _BMJ Open. _8(4), e019967 (2018). Article PubMed PubMed
Central Google Scholar * Kitchens, C. S. How I treat superficial venous thrombosis. _Blood _117 (1), 39–44 (2011). Article CAS PubMed Google Scholar * Bonkemeyer Millan, S., Gan, R.
& Townsend, P. E. Venous ulcers: diagnosis and treatment. _Am. Fam. Physician _100(5), 298–305 (2019). PubMed Google Scholar * Novais, E. A. et al. Optical coherence tomography
angiography of Chorioretinal diseases. _Ophthalmic Surg. Lasers Imaging Retina _47(9), 848–861 (2016). Article PubMed Google Scholar * Mansilha, A. & Sousa, J. Pathophysiological
mechanisms of chronic venous disease and implications for Venoactive Drug Therapy. _Int. J. Mol. Sci. _19(6). (2018). * Rabe, E. & Pannier, F. What is evidence-based in the treatment of
chronic venous insufficiency?. _Internist (Berl.) _61(12), 1230–1237 (2020). Article PubMed Google Scholar * Modi, A., Vai, S., Caramelli, D. & Lari, M. The Illumina sequencing
protocol and the NovaSeq 6000 System. _Methods Mol. Biol. _2242, 15–42 (2021). Article CAS PubMed Google Scholar * de Sena Brandine, G. & Smith, A. D. Falco: high-speed FastQC
emulation for quality control of sequencing data. _F1000Res _8, 1874 (2019). Article PubMed Google Scholar * Wingett, S. W. & Andrews, S. FastQ screen: a tool for multi-genome mapping
and quality control. _F1000Res _7, 1338 (2018). Article PubMed PubMed Central Google Scholar * Ward, C. M., To, T. H. & Pederson, S. M. ngsReports: a Bioconductor package for
managing FastQC reports and other NGS related log files. _Bioinformatics _36(8), 2587–2588 (2020). Article CAS PubMed Google Scholar * Bolger, A. M., Lohse, M. & Usadel, B.
Trimmomatic: a flexible trimmer for Illumina sequence data. _Bioinformatics _30(15), 2114–2120 (2014). Article CAS PubMed PubMed Central Google Scholar * Kim, D., Paggi, J. M., Park,
C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. _Nat. Biotechnol. _37(8), 907–915 (2019). Article CAS PubMed PubMed
Central Google Scholar * Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. _Genome Biol. _15(12), 550 (2014). Article
PubMed PubMed Central Google Scholar * Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. _OMICS _16(5),
284–287 (2012). Article CAS PubMed PubMed Central Google Scholar * Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer.
_Immunity _39(4), 782–795 (2013). Article CAS PubMed Google Scholar * Ho, J. J., Subramaniam, P. & Davis, P. G. Continuous positive airway pressure (CPAP) for respiratory distress in
preterm infants. _Cochrane Database Syst. Rev. _10(10), CD002271 (2020). PubMed Google Scholar * Dweep, H., Gretz, N. & Sticht, C. miRWalk database for miRNA-target interactions.
_Methods Mol. Biol. _1182, 289–305 (2014). Article PubMed Google Scholar * Sticht, C., De La Torre, C., Parveen, A. & Gretz, N. miRWalk: an online resource for prediction of microRNA
binding sites. _PLoS One _13(10), e0206239 (2018). Article PubMed PubMed Central Google Scholar * Li, J. H., Liu, S., Zhou, H., Qu, L. H. & Yang, J. H. starBase v2.0: decoding
miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. _Nucleic Acids Res. _42(Database issue), D92–D97 (2014). Article CAS PubMed Google Scholar *
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. _Genome Res. _13(11), 2498–2504 (2003). Article CAS PubMed PubMed Central
Google Scholar * Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. _Nucleic Acids Res. _46(D1), D1074–D1082 (2018). Article CAS PubMed Google
Scholar * Wishart, D. S. DrugBank and its relevance to pharmacogenomics. _Pharmacogenomics _9(8), 1155–1162 (2008). Article CAS PubMed Google Scholar * Xu, F. et al. SLC1A5 prefers to
play as an accomplice rather than an opponent in pancreatic adenocarcinoma. _Front. Cell. Dev. Biol. _10, 800925 (2022). Article PubMed PubMed Central Google Scholar * Wei, J., Zhu, H.,
Zhang, Q. & Zhang, Q. Prediction of Functional Genes in Primary Varicose Great Saphenous Veins Using the lncRNA-miRNA-mRNA Network. Comput Math Methods Med. 2022: 4722483. (2022). * Chu,
H. B. et al. Assessment of the infiltration of inflammatory cells in the walls of thrombotic varicose veins. _Angiology _64(1), 69–72 (2013). Article PubMed Google Scholar * Callesen, K.
T. et al. Characterization of mast cells from healthy and Varicose Human Saphenous Vein. _Biomedicines _10(5), 1062 (2022). Article CAS PubMed PubMed Central Google Scholar * Ojdana,
D. et al. Evaluation of the memory CD4 + and CD8 + T cells homeostasis during chronic venous disease of lower limbs. _Folia Histochem. Cytobiol _47(3), 471–477 (2009). PubMed Google Scholar
* Feng, Y. et al. LRRC25 functions as an inhibitor of NF-κB signaling pathway by promoting p65/RelA for autophagic degradation. _Sci. Rep. _7(1), 13448 (2017). Article ADS PubMed PubMed
Central Google Scholar * Jiang, X. et al. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. _Cancer
Cell. _13(6), 529–541 (2008). Article CAS PubMed PubMed Central Google Scholar * Maia, V. et al. PLP2-derived peptide Rb4 triggers PARP-1-mediated necrotic death in murine melanoma
cells. _Sci. Rep. _12(1), 2890 (2022). Article ADS CAS PubMed PubMed Central Google Scholar * Kogure, A., Shiratori, I., Wang, J., Lanier, L. L. & Arase, H. PANP is a novel
O-glycosylated PILRα ligand expressed in neural tissues. _Biochem. Biophys. Res. Commun. _405(3), 428–433 (2011). Article CAS PubMed PubMed Central Google Scholar * Wang, W. et al.
Biological function of long non-coding RNA (LncRNA) xist. _Front. Cell. Dev. Biol. _9, 645647 (2021). Article PubMed PubMed Central Google Scholar * Evans, N. S. & Ratchford, E. V.
Superficial vein thrombosis. _Vasc Med. _23(2), 187–189 (2018). Article PubMed Google Scholar * Dixon, A. N., Webb, J. C., Wenzel, J. L., Wolf, J. S. Jr & Osterberg, E. C. Current
management of pelvic fracture urethral injuries: to realign or not? _Transl Androl. Urol. _7(4), 593–602 (2018). Article PubMed PubMed Central Google Scholar * Zhang, C., Li, H. &
Guo, X. FOXC2-AS1 regulates phenotypic transition, proliferation and migration of human great saphenous vein smooth muscle cells. _Biol. Res. _52(1), 59 (2019). Article ADS PubMed PubMed
Central Google Scholar Download references ACKNOWLEDGEMENTS All authors would like to thank Shaanxi Provincial People’s Hospital for its support. FUNDING This study was supported by
Natural Science Foundation of Shaanxi Province, China (No. 2022SF-079) and the Talents Special Foundation of Shaanxi Provincial People’s Hospital, China (No. 2022JY-39). AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Department of Vascular Surgery, Shaanxi Provincial People’s Hospital, No.256 Youyi west Road, Xi’an, 710068, Shaanxi, China Meng-Jie Shi, Yan Yan, Fei Liu,
Jin-Xing zhao, Feng Hou, Shi-Cai He, Rui-Peng Zhang & Hui Wang Authors * Meng-Jie Shi View author publications You can also search for this author inPubMed Google Scholar * Yan Yan View
author publications You can also search for this author inPubMed Google Scholar * Fei Liu View author publications You can also search for this author inPubMed Google Scholar * Jin-Xing zhao
View author publications You can also search for this author inPubMed Google Scholar * Feng Hou View author publications You can also search for this author inPubMed Google Scholar *
Shi-Cai He View author publications You can also search for this author inPubMed Google Scholar * Rui-Peng Zhang View author publications You can also search for this author inPubMed Google
Scholar * Hui Wang View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS HW conceived and designed the study. MJS, YY, FL, JXZ, FH and SCH
analyzed and interpreted the data. MJS, YY, FL, JXZ and RPZ wrote the manuscript. MJS, YY, JXZ, FH and SCH conduced the vitro experiments. All authors have read and approved the manuscript.
CORRESPONDING AUTHOR Correspondence to Hui Wang. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ETHICAL APPROVAL AND INFORMED CONSENT This study was
approved by the Shaanxi Provincial People’s Hospital ethical review committee (No.SP20230672). All methods in this study were carried out in accordance with relevant guidelines and
regulations. CONSENT FOR PUBLICATION All patients provided written informed consent before sequencing and Western blot assays. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains
neutral with regard to jurisdictional claims in published maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL Below is the link to the electronic supplementary material.
SUPPLEMENTARY MATERIAL 1 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits
any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to
the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts
of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If
material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain
permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Shi, MJ., Yan, Y., Liu, F. _et al._ Identification of biological significance of different stages of varicose vein development based on mRNA sequencing. _Sci Rep_ 14, 22536 (2024).
https://doi.org/10.1038/s41598-024-73691-3 Download citation * Received: 09 December 2023 * Accepted: 19 September 2024 * Published: 28 September 2024 * DOI:
https://doi.org/10.1038/s41598-024-73691-3 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative KEYWORDS * Varicose vein * Shallow vein thrombosis * Gene
regulation * Function * Immune








