- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Enterohemorrhagic E. coli (EHEC) is a major cause of large outbreaks worldwide associated with hemorrhagic colitis and hemolytic uremic syndrome. While vaccine development is warranted, a
licensed vaccine, specific for human use, against EHEC is not yet available. In this study, the reverse vaccinology approach combined with genomic, transcriptional and molecular epidemiology
data was applied on the EHEC O157:H7 genome to select new potential vaccine candidates. Twenty-four potential protein antigens were identified and one of them (MC001) was successfully
expressed onto Generalized Modules for Membrane Antigens (GMMA) delivery system. GMMA expressing this vaccine candidate was immunogenic, raising a specific antibody response. Immunization
with the MC001 candidate was able to reduce the bacterial load of EHEC O157:H7 strain in feces, colon and caecum tissues after murine infection. MC001 is homologue to lipid A deacylase
enzyme (LpxR), and to our knowledge, this is the first study describing it as a potential vaccine candidate. Gene distribution and sequence variability analysis showed that MC001 is present
and conserved in EHEC and in enteropathogenic E. coli (EPEC) strains. Given the high genetic variability among and within E. coli pathotypes, the identification of such conserved antigen
suggests that its inclusion in a vaccine might represent a solution against major intestinal pathogenic strains.
Enterohemorrhagic Escherichia coli (EHEC) is an anthropozoonotic and etiological agent of diarrheal disease and hemorrhagic colitis. EHEC infections occur mainly in developed countries and
the strains most often implicated in outbreaks are the O157:H7 and the big six non-157 serotypes (O26:H11, O45:H2, O103:H2, O111:H8, O121:H19 and O145:H28)1,2,3. Ruminants are the main
reservoir of EHEC and therefore the infection mainly occurs from fecal contamination of food products4. EHEC strains are characterized by the expression of the Shiga toxin (Stx), the
hallmark of the pathotype. Furthermore, some strains also carry the enterocyte effacement (LEE) locus that encodes the Type III secretion system (T3SS) responsible for the generation of
attachment and effacing (A/E) lesion on the intestinal microvilli1.
The complications arising from EHEC include hemorrhagic colitis, the development of the hemolytic uremic syndrome (HUS) and renal failure5. Although the use of antibiotics remains the gold
standard for the treatment of bacterial diseases, they are not recommended to treat EHEC infections4,6. Antibiotic treatment could lead to cellular damages by increasing the production of
Stx, causing its release into the blood stream and further worsening the disease outcome7.
In general, the increasing burden of these E. coli diarrheal diseases, the emergence of hybrids strains, and the increasing annual cost for the health care systems reflect the need to
develop effective therapeutic and preventive strategies. Among these, vaccination is the most promising strategy to control disease not only for EHEC but also for others pathogenic E. coli
strains2,3,8,9.
So far, several vaccine candidates have been identified by different approaches. Virulence factors expressed as recombinant proteins such as Stx, intimin, E. coli secreted protein A (EspA),
and avirulent ghost cells of EHEC O157:H7 have been tested using different immunization routes and adjuvant combinations in several animal models with encouraging results10. A recent in
silico approach aimed to develop DNA based vaccine identified new EHEC antigens, including among others a putative pilin subunit, T3SS structural protein (escC) gene, and an outer membrane
protein encoded by the bacteriophage Bp933W gene lomW11,12. Additionally, previous studies have identified new promising vaccine candidates demonstrating the potential of exploiting the
reverse vaccinology concept11,12,13,14,15,16. So far, this strategy has been performed on a completely sequence genome of an extraintestinal neonatal meningitis E. coli isolate (NMEC)
leading to the identification of 230 potential antigens. Among these, a conserved zinc metallopeptidase, SslE, was one of the most protective antigens by conferring protection in three
different murine models15,17,18.
In addition to the available technologies, new vaccine development strategies have been recently explored. These innovations ideally serve to make vaccine production simpler, more cost
effective, and improve antigen presentation and immune response19. Outer membrane vesicles are one of these systems employed for vaccine development against Gram-negative bacteria. These
microorganisms release native outer membrane vesicles (NOMV) that are rich in outer membrane lipids, outer membrane and periplasmic proteins, and are subsequently presented to the immune
system in their natural conformation20. NOMV-based vaccines have been largely employed against the organism from which they are recovered21,22,23 or to express and deliver heterologous
antigens24,25,26. However, in native conditions NOMV are recovered in small quantities but E. coli strains can be genetically modified by deletion of the tolR gene to enhance the level of
vesicle production27. This system has been successfully used for expressing properly folded membrane-associated recombinant antigens and to induce functional immune responses24. Recently,
this antigen delivery approach, also known as GMMA (Generalized Modules for Membrane Antigens), has been successfully implemented for vaccine development28,29,30.
The main goal of this work was to identify novel antigens as potential vaccine candidates against infections caused by EHEC, using GMMA as delivery system. Our study led to the
identification of a new potential vaccine candidate present in EHEC O157:H7 strains able to reduce intestinal bacterial colonization in mice.
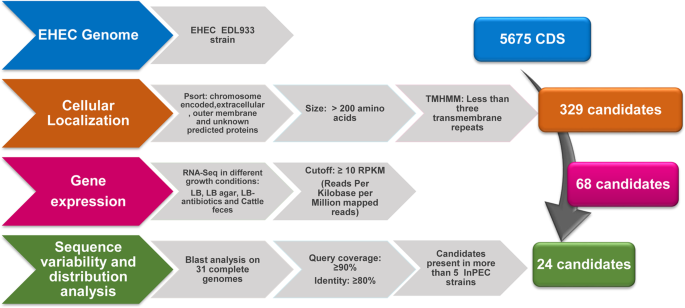
To identify potential antigens in the EHEC O157:H7 EDL933 prototype strain, the reverse vaccinology approach was applied by combining genomic analysis with transcriptional and molecular
epidemiology data as summarized in Fig. 1. The PSORT algorithm was applied to predict the subcellular localization of the 5675 coding sequences (CDS) annotated in the genome. Chromosomal
genes encoding for proteins predicted to be secreted, surface-exposed, outer membrane-associated or with an unknown subcellular localization, were selected. In addition, only genes encoding
for proteins of at least 200 amino acids in length and with fewer than 3 transmembrane domains (as determined by the TMHMM algorithm) were included. This analysis resulted in 329 potential
vaccine candidates (Table S1) and the transcription level of these genes was analyzed using the RNA-Seq database available in the NCBI Sequence Read Archive (SRA). The RNA-Seq dataset was
generated using EHEC EDL933 strain grown in LB, LB with antibiotics, LB-agar media and cattle feces31. Sixty-eight of the 329 identified CDSs showed an absolute index number of ≥10 RPKM
(reads per kilobase per million mapped reads) in at least one of the four growth conditions analyzed (Table S2). The 68 genes were then analyzed for gene distribution and variability in a
panel of 31 complete genomes. Twenty-four genes were present and conserved in more than 5 intestinal pathogenic E. coli strains (query coverage: ≥90%; sequence identity ≥80%) (Fig. 1 and
Fig. S1). The 24 genes encoding for potential vaccine antigens were cloned in E. coli, expressed and purified as recombinant His-tagged fusion proteins. Twelve were successfully purified in
their soluble form, while 12 were insoluble (Table S3). The 24 recombinant proteins were then used to immunize mice to generate antigen-specific polyclonal antibodies. These antibodies were
subsequently tested in Western Blot analyses to assess the expression level of the corresponding potential candidates in the EHEC O157:H7 strain whole cell extract, leading to the
identification of 17 expressed proteins in standard laboratory growth conditions, of which five exhibited high protein expression (Table S3). These five candidates included four
colonization/virulence factors and one putative outer membrane protein. Among these expressed proteins the putative outer membrane candidate (MC001) showed also a higher RNA expression level
in two different growth conditions (Table S3). Considering the higher level of expression and all the aforementioned criteria, we selected MC001 for further characterization.
Reverse vaccinology strategy applied for vaccine candidate selection in EHEC O157:H7. Schematic representation of the in-silico pipeline used for vaccine candidate selection. The 5675 coding
DNA sequences (CDS) encoded on the chromosome of EHEC O157:H7 EDL933 strain were analyzed to identify potential antigens based on their subcellular localization (PSORT). The selection
included proteins greater than 200 amino acids and with fewer than 3 transmembrane repeats determined by the TMHMM algorithm. RNA-Seq data were used to find expressed candidate genes. Gene
variability and distribution analysis of 68 potential antigens on 31 complete E. coli genomes was performed to select those present (query coverage: ≥ 90%) and conserved (sequence identity ≥
80%) in more than 5 different intestinal pathogenic E. coli strains.
When overexpressed in E. coli the MC001 protein was localized into the outer membrane and was purified from the recombinant strain only in its insoluble form. To keep the protein in its
natural conformation, we investigated the possibility to overexpress it in E. coli K12 outer membrane vesicles. First, to identify the E. coli strain in which the candidate could be better
expressed, native vesicles (NOMV) released by the E. coli K12 wild-type strain and by a E. coli K12 strains in which the tolR gene has been deleted (K12 ∆tolR::cat) to induce an overblebbing
phenotype (GMMA)24, were purified and characterized further. The comparison of the total protein content from vesicles derived from the two strains showed that yield of the tolR mutant
strain was 25-fold higher compared to the wild-type strain (Fig. 2C). By transmission electronic microscopy (TEM), the NOMV from the wild-type strain appeared as closed spherical particles
ranging from 20 to 100 nm in diameter (Fig. 2A), while the GMMA from tolR mutant ranged from 20 to 200 nm (Fig. 2B). Based on these data, we selected the GMMA as system for the
overexpression of the MC001 vaccine candidate. MC001 was cloned in frame onto pBAD plasmid and a FLAG-tag inserted after signal peptide sequence (Fig. 3A). The generated construct
(pBAD-MC001F) was transformed into the tolR mutant and the expression and localization of the vaccine candidate into GMMA were evaluated by western blot on the vesicle preparations using the
anti-FLAG antibody for detection. As shown in Fig. 3B, MC001 was successfully expressed and localized in the GMMA.
E. coli K12 engineering to generate Generalized Modules for Membrane Antigens (GMMA). NOMV and GMMA were isolated by ultracentrifugation from supernatants of E. coli K12 WT and K12
∆tolR::cat (A) Negative staining of native NOMV released from a wild-type E. coli K12 observed by transmission electron microscopy (TEM). NOMV from K12 WT strain appeared as closed spherical
particles and homogeneous in shape with a size ranging from 20 to 100 nm. (B) Negative staining of GMMA produced by the K12 tolR::cat (K12tolR::cat) strain analyzed by TEM GMMA from tolR
mutant (size ranging from 20 to 200 nm) (magnification 120,000x). (C) SDS-PAGE (4–12% bis-tris polyacrylamide) of membrane vesicles (NOMV and GMMA) purified from 75 ml of culture
supernatants. Total protein content was quantified and 50 μg of GMMA obtained from K12 ∆tolR::cat sample was loaded into the SDS-PAGE gel. An equivalent volumetric amount of NOMV from K12 WT
obtained from 75 mL of supernatant was loaded. The tolR mutant showed an extensive protein profile in the supernatant compared to wild type.
Antigen delivery into GMMA. (A) Schematic representation of the candidate cloning strategy. The coding sequence of the MC001 potential antigen was cloned into a pBAD vector incorporating its
native signal peptide. A FLAG-tag was inserted after the signal peptide of the construct. (B) Western blot of GMMA preparation expressing the MC001 candidate purified from the K12 tolR::cat
mutant, using an anti-FLAG antibody. Asterisks (*) indicate the expected molecular size of MC001.
To assess the immune response induced by GMMA immunization per se and the specific contribution of MC001 vaccine candidate overexpressed in GMMA, mice serum antibody levels were determined
by ELISA. Serum samples were collected prior to the first immunization (preimmune sera) and two weeks after the third immunization, before challenging mice with the EHEC O157:H7 86–24 stain.
Microtiter plates were coated with purified preparations of GMMA-K12 or with GMMA expressing the MC001 vaccine candidate. Higher total IgG levels were measured in the immunized group versus
the preimmune sera or PBS-alum immunized mice. No-significant differences in total IgG were found among the mice immunized with empty GMMA-K12 in comparison with GMMA expressing MC001 (Fig.
S2), suggesting that the presence of the antigen does not interfere with the immunogenicity of the GMMA. To test whether there was an induction of a specific immune response attributable to
the antigen expressed in GMMA, antibodies against MC001 were measured by the ELISA assay using the recombinant MC001 as coating antigen. A significant increase in antibody response was
found in sera of mice immunized with GMMA-MC001 (P = 0.0076) in comparison to GMMA-K12 (Fig. 4A). Furthermore, the sera raised against GMMA-MC001 were able to specifically detect the MC001
recombinant protein by western blot analysis (Fig. 4B).
Detection of specific antibodies raised against GMMA overexpressing the MC001 vaccine candidate. (A) Specific antibody (IgG) measurement of sera obtained by immunization of GMMA-K12 and GMMA
overexpressing MC001 was assessed by an ELISA assay using microtiter plates coated with the MC001 recombinant protein. The plots represent individual average. Data are expressed as means ±
the Standard Deviation of values from 9 mice for GMMA-MC001 and 10 mice for GMMA-K12 group and asterisks (**) indicate statistically significant differences (P value

:max_bytes(150000):strip_icc():focal(737x446:739x448)/Gwen-Stefani-081323-02-79864966d70d4838b25bec16dd2feeb2.jpg)

:max_bytes(150000):strip_icc():focal(319x0:321x2)/people_social_image-60e0c8af9eb14624a5b55f2c29dbe25b.png)
