- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The rapid spread of the severe acute respiratory syndrome coronavirus 2 led to a global overextension of healthcare. Both Chest X-rays (CXR) and blood test have been demonstrated to
have predictive value on Coronavirus Disease 2019 (COVID-19) diagnosis on different prevalence scenarios. With the objective of improving and accelerating the diagnosis of COVID-19, a multi
modal prediction algorithm (MultiCOVID) based on CXR and blood test was developed, to discriminate between COVID-19, Heart Failure and Non-COVID Pneumonia and healthy (Control) patients.
This retrospective single-center study includes CXR and blood test obtained between January 2017 and May 2020. Multi modal prediction models were generated using opensource DL algorithms.
Performance of the MultiCOVID algorithm was compared with interpretations from five experienced thoracic radiologists on 300 random test images using the McNemar–Bowker test. A total of 8578
samples from 6123 patients (mean age 66 ± 18 years of standard deviation, 3523 men) were evaluated across datasets. For the entire test set, the overall accuracy of MultiCOVID was 84%, with
a mean AUC of 0.92 (0.89–0.94). For 300 random test images, overall accuracy of MultiCOVID was significantly higher (69.6%) compared with individual radiologists (range, 43.7–58.7%) and the
consensus of all five radiologists (59.3%, _P_ < .001). Overall, we have developed a multimodal deep learning algorithm, MultiCOVID, that discriminates among COVID-19, heart failure,
non-COVID pneumonia and healthy patients using both CXR and blood test with a significantly better performance than experienced thoracic radiologists. SIMILAR CONTENT BEING VIEWED BY OTHERS
ASSESSING CLINICAL APPLICABILITY OF COVID-19 DETECTION IN CHEST RADIOGRAPHY WITH DEEP LEARNING Article Open access 21 April 2022 DEEP LEARNING MODEL FOR THE AUTOMATIC CLASSIFICATION OF
COVID-19 PNEUMONIA, NON-COVID-19 PNEUMONIA, AND THE HEALTHY: A MULTI-CENTER RETROSPECTIVE STUDY Article Open access 17 May 2022 OPEN RESOURCE OF CLINICAL DATA FROM PATIENTS WITH PNEUMONIA
FOR THE PREDICTION OF COVID-19 OUTCOMES VIA DEEP LEARNING Article Open access 18 November 2020 INTRODUCTION The outbreak of Coronavirus Disease 2019 (COVID-19), caused by severe acute
respiratory syndrome coronavirus 2 (SARS-CoV-2), stroke the worldwide population with more than 200 million cases and 4.5 million deaths by August 2021. The rapid spread of the pandemic led
to a global overexertion of health care and research facilities in order to counteract the growing rate of infection. However, a collapse of the sanitary system was imminent and inevitable
worldwide, and new technologies were needed to speed up the diagnostic process. The reference for COVID-19 diagnosis is the detection of SARS-CoV-2 viral RNA by real-time polymerase chain
reaction (RT-PCR). However, the massive requests for sample processing at the beginning of the pandemic caused serious delays to obtain results. As lung involvement is one of the main causes
of morbidity and mortality in SARS-CoV-2 infection, a quick identification of characteristic findings in chest imaging can support the diagnosis and speed up the identification of COVID-19
positive patients at the emergency units. Several studies have shown that implementation of deep learning (DL) tools to detect chest X-rays (CXR) findings typically associated with
SARS-CoV-2 infection, deliver comparable results to those acquired by interpretation of radiologists. However, most of the trained models have a drop in their prediction performance when
tested over external datasets1. In addition, one of the main hurdles to overcome when training an algorithm to detect Sars-CoV-2 infection in CXR is the similarity of findings with other
entities like bacterial pneumonias or heart failure2. On the other hand, models based on laboratory results of peripheral blood also give predictive results on diagnosis3 and prognosis4. A
key fact to highlight is how the incursion of COVID-19 caused a dramatic drop in the emergency room consultations of other pathologies. Later on, after the initial peak, the decline of the
COVID-19 prevalence made the non-COVID diseases emerge once again at the hospitals. This is relevant due to the challenge of performing an efficient differential diagnosis with selected
pathologies during a pandemic. It is well known that the predictive value of a diagnostic test is conditioned by the prevalence of the disease and that of COVID varies widely throughout the
different waves of the pandemic5. A multicategory approach that takes into account differential diagnoses that are more stable in their prevalence could reduce this variability. With the
objective of improving and accelerating the diagnosis of COVID-19, we developed a tool to assist physicians in reaching a diagnosis. This tool is a multi-modal prediction algorithm
(MultiCOVID) based on CXR and blood test with the ability to discriminate between COVID-19, Heart Failure (HF), Non-COVID Pneumonia (NCP) and healthy (Control) samples. MATERIALS AND METHODS
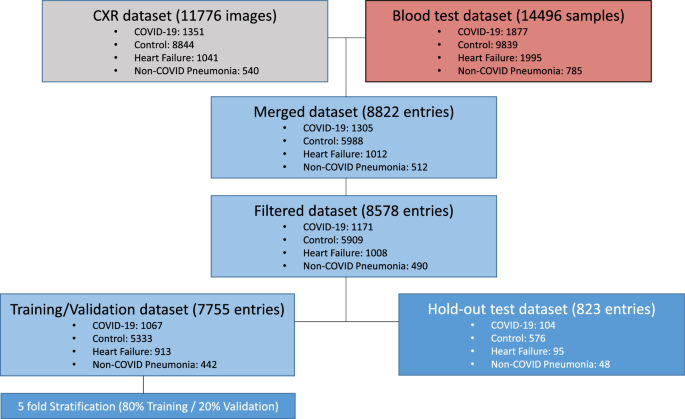
DATASET We retrospectively collected CXR images and hemogram values from 8578 samples from 6123 patients and healthy subjects (mean age 66 ± 18 years of standard deviation, 3523 men) from
Parc Salut Mar (PSMAR) Consortium, Barcelona, Spain. Four cohorts were designed: (i) 1171 samples from patients diagnosed with COVID-19 by RT-PCR from March to May 2020; (ii) 1008 samples of
patients who suffered an episode of heart failure between 2012 to 2019; (iii) 490 samples of patients diagnosed with non-COVID pneumonia (NCP) from 2018 to 2019; (iv) 5909 samples of
standard preoperatory studies of healthy subjects from 2017 to 2019 (Fig. 1). HR and NCP diagnosis were selected as defined by the International Classification of Diseases, Tenth Revision
(ICD-10) code. All the CXR images from groups i-iii were validated by two independent radiologists (MB and JM). ACQUISITION OF BLOOD SAMPLE AND IMAGE DATA We included CXR images performed in
a period ranging from 1 day before the patient’s diagnosis to 7 days after. The images were filtered to include only frontal projections regardless of the quality and the radiography system
used. Blood sample results were collected within a range of 2 days before or 7 days after the CXR acquisition date using PSMAR lab record system, except for control samples whose
measurements ranged for 2 weeks. If two or more blood test results were collected, measurements were averaged. CXR images and blood test results were combined in the same dataset and split
into train/validation set (90%), and hold-out test (10%) set. For training/validation split, we divided the dataset in training (80%) and validation (20%) sets with 5 different random seeds.
We ensured that there were no cross-over patients between groups. DEEP LEARNING MODELS Detailed description of the models, training policy and image preprocessing are provided in
Supplementary Material. In brief, segmentation model is based on a U-Net architecture6. The CXR-only classification model consists of a validated Convolutional neural network (CNN) resnet-34
architecture7. Tabular only-model is an Attention-based network (TabNet)8. Joint model is a multi-modal deep learning algorithm which merges the CXR-only and the Blood-only models and uses
both CXR image and blood tests as input values. It uses Gradient Blending in order to prevent overfitting and improve generalization9. MultiCOVID model is an ensemble predictor of 5
different Joint models that would classify independently between the different classes. Then it uses majority vote to assign a final classification. The whole pipeline development and
training was performed using fastai deep learning API10. COMPARISON WITH THORACIC RADIOLOGIST INTERPRETATIONS Hold-out test dataset consisting of 300 samples (ensuring no patient overlap
with training or validation sets) was used for expert interpretation. Each sample consisted of a CXR with matched blood results. Expert interpretations were independently provided by five
board-certified thoracic radiologists (FZ, SC, LdC, DR, AG) with 2–30 years post-residency training experience. Radiologists were able to check both non segmented images and blood test
results without any other additional information in a platform created ad-hoc for prediction. They provided a classification for each image in one of the four categories (COVID-19, control,
HF and NCP). A consensus interpretation for the radiologist was obtained by the majority vote for each paired CHX-blood test analyzed. STATISTICAL ANALYSIS A two-tailed t-test P value was
reported when clinical and population blood test differences were assessed. McNemar–Bowker test was used to compare model performance against radiologist majority vote using FDR correction.
Plotting and statistical analyses were performed using the packages ggplot, ggpubr and rcompanion in R, version 3.6 (R Core Team; R Foundation for Statistical Computing). ETHICAL APPROVAL
The study was designed to use radiology images and associated clinical/demographic/ laboratory patient information already collected for the purpose of performing clinical COVID-19 research
by Hospital del Mar. The study was conducted in accordance with the relevant institutional guidelines and regulations. The experimental protocols, data acquisition and analysis were approved
by the Parc de Salut Mar Clinical Research Ethics Committee (2020/9199/I). Informed consent was obtained, when possible, from patients or legal representatives or waived by the local Parc
de Salut Mar Clinical Research Ethics Committee (2020/9199/I) if informed consent was not available due to the pandemic situation. RESULTS PATIENT CHARACTERISTICS A total of 8578 samples
were evaluated across datasets. Patient characteristics and blood test parameters are shown in Table 1. A highly significant difference in age was found between the cohort of patients with
heart failure (82.8 ± 10 years) and the other three cohorts (66.0 ± 16 years for COVID-19 samples, 63.2 ± 18 years for control samples and 67.8 ± 17 years for NCP samples, _P_ < 0.001 for
each comparison) and was not considered as a valid variable for further classification. WHOLE CXR MODELS LEARN SPURIOUS CHARACTERISTICS FOR CLASSIFICATION Previous studies have demonstrated
that deep learning (DL)-based algorithms should be rigorously evaluated due to their ability to learn non relevant features in order to increase its prediction accuracy1. For this reason,
we first developed a segmentation algorithm able to segment lung parenchyma at a 95%-pixel accuracy. Then, after segmentation, we evaluated the accuracy of the algorithms for three
complementary datasets: non-segmented images, segmented regions and excluded regions. After a few training epochs the three different models achieved nonrandom accuracies between 67 and 74%
(Fig. 2A). However, attention map exploration on the images showed that the different models based their predictions not only inside but also outside of the lung parenchyma (Fig. 2B). These
observations showed that, although there are important features outside the lung parenchyma that may help the model to classify between the different entities (eg. heart size), there are
other elements (eg. oxygen nasal cannulas or intravenous (IV) catheters) that might confound the model. Thus, we decided to first segment all the CXR before training our models for
prediction of diagnosis. In order to accomplish this task, we generated a 785-radiology level lung segmentation dataset and trained a U-net model to regenerate the whole CXR dataset keeping
only the lung parenchyma. PERFORMANCE OF SINGLE AND MULTIMODAL MODELS In order to evaluate the prediction capacity of both segmented CXR and blood sample data, we built different DL models
using both sources alone or in combination. Metrics comparison of all the single vision (CXR-only) and tabular (Blood-only) models are detailed in Supplementary Material. As expected,
CXR-only models had a more robust prediction of all 4 categories tested compared to Blood-only models (Fig. 3). This difference is stronger in the classes with less samples (HF, and NCP)
where CXR-only models could identify features in the CXR images which are characteristic of these two entities whereas this was not possible with Blood-only models. Model interpretability of
Blood-only models by analyzing feature importance using Shapley Additive explanations12 showed that patient classification was related to two different axes: the immune compartment and the
red blood cell (RBC) compartment, respectively (Fig. 4A). The first axis seems to be strongly associated with COVID-19 classification and shows a specific signature looking at the blood
counts (Fig. 4B-top). However, the second axis seems to subdivide patients between COVID-19/Control and HF/NCP, although COVID-19 blood counts seems to be statistically different from
Control samples, too (Fig. 4B-bottom). The combination of CXR and blood tests using multimodal models that combine inputs from tabular and image data to perform a global prediction, slightly
increased the prediction capacity of the single models even when DL tabular models are worse than machine learning (ML—XGBoost) models alone (Supplementary Table 1). This underpins the
concept that adding new sources of information to the data could increase the ability of the models to generate better predictions 13. Moreover, the joint approach used for building
MultiCOVID algorithm resulted on an improved performance in the majority of the metrics analyzed (Fig. 3 and Supplementary Table 1). COMPARISON WITH EXPERT THORACIC RADIOLOGISTS Finally, we
compared the performance of MultiCOVID algorithm with the interpretation of expert chest radiologists. This comparison was performed with 300 CXR randomly selected from the hold-out test set
that were independently reviewed by 5 radiologists together with the blood test results. The independent results from radiologists showed an accuracy ranging from 43.7 to 58.7%. This value
rose to 59.3% (178/300) when the consensus interpretation of all 5 radiologists based on the majority vote was considered. Of note, the overall accuracy achieved by MultiCOVID was 69.6%
(209/300) that was significantly higher than consensus interpretation (_P_ < 0.001). In addition, for COVID-19 prediction individually, MultiCOVID showed similar sensitivity to the
radiologists’ consensus but with a much higher specificity, leading to significantly better performance when discerning between COVID-19 versus Control and COVID-19 vs HF patients (_P_ <
0.05 for both comparisons; Fig. 5). DISCUSSION Diagnosis of COVID-19 is an evolving challenge. During the beginning of the pandemic and the successive peaks with high prevalence rates, a
prompt and effective diagnosis was critical for proper patient isolation and evaluation. However, since the prevalence of the COVID-19 cases oscillated, showing fewer cases between waves,
and more non-COVID cases, it was important to differentiate patients with other diseases than COVID-19 presenting similar visual characteristics in the CXR. During patient assessment in the
emergency room, clinicians take into account different inputs for a proper diagnosis. First, the anamnesis, symptoms, vitals and physical findings guide the physician to an initial
assumption. Based on this information, additional tests are requested (CXR, blood test, ECG and SARS-CoV-2 detection). The integration of these results allows the team to diagnose a patient
accurately. However, this process is time consuming and sometimes findings are difficult to interpret, leading to misdiagnosis. To improve this diagnostic process, we have developed and
trained a multimodal deep learning algorithm based in a multiple input approach combining CXR images together with blood sample data to identify COVID-19 diagnosis with high sensitivity.
This way we were able to manage the increased complexity of the dataset. These data from multiple sources are somehow correlated and complementary to each other and could reflect patterns
that are not present in single models alone13. Hence, MultiCOVID is fed by two of the most common and fast clinical tests requested in the emergency room (CXR and Blood test) and can predict
the presence of three different diseases (COVID-19, heart failure and non-COVID pneumonia) with similar CXR characteristics. Analysis of single models shows the importance of model
interpretation. While CXR-only models could identify patterns outside the lung parenchyma that could diminish its generalization capacity9, Blood-only models could point to interesting
population of cells that are differently represented in COVID-19 patients, leveraging its prediction capacity. In this context, the immune compartment plays an important role in the COVID-19
response, and it has been already published that COVID-19 patients present fewer overall leukocytes counts and, more concretely, eosinophil counts14, 15. Furthermore, oxygen transport seems
to be somehow affected, modulating the red cell population. In this regard, in our work we found significant differences in the erythrocyte count and the hemoglobin concentration. Although
most of the studies correlate the reduction of this values to severe COVID-19 patients16, this is the first dataset to compare them in these four different categories at the time of
diagnosis. Moreover, although a huge amount of literature about COVID-19 diagnosis and prognosis has been published using only blood tests17,18,19,20 or CXR21,22,23,24,25,26,27,28 this is
the first study that combines both parameters and compares its prediction capacity at diagnosis. Of note, only one previously published study integrates both blood test and CXR severity
scores in order to determine in-hospital death of COVID-19 patients29. Hence, it is clear that merging both sources of data leads to a better prediction performance when compared with the
two single models alone and that this difference is more pronounced where the number of cases is scarce. It is important to stress that this combination of data sources addresses the
variable prevalence of COVID-19 cases during the pandemic, which is an issue that could not be solved in previous studies23, 24. Our study has several limitations. First, the algorithm was
evaluated on a single center; thus, there was likely some degree of bias. Additionally, the sample collection was performed in different time periods for each group of patients, which could
present some kind of differences in the CXR image acquisition although this was partially solved using the lung segmentation model which removes the noise signal present outside the lung
parenchyma. And finally, model performance could be influenced by potential shifts in the disease landscape due to COVID-19 variants and vaccination efforts, which could influence the
generalizability and interpretation of our findings. CONCLUSIONS We have developed a multimodal deep learning algorithm, MultiCOVID, that discriminates among COVID-19, heart failure,
non-COVID pneumonia and healthy patients using both CXR and blood test with a significantly better performance than experienced thoracic radiologists. Our approach and results suggest an
innovative scenario where COVID-19 prediction could be identified from other similar diseases and facilitate triage within the emergency room in a COVID-19 low prevalence situation. DATA
AVAILABILITY Our code base is provided on GitHub at https://github.com/Tato14/MultiCOVID, including weights for each of the individually trained neural network architectures and respective
model weights for the weighted ensemble model. The datasets used and analyzed during the current study will be available from the corresponding author on reasonable request. In order to
correct samples bias11, additional metadata information present in the DICOM image headers from the CXR would be also available upon request. ABBREVIATIONS * DL: Deep learning * CXR: Chest
X-rays * AUC: Area under the receiver operating characteristic curve * COVID-19: Coronavirus disease 2019 * RT-PCR: Reverse-transcription polymerase chain reaction * SARS-CoV-2: Severe acute
respiratory syndrome coronavirus 2 * HF: Heart failure * NCP: Non-COVID pneumonia REFERENCES * DeGrave, A. J., Janizek, J. D. & Lee, S.-I. AI for radiographic COVID-19 detection selects
shortcuts over signal. _Nat. Mach. Intell._ 3, 610–619 (2021). Article Google Scholar * Cleverley, J., Piper, J. & Jones, M. M. The role of chest radiography in confirming covid-19
pneumonia. _BMJ_ https://doi.org/10.1136/bmj.m2426 (2020). Article PubMed Google Scholar * Avila, E., Kahmann, A., Alho, C. & Dorn, M. Hemogram data as a tool for decision-making in
COVID-19 management: Applications to resource scarcity scenarios. _PeerJ_ 8, e9482 (2020). Article PubMed PubMed Central Google Scholar * Razavian, N. _et al._ A validated, real-time
prediction model for favorable outcomes in hospitalized COVID-19 patients. _npj Digit. Med._ 3, 130 (2020). Article PubMed PubMed Central Google Scholar * Trevethan, R. Sensitivity,
specificity, and predictive values: Foundations, pliabilities, and pitfalls in research and practice. _Front. Public Heal._ 5, 307 (2017). Article Google Scholar * Ronneberger, O.,
Fischer, P. & Brox, T. _U-Net: Convolutional Networks for Biomedical Image Segmentation_ 234–241 (2015). https://doi.org/10.1007/978-3-319-24574-4_28. * He, K., Zhang, X., Ren, S. &
Sun, J. Deep residual learning for image recognition. in _2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR)_ 770–778 (IEEE, 2016). https://doi.org/10.1109/CVPR.2016.90.
* Arik, S. Ö. & Pfister, T. TabNet: Attentive interpretable tabular learning. _Proc. AAAI Conf. Artif. Intell._ 35(8), 6679–6687 (2021). Google Scholar * Wang, W., Tran, D. &
Feiszli, M. What makes training multi-modal classification networks hard?. In _Proceedings / IEEE Computer Society Conference on Computer Vision and Pattern Recognition_ 12692–12702 (2019)
https://doi.org/10.1109/CVPR42600.2020.01271. * Howard, J. & Gugger, S. Fastai: A layered API for deep learning. _Information_ 11, 108 (2020). Article Google Scholar * Garcia Santa
Cruz, B., Bossa, M. N., Sölter, J. & Husch, A. D. Public Covid-19 X-ray datasets and their impact on model bias—A systematic review of a significant problem. _Med. Image Anal._ 74,
102225 (2021). Article PubMed PubMed Central Google Scholar * Lundberg, S. M. _et al._ From local explanations to global understanding with explainable AI for trees. _Nat. Mach. Intell._
2, 56–67 (2020). Article PubMed PubMed Central Google Scholar * Ngiam, J. _et al._ Multimodal deep learning. _ICML_ (2011). * Tan, Y., Zhou, J., Zhou, Q., Hu, L. & Long, Y. Role of
eosinophils in the diagnosis and prognostic evaluation of COVID-19. _J. Med. Virol._ 93, 1105–1110 (2021). Article CAS PubMed Google Scholar * Rahman, A. _et al._ Hematological
abnormalities in COVID-19: A narrative review. _Am. J. Trop. Med. Hyg._ 104, 1188–1201 (2021). Article CAS PubMed PubMed Central Google Scholar * Lippi, G. & Mattiuzzi, C.
Hemoglobin value may be decreased in patients with severe coronavirus disease 2019. _Hematol. Transfus. Cell Ther._ 42, 116–117 (2020). Article PubMed PubMed Central Google Scholar *
Kukar, M. _et al._ COVID-19 diagnosis by routine blood tests using machine learning. _Sci. Rep._ 11, 10738 (2021). Article ADS CAS PubMed PubMed Central Google Scholar * Bayat, V. _et
al._ A Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) prediction model from standard laboratory tests. _Clin. Infect. Dis._ 73, e2901–e2907 (2021). Article CAS PubMed Google
Scholar * Soltan, A. A. S. _et al._ Rapid triage for COVID-19 using routine clinical data for patients attending hospital: Development and prospective validation of an artificial
intelligence screening test. _Lancet Digit. Heal._ 3, e78–e87 (2021). Article CAS Google Scholar * Chen, J. _et al._ Distinguishing between COVID-19 and influenza during the early stages
by measurement of peripheral blood parameters. _J. Med. Virol._ 93, 1029–1037 (2021). Article CAS PubMed Google Scholar * Hwang, E. J. _et al._ Deep learning for chest radiograph
diagnosis in the emergency department. _Radiology_ 293, 573–580 (2019). Article PubMed Google Scholar * Wang, L., Lin, Z. Q. & Wong, A. COVID-Net: A tailored deep convolutional neural
network design for detection of COVID-19 cases from chest X-ray images. _Sci. Rep._ 10, 19549 (2020). Article ADS CAS PubMed PubMed Central Google Scholar * Wehbe, R. M. _et al._
DeepCOVID-XR: An artificial intelligence algorithm to detect COVID-19 on chest radiographs trained and tested on a large U.S. clinical data set. _Radiology_ 299, E167–E176 (2021). Article
PubMed Google Scholar * Zhang, R. _et al._ Diagnosis of coronavirus disease 2019 pneumonia by using chest radiography: Value of artificial intelligence. _Radiology_ 298, E88–E97 (2021).
Article PubMed Google Scholar * Baikpour, M. _et al._ Role of a chest x-ray severity score in a multivariable predictive model for mortality in patients with COVID-19: A single-center,
retrospective study. _J. Clin. Med._ 11, 2157 (2022). Article CAS PubMed PubMed Central Google Scholar * Nishio, M. _et al._ Deep learning model for the automatic classification of
COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy: A multi-center retrospective study. _Sci. Rep._ 12, 8214 (2022). Article ADS CAS PubMed PubMed Central Google Scholar *
Sun, Y. _et al._ Use of machine learning to assess the prognostic utility of radiomic features for in-hospital COVID-19 mortality. _Sci. Rep._ 13, 7318 (2023). Article ADS CAS PubMed
PubMed Central Google Scholar * Nishio, M., Noguchi, S., Matsuo, H. & Murakami, T. Automatic classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest
X-ray image: Combination of data augmentation methods. _Sci. Rep._ 10, 17532 (2020). Article ADS CAS PubMed PubMed Central Google Scholar * Garrafa, E. _et al._ Early prediction of
in-hospital death of COVID-19 patients: A machine-learning model based on age, blood analyses, and chest x-ray score. _Elife_ 10, e70640 (2021). Article CAS PubMed PubMed Central Google
Scholar Download references AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Cancer Research Program, IMIM (Hospital del Mar Medical Research Institute), Barcelona, Spain Max Hardy-Werbin,
Nieves Garcia-Gisbert, Beatriz Bellosillo & Joan Gibert * Radiology Department, Hospital del Mar, Barcelona, Spain José Maria Maiques, Marcos Busto, Flavio Zuccarino, Santiago
Carbullanca, Luis Alexander Del Carpio, Didac Ramal & Ángel Gayete * Emergency Department, Hospital del Mar, Barcelona, Spain Max Hardy-Werbin, Isabel Cirera & Alfons Aguirre *
Innovation and Digital Transformation Department, Hospital del Mar, Barcelona, Spain Jordi Martínez-Roldan * Information Systems Department, Hospital del Mar, Barcelona, Spain Albert
Marquez-Colome * Pathology Department, Hospital del Mar, Barcelona, Spain Beatriz Bellosillo & Joan Gibert Authors * Max Hardy-Werbin View author publications You can also search for
this author inPubMed Google Scholar * José Maria Maiques View author publications You can also search for this author inPubMed Google Scholar * Marcos Busto View author publications You can
also search for this author inPubMed Google Scholar * Isabel Cirera View author publications You can also search for this author inPubMed Google Scholar * Alfons Aguirre View author
publications You can also search for this author inPubMed Google Scholar * Nieves Garcia-Gisbert View author publications You can also search for this author inPubMed Google Scholar * Flavio
Zuccarino View author publications You can also search for this author inPubMed Google Scholar * Santiago Carbullanca View author publications You can also search for this author inPubMed
Google Scholar * Luis Alexander Del Carpio View author publications You can also search for this author inPubMed Google Scholar * Didac Ramal View author publications You can also search for
this author inPubMed Google Scholar * Ángel Gayete View author publications You can also search for this author inPubMed Google Scholar * Jordi Martínez-Roldan View author publications You
can also search for this author inPubMed Google Scholar * Albert Marquez-Colome View author publications You can also search for this author inPubMed Google Scholar * Beatriz Bellosillo View
author publications You can also search for this author inPubMed Google Scholar * Joan Gibert View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS M.H.-W.: Data curation , Validation, Formal analysis, Investigation, Project administration, Supervision, Roles/Writing—original draft, Writing—review & editing; J.M.M.:
Data curation , Formal analysis, Investigation, Validation, Roles/Writing—original draft, Writing—review & editing; M.B.: Data curation, Formal analysis, Investigation Validation,
Roles/Writing—original draft, Writing—review & editing; I.C.: Data curation, Validation, Roles/Writing—original draft, Writing—review & editing; A.A.: Data curation, Validation,
Roles/Writing—original draft, Writing—review & editing; N.G.-G.: Investigation, Visualization, Project administration, Roles/Writing—original draft, Writing—review & editin; F.Z.:
Validation, Roles/Writing—original draft, Writing—review & editing; S.C.: Validation, Roles/Writing—original draft, Writing—review & editing, L.A.D.C.: Validation,
Roles/Writing—original draft, Writing—review & editing, D.R.: Validation, Roles/Writing—original draft, Writing—review & editing; Á.G.: Validation, Roles/Writing—original draft,
Writing—review & editin; J.M.-R.: Project administration, Supervision, Roles/Writing—original draft, Writing—review & editing; A.M.-C.: Data curation, Project administration,
Supervision, Roles/Writing—original draft, Writing—review & editing; B.B.: Data curation, Formal analysis, Investigation, Project administration, Supervision, Roles/Writing—original
draft, Writing—review & editin; J.G.: Data curation, Formal analysis; Investigation, Visualization, Project administration, Supervision, Roles/Writing—original draft, Writing—review
& editing. CORRESPONDING AUTHOR Correspondence to Joan Gibert. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER'S
NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION 1.
SUPPLEMENTARY TABLE 1. RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation,
distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and
indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit
line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use,
you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS
ARTICLE CITE THIS ARTICLE Hardy-Werbin, M., Maiques, J.M., Busto, M. _et al._ MultiCOVID: a multi modal deep learning approach for COVID-19 diagnosis. _Sci Rep_ 13, 18761 (2023).
https://doi.org/10.1038/s41598-023-46126-8 Download citation * Received: 10 February 2023 * Accepted: 27 October 2023 * Published: 31 October 2023 * DOI:
https://doi.org/10.1038/s41598-023-46126-8 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative



