- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The OVATE gene family plays an important role in regulating the development of plant organs and resisting stress, but its expression characteristics and functions in _sorghum_ have
not been revealed. In this study, we identified 26 OVATE genes in the _sorghum_ BTx623 genome, which were divided into four groups and distributed unevenly across 9 chromosomes. Evolutionary
analysis showed that after differentiation between _sorghum_ and _Arabidopsis_, the OVATE gene family may have experienced unique expansion events, and all OVATE family members were
negatively selected. Transcriptome sequencing and RT-qPCR results showed that OVATE genes in _sorghum_ showed diverse expression characteristics, such as gene _SORBl_3001G468900_ and
_SORBl_3009G173400_ were significantly expressed in seeds, while _SORBI_3005G042700_ and _SORBI_3002G417700_ were only highly expressed in L1. Meantime, in the promoter region, a large
number of hormone-associated cis-acting elements were identified, and these results suggest that members of the OVATE gene family may be involved in regulating specific development of
_sorghum_ leaves and seeds. This study improves the understanding of the OVATE gene family of _sorghum_ and provides important clues for further exploration of the function of the OVATE gene
family. SIMILAR CONTENT BEING VIEWED BY OTHERS COMPARATIVE ANALYSIS OF THE TRANSCRIPTOMES OF TWO RICE SUBSPECIES DURING DOMESTICATION Article Open access 11 February 2021 GENOME-WIDE
IDENTIFICATION, PHYLOGENETIC, AND EXPRESSION ANALYSIS UNDER ABIOTIC STRESS CONDITIONS OF WHIRLY (WHY) GENE FAMILY IN _MEDICAGO SATIVA _L. Article Open access 04 November 2022 GENOME-WIDE
IDENTIFICATION, CHARACTERIZATION AND GENE EXPRESSION OF BES1 TRANSCRIPTION FACTOR FAMILY IN GRAPEVINE (_VITIS VINIFERA_ L.) Article Open access 05 January 2023 INTRODUCTION For many food and
industrial crops, the growth and development of grains and seeds are of special importance for their economic value and food security. _Sorghum bicolor_ (L.), as an important food and
bioenergy crop, is widely planted in many areas around the world1. The harvested _sorghum_ seeds and stalks can be used not only for brewing wine and sugar, but also as animal feed to
improve the industrial value of _sorghum_2. Meanwhile, due to its excellent drought resistance and salinity tolerance, _sorghum_ has become a representative of C4 model crops and has been
applied in many stress tolerance studies 3. With the rapid development of the brewing and clean bioenergy industries, the market demand for _sorghum_ is gradually increasing. However,
compared with field crops such as maize and rice, the study of seed development and yield traits of _sorghum_ still needs to be strengthened4,5,6,7. Advances in sequencing technology have
greatly contributed to the development of plant science, and s_orghum_ research has benefited from it no exception8,9. In 2009, Paterson et al.10 assembled the first sorghum genome with a
size of about 730 Mb, and found that gene loss occurred after ancient polyploidy. After that, several _sorghum_ genomes of different germplasm were published one after another and in-depth
research was carried out at the population level based on these genomes11,12,13. For example, Morris et al.14 analyzed the population structure of _sorghum_ based on more than 260,000 SNPs
identified from 971 germplasm, and revealed the potential genetic loci regulating important agronomic traits through genome-wide association; Tao et al.15 constructed the first _sorghum_
pan-genome map by assembling and integrating 16 _sorghum_ genome data and discovered a large number of presence/absence variants involved in _sorghum_ domestication and improvement. The
publication of these basic data provides a strong support for further study on stress resistance and quality traits of _sorghum_. At present, some studies focus on the analysis of the stress
resistance mechanism of _sorghum_ and the identification of seed development regulatory loci. For example, Calone et al.16 studied the mechanism of salt tolerance and Na in _sorghum_
through experiments; Zhou et al.17 revealed the effects of different nitrogen application rates and planting densities on _sorghum_ grain yield and quality and Ahn et al.1 identified some
SNP loci that regulate the morphological development of _sorghum_ seeds through genome-wide association analysis. The OVATE gene family (OFP), as a new type of plant transcription factor
first discovered in tomato, has been proved to have various functions such as regulating plant organ development and participating in plant resistance to stress18,19. Studies in peppers show
that _CaOFP20_ can regulate the shape of peppers as an inhibitor of fruit elongation and An et al.'s research indicates that members of ovate family may be the potential regulatory
factors for the difference of tea leaf area among different tea varieties18,20; Zhou and Guan et al.21,22 respectively found that an inversion about 1.7 Mb in length downstream of OVATE
family members in peach trees can regulate the shape of peach fruits. In addition, studies in rice and _Arabidopsis_ have proved that _OsOFP6_ and _AtOFP8_ can resist drought and freezing
stress by regulating the level of H2O2 in plants or wax synthesis on the surface of leaves23,24. At the same time, Li and Hackbusch found that that members of the OVATE gene family can
regulate plant development by combining with TALE or KNOX II family proteins to form complexes25,26. In mango, it was found that 25 OVATE gene family members were mainly highly expressed in
flowers and immature fruits, and six family members associated with the shape of mango fruit were identified27; While most of the OVATE family members in cotton have no introns and play an
important role in fiber and ovule development28. At present, there are relatively few studies on the OVATE family and most of them focus on the research of its regulatory mechanism for
tissue development. The microarray expression analysis shows that members of the OVATE family may be involved in salt tolerance, drought resistance and pest resistance29. The above results
indicate that the OVATE gene family plays an important role in the process of plant growth and development. However, the number and whether it has the function of regulating tissue
development in _sorghum_ remain unclear. In the current study, we have comprehensively identified the members of the OVATE gene family in _sorghum_, and analyzed their gene structure,
protein motif and cis-acting elements in the promoter region. Meanwhile, its evolutionary characteristics were analyzed by phylogenetic and genome-wide synteny analysis. On this basis,
transcriptome sequencing and RT-qPCR analysis were used to determine the expression levels of the OVATE gene family members in sorghum to reveal the candidate family members that have the
potential to regulate tissue development. Our research not only provides a global perspective for the functional evolution of OVATE gene family in _sorghum_ genome, but also provides
important clues for further understanding its regulatory role in _sorghum_ growth and development through quantitative expression analysis. MATERIALS AND METHODS PLANT MATERIALS The
“JingDuXiaoBaiRen” _sorghum_ variety is planted in the Moutai College Germplasm Resource Garden in Guizhou Province, China (27.74°N, 106.33°E). Three biological replicates of seeds (S1 and
S2 represent seed formation and filling stages, respectively) and leaves (L1 and L2 sampling times correspond to seeds) samples at different developmental stages were collected.
Subsequently, these samples are placed in a -80 ℃ refrigerator until they are used. TRANSCRIPTOME SEQUENCING AND DIFFERENTIALLY EXPRESSED GENE ANALYSIS Prior to transcriptome sequencing, the
total RNA of all samples was extracted using the Total RNA Purification Kit (cat DP441, Tiangen, China) according to the manufacturer's protocol. Qualified RNA samples were used for
sequencing, and the average sequencing depth was more than 20 × . At the same time, the longest CDS sequence of each gene in the reference genome BTx623 was extracted according to the gff
file and indexed using SMALT software30. After that, the following parameters of the SMALT software are used to align clean reads with the reference database: -m 35 -j 20. After calculating
the gene expression level by selectSmart.pl script, the R package DEGseq was used for the analysis of differentially expressed genes (fold_change > 2 and the q-value < 0.05)31. In
addition, expression level heatmaps of OVATE gene family members were visualized by TBtools software. IDENTIFICATION OF OVATE GENE FAMILY IN _SORGHUM_ BTx623 (Sorghum_bicolor_NCBIv3) and Rio
(Sorghum_rio.JGI-v2.0) _sorghum_ genome and protein sequence files were downloaded from SORGHUMBASE (https://www.sorghumbase.org/) and used as reference sequences, while the gff files
corresponding to the two genomes were also downloaded at the same time32. To identify the _sorghum_ OVATE gene family, the HMM (The Hidden Markov Model) model was first downloaded from the
Protein Family Database (Pfam) (http://pfam.sanger.ac.uk/) website. The HMMsearch software was then used to identify members of the _sorghum_ gene family. The specific identification method
refers to our previous research18. At the same time, in order to ensure the reliability of the identification results, the OVATE sequence of _Arabidopsis thaliana_ was downloaded and aligned
with the candidate OVATE sequence of sorghum, and the sequence with a coincidence rate of more than 75% was finally retained. CHROMOSOME DISTRIBUTION AND SYNTENY ANALYSIS After obtaining
the location information of OVATE gene family members from the gff file and calculating the length of different chromosomes, the MG2C website (http://mg2c.iask.in/mg2c_v2.1/) was used to
visualized the chromosome distribution of the OVATE gene family. After downloading _Arabidopsis_ (https://www.arabidopsis.org/) and _maize_ (https://maizegdb.org/) genomes, gene duplication
and synteny analysis were performed with reference to previous studies20,33,34. According to the above results, the collinearity gene pairs were extracted, and the CDS and protein sequences
of the OVATE gene family were obtained at the same time, and then the Ka/Ks values were calculated under the default parameters using the Simple Ka/Ks Calculator tool built into TBtools.
SEQUENCE ANALYSIS AND PHYLOGENETIC TREE CONSTRUCTION The MW (molecular weight) and pI (isoelectric point) of the members of the OVATE gene family in sorghum were analyzed by ExPasy site
(https://web.expasy.org/). The cis-acting elements were analyzed based on the PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) online analysis tool, and subcellular
localization results for all OVATE gene family members were predicted based on Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/). At the same time, the built-in MEME program
of TBtools is used for motifs analysis. Finally, information such as motifs, cis-acting elements and gene structure (based on the gff file) are displayed graphically through TBtools34. The
downloaded protein sequences of the OVATE gene family from maize and Arabidopsis were aligned with the Sorghum BTx623 and Rio OVATE gene family sequences using the ClustalW software
integrated into MEGA 11. The NJ phylogenetic tree is then constructed with a bootstrap value of 100035. VERIFICATION OF EXPRESSION LEVELS OF OVATE GENE FAMILY MEMBERS BY RT-QPCR For RT-qPCR,
first-strand cDNA was synthesized from total RNA with the PrimeScript RT Reagent Kit (cat RR036A, Takara, Japan) using the manufacturer’s protocols. A 10 μL RT-qPCR amplification system was
performed with the following parameters: 95 °C predenaturation for 3 min, 95 °C denaturation for 10 s, 64 °C annealing for 30 s, for a total of 45 cycles. At the same time, each biological
replicate included three technical replicates, and EIF4α was used as the internal reference gene36. The relative gene expression values were analyzed using the 2-△Ct method. All primer
sequences are listed in Table S1. All experimental methods were carried out in accordance with the relevant guidelines and regulations. DATA ANALYSIS The data were analyzed for data
processing and significant differences using SPSS 22.0 and plotted using Python 3.6 and TBtools. The data obtained were shown below mean ± standard deviation (SD). Data were all analyzed
using Student’s t-test and one-way ANOVA. In the representation of the results, different letters were used to indicate that the differences were statistically significant (P < 0.05).
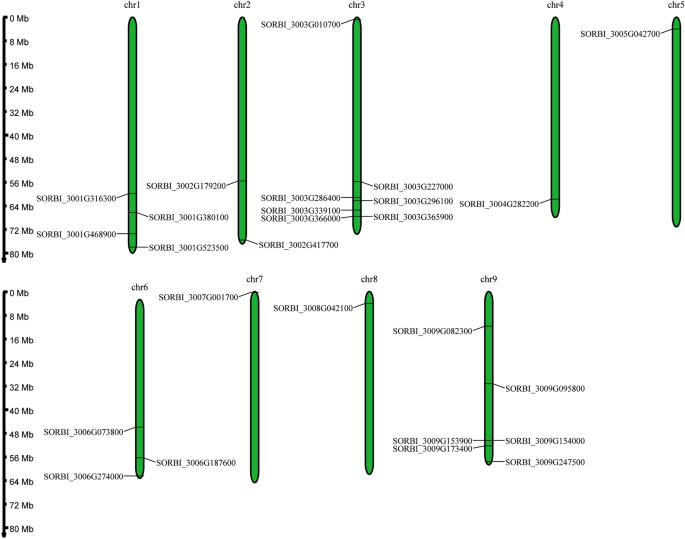
RESULTS IDENTIFICATION OF OVATE GENE FAMILY IN _SORGHUM_ A total of 26 OVATE gene family members were identified from the _sorghum_ genome (BTx623), which were unevenly distributed on all
chromosomes except chromosome 10 (Fig. 1). Among them, chromosomes 4, 5, 7 and 8 each contain an OVATE gene. For the remaining chromosomes, the number of OVATE family members ranged from 2
(chromosome 2) to 7 (chromosome 3). The sequence analysis showed that the protein length of OVATE gene family ranged from 229 amino acids (_SORBI_3009G153900_) to 543 amino acids
(_SORBI_3003G339100_), corresponding to the minimum 24.68 kDa and the maximum 60.06 kDa of OVATE family members (with an average of 35.63 kDa) (Table S2). Isoelectric point calculations
found that the pI values of different OVATE proteins varied between 4.49 (_SORBI_3005G042700_) and 11.14 (_SORBI_3003G296100_) with an average of 8.64. At the same time, bioinformatics
prediction results show that most OVATE genes are located in nucleus, but some members are also predicted to be located in cell membrane (_SORBI_3003G365900_ and _SORBI_3005G042700_) and
chloroplast (_SORBI_3001G316300_ and _SORBI_3001G523500_) (Table S2). The differences in MW and pI indicate that these genes may have functional differentiation in _sorghum_. PHYLOGENETIC
ANALYSIS OF THE OVATE GENE FAMILY In order to reveal the evolutionary relationship between the _sorghum_ OVATE gene family and other species, the OVATE proteins of rice, maize, _Arabidopsis_
and sweet _sorghum_ varieties Rio were collected together with the sequences identified in this study to construct phylogenetic trees (Table S3). The results are shown in Fig. 2. All
sequences were divided into four groups, of which Group contained only rice and _Arabidopsis_ OVATE proteins. In the Group II, there were 8 members of the OVATE family from BTx623 and 8 from
the Rio genome, while in the Group III, there were 5 OVATE family members from both _sorghum_ genomes. The Group IV contained the largest number of sorghum OVATE gene family members,
reaching 25, of which 13 were from BTx623 and 12 from Rio. The highly similar clustering results showed that there was no significant differentiation of the OVATE gene family in grain
_sorghum_ and sweet _sorghum_. PROTEIN MOTIF, _CIS_-ACTING ELEMENTS AND GENE STRUCTURE ANALYSIS We analyzed the protein-conserved motifs of all 26 members of the OVATE gene family. The
result is shown in Fig. 3A. A total of 10 conserved motifs were identified, with each protein sequence containing 3 to 7 motifs. Among them, motif 1 and motif 2 were the most conserved and
identified in all protein sequences, while _SORBI_3003G296100_ contained the most motif types. Since cis-acting elements play an important role in gene transcription and expression, we
performed cis-acting element analysis on the first 2000 bp sequence of promoters of OVATE family members (Fig. 3B). Interestingly, we found a large number of hormone-related cis-acting
elements, including GA, MeJA, ABA and SA response elements. At the same time, some cis-acting elements related to drought and low temperature stress and involved in the regulation of seed
development were identified. These results suggest that members of the OVATE gene family of _sorghum_ are not only able to participate in resisting various environmental stresses, but may
also regulate the development of organs such as _sorghum_ seeds. At the same time, it should be noted that a large number of light-responsive elements are also found in the promoter region.
Gene structure analysis showed that except for the gene _SORBI_3008G042100_ containing 3 exons, other members of the OAVET family contained 1 to 2 exons (Fig. 3C). SEGMENT DUPLICATION AND
COLLINEARITY ANALYSIS Many studies have shown that segment duplication may drive the expansion of gene families in the plant genome33,37. Therefore, to explore the mechanism of expansion of
the OVATE gene family in _sorghum_, we performed segment duplication event analysis. As shown in Fig. 4, the duplication event exists widely on 9 chromosomes. Twenty genes from 26 OVATE
family members participated in the formation of 19 gene pairs (Table S4). Among them, some genes such as _SORBI_3001G380100_ formed multiple gene pairs. Most gene pairs are located on
different chromosomes, which indicates that segment duplication is an important driving force for the expansion of sorghum OVATE gene family. However, in the process of expansion, their
evolutionary direction is still unknown. In order to determine the effect of selection pressure on these genes, we calculated the Ka/Ks values of all homologous genes (Table S5). The results
show that all Ka/Ks values are less than 1, which indicates that they have experienced a strong purifying or negative selection35. Furthermore, we performed collinearity analysis to reveal
the origin and evolutionary relationship of OVATE gene family in different species (Fig. 5). The results showed that 63 homologous gene pairs were identified between _sorghum_ and _maize_
genomes, and some _sorghum_ genes were found to participate in the formation of gene pairs many times. However, only ten gene pairs of collinearity were found between _sorghum_ and
_Arabidopsis_ genome. These results suggest that there is a closer genetic relationship between _sorghum_ and _maize_, and some members of OVATE family have expanded in _maize_. ANALYSIS OF
OVATE GENE EXPRESSION USING TRANSCRIPTOME DATA In order to reveal the expression characteristics of OVATE gene family in _sorghum_, we selected leaves and seeds of three different
development stages for transcriptome sequencing. Transcriptome analysis showed that there were a large number of differentially expressed genes in different tissues (Fig. 6). As shown in
Fig. 6A, there are the most differentially expressed genes in the two developmental stages of S2 and L2, with a number of 14,116, and the differentially expressed genes in different
developmental stages of leaf are only 940 (L2vL1); between leaves and seeds, the differentially expressed genes shared by different developmental stages reached more than 5000. Meantime, the
differentially expressed genes in different stages of seed development reached 7445 (S2vS1), which was much higher than that in leaves (L2vL1). In order to further explore the functional
characteristics of the difference genes, we performed a KEGG enrichment analysis of the difference genes between seeds and leaves. The results showed that for S1vS2, the differential genes
were mainly enriched in the biosynthesis of secondary metabolites such as flavonoids and polyphenols (Fig. 6B). However, between seeds and leaves, a large number of differential genes are
enriched not only in metabolism-related pathways, but also in carbon metabolism and photosynthesis-related pathways (Fig. 6C). Furthermore, based on the transcriptome sequencing data, we
analyzed the expression levels of OVATE gene family members in different tissues. The results are shown in Fig. 7, most members of the OVATE gene family did not have different expression in
_sorghum_ seeds and leaves, while some genes such as _SORBI_3003G010700_ and _SORBI_3009G173400_ were expressed at higher levels in _sorghum_ seeds, but some genes were expressed at
significantly higher levels in leaves than seeds (such as _SORBI_3005G042700_). VERIFICATION OF EXPRESSION OF MEMBERS OF THE OVATE GENE FAMILY OF _SORGHUM_ BY RT-QPCR To verify the
expression levels of OVATE gene family members in _sorghum_ seeds and leaves, we designed 26 pairs of primers for RT-qPCR amplification. Due to the imbalance in sequence GC content and the
presence of repetitive sequences, only 8 genes were successfully validated in the end (some family members do not express or have extremely low expression levels and are difficult to be
verified) (Fig. 7). The results showed that these genes had multiple expression patterns in the process of _sorghum_ tissue development (Fig. 8). For example, except for _SORBI_3003G339100_
and _SORBl_3006G187600_, the expression levels of all genes in S1 and L1 are significantly different. Among them, with the development of leaves and seeds, the expression level of some genes
gradually decreased (_SORBI_3002G417700_ and _SORBl_3005G042700_). This indicates that members of OVATE gene family may be involved in regulating the development of _sorghum_ leaves and
seeds. At the same time, the similar expression level of RT-qPCR and transcriptome proves the reliability of the above results. DISCUSSION Exploring the growth and development of seeds plays
an important role in improving the yield and economic value of _sorghum_. In previous studies, some researchers have conducted in-depth research on the development regulation of _sorghum_
seeds. For example, Tao et al.38 identified 114 candidate genes associated with _sorghum_ seed size, of which 63 showed a signal of purification selection during domestication; Zhang et
al.39 identified 73 QTLs related to grain color and tannin content in China _sorghum_ materials through genome-wide association study, and found a new recessive allelic variant of _TAN2_
gene. Many studies in other plants show that OVATE gene family plays an important role in regulating the development of plant organs and resisting stress. Studies have shown that _OsOFP6_ in
rice not only participates in the growth regulation of rice plant development, but also may enhance the drought tolerance of rice40; In tomato, OVATE can not only regulate the shape of
fruit, but also affect the development of flower organs and pollen by regulating the signal transduction of BR and GA41. However, the biological function of OVATE gene family in sorghum has
not been revealed. In this study, we identified 26 OVATE gene family members from the sorghum genome, which are more numerous than _Arabidopsis thaliana_ but lower than rice, and equal to
the number of OVATE gene family members in tea plants18,42,43. These genes are unevenly distributed across nine chromosomes, with numbers ranging from 1 (chr4, chr5, chr7, chr8) to 7 (chr3).
Phylogenetic analysis showed that 26 OVATE genes were classified into four groups, and OVATE gene family members showed high homology in grain _sorghum_ and sweet, suggesting that the OVATE
gene family had a low degree of differentiation among different _sorghum_ lines. The motif analysis found that motif 1 and motif 2 were identified in all members of the OVATE gene family,
but some specific motifs were only identified in some genes. For example, motif 5 and motif 7 are only found in the four genes of _SORBI_3001G316300_, _SORBI_3001G523500_,
_SORBI_3004G282200_ and _SORBI_3006G187600_, while motif 10 is only identified in _SORBI_3008G042100_, _SORBI_3005G042700_ and _SORBI_3006G274000_. These findings suggest that these motifs
may be important for functional diversity differentiation of the OVATE gene in _sorghum_20,37. In addition to the common light-responsive elements, a large number of hormone-related
cis-acting elements were found in the promoter sequence of 2000 bp upstream of the OVATE genes. At the same time, in addition to some cis-acting elements associated with abiotic stresses
such as low temperature and drought, seed-specific regulation related elements were also identified. These results suggest that members of the OVATE gene family in _sorghum_ may be
responsible for multiple biological functions. Segmental and tandem duplication are important drivers of gene family expansion44. Evolutionary analysis within the genome showed that members
of the OVATE gene family formed 19 gene pairs, some of which were involved in the formation of gene pairs multiple times. These results suggest that segmental duplication events play an
important role in the expansion of the OVATE gene family in _sorghum_45,46. Collinear analysis showed more collinear gene pairs between maize and sorghum compared to _Arabidopsis_,
suggesting that the expansion of the OVATE gene family may have occurred after the differentiation of sorghum and _Arabidopsis_. In addition, Ka/Ks analysis showed that members of the OVATE
gene family had a higher frequency of harmful mutations, and they were in the state of purification or negative selection35,42,47. To explore the expression characteristics of OVATE gene
family members during _sorghum_ leaf and seed development, we performed transcriptome sequencing of leaves and seeds in two developmental stages. The results of transcriptome analysis showed
that there were a large number of differentially expressed genes between sorghum leaves and seeds at different stages of development. For example, more than 5,000 differential genes are
shared between S1vL1, S1vL2, S2vL1, and S2vL2. KEGG analysis showed that metabolite-related pathways were enriched at different stages of seed development and between seed and leaf.
Furthermore, we focused on the expression analysis of members in the OVATE gene family in different tissues of _sorghum_. The results showed that members of the OVATE gene family exhibited
diverse expression characteristics during the development of leaves and seeds, suggesting their potential for diversified regulatory functions19,29,48. It needs to be pointed out that the
transcriptome analysis results show that some members of the OVATE family do not express or have extremely low expression levels in _sorghum_, which indicates that these genes may not
undertake regulatory functions in _sorghum_ leaves and seeds. In addition, due to a large number of repetitive sequences, it is very difficult to design appropriate primer pairs. Finally,
only the expression levels of 8 OVATE gene members were successfully verified by RT-qPCR analysis, proving that the expression levels of these genes are tissue-specific. For example,
_SORBl_3001G468900_, _SORBI_3009G173400_ and _SORBl_3001G380100_ are expressed significantly higher in seeds than in leaves, while gene _SORBl_3005G042700_ and _SORBl_3004G282200_ are highly
expressed in leaves (L1 stage). Meantime, the _SORBI_3002G417700_, _SORBI_3001G380100_, _SORBI_3009G173400_, _SORBI_3003G339100_ and _OFP4_ (_AT1G06920_) in _Arabidopsis thaliana_ are
highly homologous, and they are all clustered in Group III, while the latter has been confirmed to have the function of regulating cell wall formation25. Homology analysis found that the
genes _SORBI3009G173400_ and _SORBI3003G339100_ in _sorghum_ have relatively high homology with _SlOFP20_ in tomatoes that regulates floral organ and pollen development41. These results
strongly suggest that the eight genes verified by RT-qPCR may play an important role in the development process of _sorghum_ tissues and organs. CONCLUSIONS In this study, we performed a
comprehensive identification of the OVATE gene family based on the BTx623 _sorghum_ genome. The 26 OVATE gene family members are spread across 9 chromosomes and are divided into four groups.
Promoter sequence analysis showed that a large number of hormone-related cis-acting elements were identified, suggesting that members of the OVATE gene family may be involved in the
development process of _sorghum_ tissues and organs. At the same time, evolutionary analysis showed that the OVATE family members in _sorghum_ had expanded after differentiation with
_Arabidopsis_, and all members were subject to purification selection. RNA sequencing and RT-qPCR analysis showed that members of the OVATE gene family exhibited diverse expression
characteristics in sorghum leaves and seeds. These results will provide fundamental support for our deeper understanding of the biological function of the OVATE gene family in _sorghum_ and
promote the _sorghum_ breeding process. DATA AVAILABILITY Transcriptome sequencing data was uploaded to the National Bioinformatics Center of China (CNCB), the BioProject accession number is
PRJCA020623 (https://ngdc.cncb.ac.cn/gsa/browse/CRA013087). REFERENCES * Ahn, E. _et al._ Genome-Wide Association Study of Seed Morphology Traits in Senegalese Sorghum Cultivars. _Plants_
12, 2344 (2023). Article CAS PubMed PubMed Central Google Scholar * Zhang, Y. _et al._ The Identification of a Yield-Related Gene Controlling Multiple Traits Using GWAS in Sorghum
(Sorghum bicolor L.). _Plants_ 12, 1557 (2023). Article CAS PubMed PubMed Central Google Scholar * Jeon, D., Kim, J.-B., Kang, B.-C. & Kim, C. Deciphering the Genetic Mechanisms of
Salt Tolerance in Sorghum bicolor L.: Key Genes and SNP Associations from Comparative Transcriptomic Analyses. _Plants_ 12, 2639 (2023). Article CAS PubMed PubMed Central Google Scholar
* Wang, X. _et al._ Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. _Nat. Genet._ 48, 1233–1241 (2016). Article CAS PubMed Google Scholar * Wen, W. _et
al._ Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. _Nat. Commun._ 5, 3438 (2014). Article ADS PubMed Google Scholar * Wang, Q. _et
al._ Genetic Architecture of Natural Variation in Rice Chlorophyll Content Revealed by a Genome-Wide Association Study. _Mol. Plant._ 8, 946–957 (2015). Article CAS PubMed Google Scholar
* Wang, H. _et al._ The Power of Inbreeding: NGS-Based GWAS of Rice Reveals Convergent Evolution during Rice Domestication. _Mol. Plant._ 9, 975–985 (2016). Article CAS PubMed Google
Scholar * Xia, E. _et al._ The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into genome evolution and adaptation of tea plants. _Mol. Plant_
https://doi.org/10.1016/j.molp.2020.04.010 (2020). Article PubMed Google Scholar * Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression
analysis of RNA-seq experiments with HISAT StringTie and Ballgown. _Nat. Protoc._ 11, 1650–1667 (2016). Article CAS PubMed PubMed Central Google Scholar * Paterson, A. H. _et al._ The
Sorghum bicolor genome and the diversification of grasses. _Nature_ 457, 551–556 (2009). Article ADS CAS PubMed Google Scholar * McCormick, R. F. _et al._ The Sorghum bicolor reference
genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. _Plant J._ 93, 338–354 (2017). Article PubMed Google Scholar * Deschamps, S. _et
al._ A chromosome-scale assembly of the sorghum genome using nanopore sequencing and optical mapping. _Nat. Commun._ https://doi.org/10.1038/s41467-018-07271-1 (2018). Article PubMed
PubMed Central Google Scholar * Cooper, E. A. _et al._ A new reference genome for Sorghum bicolor reveals high levels of sequence similarity between sweet and grain genotypes: implications
for the genetics of sugar metabolism. _BMC Genomics_ https://doi.org/10.1186/s12864-019-5734-x (2019). Article PubMed PubMed Central Google Scholar * Morris, G. P. _et al._ Population
genomic and genome-wide association studies of agroclimatic traits in sorghum. _Proc. Natl. Acad. Sci. U S A_ 110, 453–458 (2013). Article ADS CAS PubMed Google Scholar * Tao, Y. _et
al._ Extensive variation within the pan-genome of cultivated and wild sorghum. _Nat. Plants_ 7, 766–773 (2021). Article CAS PubMed Google Scholar * Calone, R. _et al._ Salt Tolerance and
Na Allocation in Sorghum bicolor under Variable Soil and Water Salinity. _Plants_ https://doi.org/10.3390/plants9050561 (2020). Article PubMed PubMed Central Google Scholar * Zhou, Y.
_et al._ Yield and Quality in Main and Ratoon Crops of Grain Sorghum Under Different Nitrogen Rates and Planting Densities. _Front. Plant Sci._ https://doi.org/10.3389/fpls.2021.778663
(2022). Article PubMed PubMed Central Google Scholar * An, Y., Xia, X., Jing, T. & Zhang, F. Identification of gene family members and a key structural variation reveal important
roles of OVATE genes in regulating tea (Camellia sinensis) leaf development. _Front. Plant Sci._ https://doi.org/10.3389/fpls.2022.1008408 (2022). Article PubMed PubMed Central Google
Scholar * Wang, S., Chang, Y. & Ellis, B. Overview of ovate family proteins, A novel class of plant-specific growth regulators. _Front. Plant Sci._
https://doi.org/10.3389/fpls.2016.00417 (2016). Article PubMed PubMed Central Google Scholar * Luo, Y. _et al._ Genome-wide analysis of OFP gene family in pepper (Capsicum annuum L.).
_Front. Genet._ https://doi.org/10.3389/fgene.2022.941954 (2022). Article PubMed PubMed Central Google Scholar * Zhou, H. _et al._ A 1.7-Mb chromosomal inversion downstream of a PpOFP1
gene is responsible for flat fruit shape in peach. _Plant Biotechnol. J._ 19, 2 (2020). Google Scholar * Guan, J. _et al._ Genome structure variation analyses of peach reveal population
dynamics and a 1.67 Mb causal inversion for fruit shape. _Genome Biol._ 22, 13 (2021). Article CAS PubMed PubMed Central Google Scholar * Sun, X., Ma, Y., Yang, C. & Li, J. Rice
OVATE family protein 6 regulates leaf angle by modulating secondary cell wall biosynthesis. _Plant Mol. Biol._ 104, 249–261 (2020). Article CAS PubMed Google Scholar * Tang, Y. _et al._
Expression of ovate family protein 8 affects epicuticular waxes accumulation in Arabidopsis thaliana. _Bot. Stud._ https://doi.org/10.1186/s40529-018-0228-8 (2018). Article PubMed PubMed
Central Google Scholar * Li, E., Wang, S., Liu, Y., Chen, J.-G. & Douglas, C. J. OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in
Arabidopsis thaliana. _Plant J._ 67, 328–341 (2011). Article CAS PubMed Google Scholar * Uhrig, J.H.K.R.J.M.F.S.a.J.F. A central role of Arabidopsis thaliana ovate family proteins in
networking and subcellular localization of 3-aa loop extension homeodomain proteins. PNAS (2004). * Wu, Q. _et al._ Identification and Comprehensive Analysis of OFP Genes for Fruit Shape
Influence in Mango. _Genes_ 15, 823 (2024). Article CAS Google Scholar * Zhang, L., Tehseen Azhar, M., Che, J. & Shang, H. Genome-wide identification, expression and evolution
analysis of OVATE family proteins in cotton (Gossypium spp.). _Gene_ 834, 146653 (2022). Article CAS PubMed Google Scholar * Ahmad, S., Ahmad, N. & Bayar, J. Genome-wide
identification, characterization, and expression analysis of the Ovate family protein in Oryza sativa under biotic and abiotic stresses. _Plant Stress_ 10, 100228 (2023). Article CAS
Google Scholar * Hou, Y., Wu, A., He, Y., Li, F. & Wei, C. Genome-wide characterization of the basic leucine zipper transcription factors in Camellia sinensis. _Tree Genet. Genom._
https://doi.org/10.1007/s11295-018-1242-4 (2018). Article Google Scholar * Wang, L., Feng, Z., Wang, X., Wang, X. & Zhang, X. DEGseq: an R package for identifying differentially
expressed genes from RNA-seq data. _Bioinformatics_ 26, 136–138 (2010). Article PubMed Google Scholar * Gladman, N. _et al._ SorghumBase: a web-based portal for sorghum genetic
information and community advancement. _Planta_ https://doi.org/10.1007/s00425-022-03821-6 (2022). Article PubMed PubMed Central Google Scholar * An, Y. _et al._ Genome-wide
identification of the PYL gene family of tea plants (Camellia sinensis) revealed its expression profiles under different stress and tissues. _BMC Genom._
https://doi.org/10.1186/s12864-023-09464-5 (2023). Article Google Scholar * Chen, C. _et al._ TBtools - an integrative toolkit developed for interactive analyses of big biological data.
_Mol. Plant._ https://doi.org/10.1016/j.molp.2020.06.009 (2020). Article PubMed Google Scholar * Shen, C. & Li, X. Genome-wide identification and expression pattern profiling of the
ATP-binding cassette gene family in tea plant (Camellia sinensis). _Plant Physiol. Biochem._ 202, 107930 (2023). Article CAS PubMed Google Scholar * Sudhakar Reddy, P. _et al._
Evaluation of Sorghum [Sorghum bicolor (L.)] Reference Genes in Various Tissues and under Abiotic Stress Conditions for Quantitative Real-Time PCR Data Normalization. _Front. Plant Sci._
https://doi.org/10.3389/fpls.2016.00529 (2016). Article PubMed PubMed Central Google Scholar * Huang, D., Mao, Y., Guo, G., Ni, D. & Chen, L. Genome-wide identification of PME gene
family and expression of candidate genes associated with aluminum tolerance in tea plant (Camellia sinensis). _BMC Plant Biol._ https://doi.org/10.1186/s12870-022-03686-7 (2022). Article
PubMed PubMed Central Google Scholar * Tao, Y. _et al._ Whole-Genome Analysis of Candidate genes Associated with Seed Size and Weight in Sorghum bicolor Reveals Signatures of Artificial
Selection and Insights into Parallel Domestication in Cereal Crops. _Front. Plant Sci._ https://doi.org/10.3389/fpls.2017.01237 (2017). Article PubMed PubMed Central Google Scholar *
Zhang, L. _et al._ GWAS of grain color and tannin content in Chinese sorghum based on whole-genome sequencing. _Theor. Appl. Gene._ https://doi.org/10.1007/s00122-023-04307-z (2023). Article
ADS Google Scholar * Ma, Y. _et al._ Rice OVATE family protein 6 regulates plant development and confers resistance to drought and cold stresses. _J. Exp. Bot._ 68, 4885–4898 (2017).
Article CAS PubMed Google Scholar * Zhou, S. _et al._ Overexpression of SlOFP20 affects floral organ and pollen development. _Hortic. Res._ https://doi.org/10.1038/s41438-019-0207-6
(2019). Article PubMed PubMed Central Google Scholar * Liu, D. _et al._ Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. _Ann.
Bot._ 113, 1219–1233 (2014). Article CAS PubMed PubMed Central Google Scholar * Beemster, G. T. S. _et al._ Expression Pattern and Subcellular Localization of the Ovate Protein Family
in Rice. _Plos One_ 10, e0118966 (2015). Article Google Scholar * Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D. & May, G. The roles of segmental and tandem gene duplication in
the evolution of large gene families in Arabidopsis thaliana. _BMC Plant Biol._ https://doi.org/10.1371/journal.pone.0118966 (2004). Article PubMed PubMed Central Google Scholar * Yan
Zhu, N. W. _et al._ Soybean (Glycine max) expansin gene superfamily origins: Segmental and tandem duplication events followed by divergent selection among subfamilies. _BMC Plant. Biol._
https://doi.org/10.1186/1471-2229-14-93 (2014). Article PubMed Google Scholar * Vatansever, R. _et al._ Genome-wide identification and expression analysis of sulfate transporter (SULTR)
genes in potato (Solanum tuberosum L.). _Planta_ 244, 1167–1183 (2016). Article CAS PubMed Google Scholar * Rasool, F. _et al._ Phenylalanine Ammonia-Lyase (PAL) Genes Family in Wheat
(Triticum aestivum L.): Genome-Wide Characterization and Expression Profiling. _Agronomy_ 11, 2511 (2021). Article CAS Google Scholar * Han, L.-J. _et al._ Genome-wide analysis of OVATE
family proteins in cucumber (Cucumissativus L.). _J. Integr. Agric._ 21, 1321–1331 (2022). Article Google Scholar Download references ACKNOWLEDGEMENTS This research was jointly supported
by grants from the Zunyi Technology and Big data Bureau, Moutai institute Joint Science and Technology Research and Development Project (ZSKHHZ [2022] No.166 and ZSKHHZ [2021] No.327); the
National Natural Science Foundation of China (32160441); the Natural Science Foundation of Guizhou Province (ZK[2023]453); research Foundation for Scientific Scholars of Moutai Institute
(mygccrc[2022]074, mygccrc[2022]088, mygccrc[2022]089, mygccrc[2022]093). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Food Science and Engineering, Moutai Institute, Renhuai,
China Yanlin An, Xiaoqin Zhang, Li Liu, Sixia Jiang & Feng Zhang * State Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei, China Tingting Jing *
College of Plant Protection, Nanjing Agricultural University, Nanjing, 210095, China Xiaobo Xia Authors * Yanlin An View author publications You can also search for this author inPubMed
Google Scholar * Xiaobo Xia View author publications You can also search for this author inPubMed Google Scholar * Xiaoqin Zhang View author publications You can also search for this author
inPubMed Google Scholar * Li Liu View author publications You can also search for this author inPubMed Google Scholar * Sixia Jiang View author publications You can also search for this
author inPubMed Google Scholar * Tingting Jing View author publications You can also search for this author inPubMed Google Scholar * Feng Zhang View author publications You can also search
for this author inPubMed Google Scholar CONTRIBUTIONS F.Z. and T.J. designed the experiments. X.X., X.Z. and S.J. performed the computational analysis. L.L. assisted with experiments in data
collection and analysis. Y.A. participated in the design and supervised the study. Y.A. drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
CORRESPONDING AUTHORS Correspondence to Tingting Jing or Feng Zhang. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION
PUBLISHER'S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY TABLE S1.
SUPPLEMENTARY TABLE S2. SUPPLEMENTARY TABLE S3. SUPPLEMENTARY TABLE S4. SUPPLEMENTARY TABLE S5. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons
Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original
author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the
article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your
intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence,
visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE An, Y., Xia, X., Zhang, X. _et al._ Genome-wide identification of the
_sorghum_ OVATE gene family and revelation of its expression characteristics in _sorghum_ seeds and leaves. _Sci Rep_ 14, 15123 (2024). https://doi.org/10.1038/s41598-024-66103-z Download
citation * Received: 09 January 2024 * Accepted: 27 June 2024 * Published: 02 July 2024 * DOI: https://doi.org/10.1038/s41598-024-66103-z SHARE THIS ARTICLE Anyone you share the following
link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature
SharedIt content-sharing initiative KEYWORDS * _Sorghum_ * OVATE * Gene family * Transcriptome sequencing