- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Download PDF Article Open access Published: 30 April 2019 A novel multidomain acyl-CoA carboxylase in Saccharopolyspora erythraea provides malonyl-CoA for de novo fatty acid biosynthesis
Andrea L. Livieri1, Laura Navone2 nAff3, Esteban Marcellin2, Hugo Gramajo1 & …Eduardo Rodriguez1 Show authors Scientific Reports volume 9, Article number: 6725 (2019) Cite this article
3862 Accesses
Metrics details
Subjects EvolutionMicrobiology AbstractAcetyl-CoA carboxylases (ACCs) are enzyme complexes generally composed of three catalytic domains and distributed in all organisms. In prokaryotes and plastids of most plants, these domains
are encoded in distinct subunits forming heteromeric complexes. Distinctively, cytosolic ACCs from eukaryotes and plastids of graminaceous monocots, are organized in a single multidomain
polypeptide. Until now, no multidomain ACCs had been discovered in bacteria. Here, we show that a putative multidomain ACC in Saccharopolyspora erythraea is encoded by the sace_4237 gene,
representing the first prokaryotic ACC homodimeric multidomain complex described. The SACE_4237 complex has both acetyl-CoA and propionyl-CoA carboxylase activities. Importantly, we
demonstrate that sace_4237 is essential for S. erythraea survival as determined by the construction of a sace_4237 conditional mutant. Altogether, our results show that this prokaryotic
homodimeric multidomain ACC provides malonyl-CoA for de novo fatty acid biosynthesis. Furthermore, the data presented here suggests that evolution of these enzyme complexes, from single
domain subunits to eukaryotic multidomain ACCs, occurred in bacteria through domain fusion.
Similar content being viewed by others Plastid ancestors lacked a complete Entner-Doudoroffpathway, limiting plants to glycolysis and the pentose phosphate pathway Article Open access 06 February 2024 An unusual glycerol-3-phosphate dehydrogenase in Sulfolobus acidocaldarius
elucidates the diversity of glycerol metabolism across Archaea Article Open access 01 April 2025 Animal biosynthesis of complex polyketides in a photosynthetic partnership Article Open
access 08 June 2020 Introduction
Acetyl-CoA carboxylases (ACC)s catalyse the carboxylation of acetyl-CoA to malonyl-CoA (E.C. 6.4.1.2), the first step in fatty acid biosynthesis in prokaryotes and eukaryotes1,2.
Carboxylation of acetyl-CoA to malonyl-CoA is the rate-limiting step in de novo fatty-acid biosynthesis and ACCs have long been used as targets to control metabolic disorders such as
obesity, metabolic syndrome and infectious diseases2,3. Recently, up-regulation of ACC was found in human cancerogenic tumors, suggesting this enzyme as a potential target for the treatment
of cancer4. In archaea, ACCs are part of the 3-hydroxypropionate pathway, involved in autotrophic carbon fixation5. ACCs belong to the biotin-dependent carboxylase protein family. The
two-step reaction mechanism of ACCs involves an ATP-dependent formation of carboxybiotin followed by the transfer of the carboxyl moiety to acetyl-CoA. Both steps are performed by three main
functional components, a biotin carboxyl carrier protein (BCCP), a biotin carboxylase domain (BC) and a carboxyl transferase domain (CT). Domain arrangement varies amongst different
biotin-dependent carboxylases and from one organism to another1. Structural studies have described new non-catalytic domains6,7.
In bacteria, the ACC model derives from the Escherichia coli complex. Cronan et al. (1972) described the role of ACCs in fatty acid biosynthesis by isolating thermosensitive E. coli mutants
capable of growing at restricted temperatures only when the media was supplemented with saturated and unsaturated fatty acids8. Further characterization of the conditional mutants allowed
for the identification of four genes encoding different components of the multi-subunit ACC from E. coli, which later became a “model bacterial ACC”9,10. The E. coli ACC contains three
domains encoded in four proteins: the BC domain, the BCCP component and two independent peptides (α and ß) that form the functional CT domain.
More recently, various studies have described distinct ACC arrangements in actinomycetes, including Streptomyces, Corynebacterium and Mycobacterium spp., which differ from the E. coli
configuration11,12,13 and disproves the universal ACC E. coli model in bacteria. For example, the actinobacteria ACCs are heteromeric multisubunit complexes formed by two main subunits, the
α subunit containing the BC and BCCP domains, and the β subunit containing the CT domain. Distinctly, these enzymes are able to carboxylate acetyl-CoA and propionyl-CoA and are referred to
as acyl-CoA carboxylases. Acyl-CoA carboxylases were first studied in Streptomyces coelicolor through the construction of a conditional mutant of the gene encoding AccB, the CT domain of the
ACC complex. Throughout this study, the essential role of the enzyme for bacterial growth, as well as its role in actinorhodin polyketide production, was demonstrated11. Further studies in
mycolic acid-containing bacteria such as Mycobacterium tuberculosis and Corynebacterium glutamicum allowed identifying the components of others essential ACC complexes13,14,15.
Strikingly, a large group of actinobacteria do not contain orthologues to the essential CT domain found in S. coelicolor, C. glutamicum or M. tuberculosis; neither to the multisubunit ACC
from E. coli. These distinctions make predictions difficult in these microorganisms. For example in Saccharopolyspora erythraea, the producer of erythromycin A and flaviolin, a vast array of
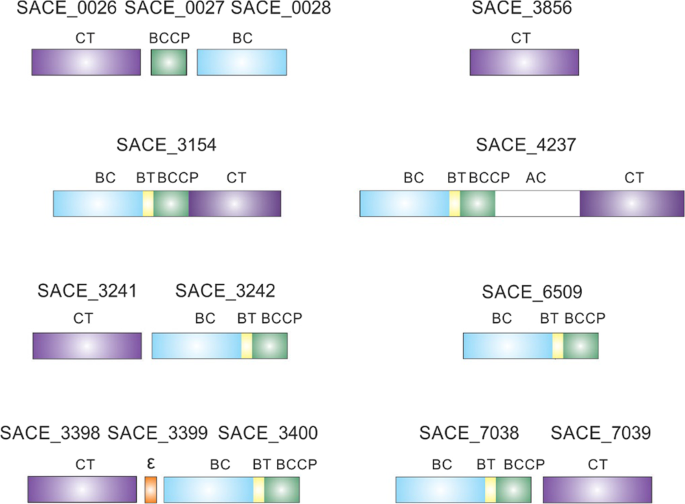
putative biotin-dependent carboxylases are annotated in the genome, including a putative multidomain carboxylase enzyme16,17,18 (Fig. 1).
Figure 1Schematic representation of acyl-CoA carboxylases found in S. erythraea. Indicated domains: BC, biotin carboxylase; BCCP, biotin carboxyl carrier protein; CT, carboxyltransferase; Ɛ, epsilon
subunit; AC, ACC central; BT, domain that mediates BC–CT interactions.
Full size imageHere, we characterised SACE_4237 from S. erythraea, previously annotated as a putative multidomain carboxylase, using biochemical assays and by constructing a conditional knockout strain. We
show that SACE_4237 is an essential multidomain acyl-CoA carboxylase, representing the first of its kind in bacteria. These findings open new avenues to find a possible new ancestor of the
eukaryotic multidomain ACC.
ResultsPhylogenetic analysisSeveral studies have shown that the acyl-CoA substrate specificity for the biotin-dependent carboxylase complexes is determined by the CT domain19. To predict the substrate specificity of
the putative ACCs annotated in the S. erythraea NRRL23338 genome, and to identify the essential ACC in this organism (throughout this paper “essential” or “non-essential” refers to the need,
or not, of the corresponding enzyme activity for cell viability), we performed a phylogenetic analysis of the CT domains16. We also compared them to the CT domains of characterized ACCs
from different organisms11,12,13,14,20,21,22 using the MEGA X software (Fig. 2)23.
Figure 2Phylogenetic analysis of CT domains of acyl-CoA carboxylase complexes. The maximum likelihood phylogenetic tree was constructed using the MEGA X software. The bootstap percentage support
(1000 replicates) are indicated in the different branches. The tree is organized into distinctive groups (I–V) of closely-related CT domains. The lengths of the branches are proportional to
the inferred evolutionary distances. The relative number of substitutions per site is indicated by the bar at the bottom left. The CT domains of S. erythraea putative acyl-CoA carboxylases
are highlighted in red.
Full size imageThe analyses showed that the tree is divided into five groups branching at deep nodes. Furthermore, it showed that the CT domains are grouped with different known carboxylases, suggesting
diverse substrate specificity (Fig. 2). Accordingly, group I contains CT domains that correspond to acyl-CoA carboxylases formed by two functional subunits. The group includes essential ACCs
from Streptomyces11, Corynebacterium and Mycobacterium12,13, non-essential propionyl-CoA carboxylase (PCC) complexes from actinobacteria15 and CT domains of acyl-CoA carboxylases from
archaea5. In this group, SACE_3398 clusters with non-essential CT subunits from PCC complexes of other actinobacteria. In addition, earlier studies of acyl-CoA carboxylases in S. erythraea
have demonstrated that SACE_3398 is part of a non-essential PCC complex17.
Group II comprises enzymes that carboxylate the ɣ-carbon of certain α-β unsaturated acids, instead of the α-carbon of saturated acids (e.g. acetyl-CoA, propionyl-CoA). This group contains
SACE_3241 and SACE_7039 that are closer related to CT domains of geranyl-CoA and methylcrotonyl-CoA carboxylases, respectively21.
The CT domain of SACE_3154 clusters in group III together with other CT domains from long-chain acyl-CoA carboxylases (LCC) that are formed by a unique polypeptide chain and that have been
previously described in mycobacteria and Pseudomonas22. These bacterial LCCs are rare and have been proposed to take role in carbon and nitrogen metabolism.
Group IV includes CT domains from carboxylases belonging to multi-subunit ACCs from Gram-negative bacteria and Firmicutes. The group also comprises CT domains from carboxylase complexes of
actinobacteria that are formed by two or three subunits and CT domains that catalyse the reverse reaction; namely they are malonyl-CoA decarboxylase enzymes involved in malonate
assimilation24. Interestingly, some of these CT subunits are encoded by genes that usually map near or inside polyketide gene clusters. As shown in Fig. 2, SACE_0026 is an ortholog of PrmP3
and it is encoded by the sace_0026 gene belonging to a putative polyunsaturated fatty acid biosynthetic gene cluster, suggesting that it is not essential for bacterial growth16. On the other
hand, SACE_3856 is an ortholog of MatA, a malonyl-CoA decarboxylase enzyme from S. coelicolor and Rhizobium trifolli24.
Finally, the CT domain of SACE_4237 emerged in close affiliation with other CT domains of putative multidomain ACCs. All these orthologues clustered (bootstrap support of 100%) with the CT
of multidomain ACCs from eukaryotes (group V, Fig. 2). This result suggests that the actinobacterial and the eukaryotic CT domains from group V come from a common ancestor. It is worth
mentioning that all actinobacterial CT domains from group V, including SACE_4237, form part of large multidomain proteins of approximately 200 kDa. All have end-to-end homology with the
260–280 kDa eukaryotic ACCs, including the conserved central region. Considering that none of the putative CT domains of S. eryhtraea analysed above show evidence of being an essential ACC,
led us to hypothesize that SACE_4237 could be the enzyme that provides malonyl-CoA for fatty acid biosynthesis.
In vivo and in vitro characterization of SACE_4237In order to determine the role of SACE_4237 as an ACC, we complemented the E. coli L8 strain with SACE_42378. The L8 strain is a temperature-sensitive (ts) mutant in the accB gene, which
causes the deficient biotinylation of the BCCP subunit, resulting in cell death above 37 °C. The sace_4237 gene was PCR amplified and cloned into the pSK-bluescript vector under the control
of the pLac promoter, to achieve the plasmid pSK-SACE_4237. This plasmid, as well as the empty vector, were transformed into the E. coli L8 strain, respectively. Transformants were tested
for growth at different temperatures. As illustrated in Fig. 3, SACE_4237 complemented the growth of L8 mutant strain at 37 °C and 42 °C when IPTG was added. No growth was observed when the
mutant strain was transformed with the empty vector, or when the complemented strain was grown in the absence of IPTG (data not shown). The results show that SACE_4237 can complement the
deficiency in ACC activity in the E. coli L8 strain and suggest that the multidomain SACE_4237 could be the essential ACC in S. erythraea.
Figure 3Complementation test for E. coli L8 strain. (A) L8 strain transformed with pSK bluescript empty-vector. (B) L8 strain transformed with pSK-SACE4237 plasmid. LB-agar plates supplemented with
0.5 mM of IPTG grown for 48 h at 30, 37 or 42 °C.
Full size imageTo biochemical characterize the protein SACE_4237, the gene was cloned under the strong pBAD promoter, to yield pBAD-SACE_4237. Optimal expression conditions were obtained using E. coli
protease deficient BL21 strain containing the multicopy plasmid pCY21625 which provides a biotin ligase protein to improve SACE_4237 biotinylation. The biochemical studies were carried out
by measuring the carboxylation of acetyl-CoA with the purified protein. The two elution peaks from the size exclusion chromatography, corresponding to pure SACE_4237, were assayed for ACC
activity (Fig. 4). Carboxylation of acetyl-CoA was only detected with the first elution peak corresponding to a homodimeric complex of approximately 400 kDa, while the monomeric form of the
protein (second elution peak) was inactive (Fig. 4; Table 1). Furthermore, carboxylation of propionyl-CoA and palmitoyl-CoA, by SACE_4237 were also tested. SACE_4237 was able to carboxylate
propionyl-CoA, while no activity was detected using palmitoyl-CoA as a substrate. The kinetic parameters for the enzyme indicate that SACE_4237 has a slightly higher catalytic efficiency for
acetyl-CoA compared to propionyl-CoA (Table 1). Nevertheless, the Km for propionyl-CoA was lower than the Km for acetyl-CoA. The assays confirmed that SACE_4237 behaves as an acyl-CoA
carboxylase.
Figure 4Determination of the oligomeric state of SACE_4237. The purified protein was analyzed by size exclusion chromatography (Superdex 200 10/300 GL, GE). The protein profiles were followed by
measuring absorbance at 280 nm (in milli-absorbance units [mAU]). The inset shows the calibration curve obtained with molecular mass standards; MW, molecular weight; Ve, elution volume; Vo,
void volume. The calibration curve was used to calculate the molecular weight of the elution peaks 1 and 2, equivalent to 404 and 186.9 kDa, respectively. These peaks correspond to the
dimeric and monomeric form of SACE_4237, respectively.
Full size imageTable 1 Kinetic parameters.Full size tableThe physiological role of SACE_4237In order to determine the physiological role of SACE_4237 in S. erythraea, we constructed a conditional mutant by exchanging the sace_4237 native promoter for the thiostrepton (Th) inducible
ptipA promoter11. Homologous recombination was confirmed by PCR and the resulting strain named hereafter AL1. AL1 showed growth dependence to Th in solid and liquid media indicating that
the expression of sace_4237 was essential for cell viability (Fig. 5A). The result suggests that SACE_4237 is the main ACC responsible for making malonyl-CoA for de novo fatty acid
biosynthesis in S. erythraea. To confirm this result, the AL1 strain was conjugated with a plasmid carrying the entire sace_4237 gene under the permE* promoter, for the constitutive
expression of the essential ACC. All kanamycin resistant transconjugants were able to grow in the absence of Th (see Supplementary Fig. S2), implying that the constitutive expression of
SACE_4237 was complementing the deficiency of the ACC activity of the conditional AL1 mutant.
Figure 5Analysis of S. erythraea NRRL23338 and AL1 conditional mutant strains on different growth conditions. (A) Growth curves on R5 medium of S. erythraea NRRL23338 supplemented or not with oleic
acid (OA) and S. erythraea AL1 induced or not with Th or supplemented with oleic acid. (B) SDS-PAGE of total protein extracts. (C) Western-blot of biotinylated proteins. MWM, standard
molecular weight markers. (D) Erythromicin A production under different growth conditions. SACE, S. erythraea NRRL23338; SACE OA, S. erythraea NRRL23338 supplemented with oleic acid; AL1 Th,
S. erythraea AL1 supplemented with Th; AL1 OA, S. erythraea AL1 supplemented with oleic acid. Data are represented as mean values ± standard deviation (n = 3). *Statistically significant
differences (p ≤ 0.05) between S. erythraea NRRL23338 and S. erythraea AL1 strains. Analysis was performed using one-way ANOVA.
Full size imageTo demonstrate that SACE_4237 is the main ACC responsible for making malonyl-CoA for de novo fatty acid biosynthesis, the AL1 conditional mutant was grown without Th and supplemented with
oleic acid to bypass the absence of fatty acid biosynthesis. The results presented in Fig. 5A show that the exogenous addition of oleic acid enables growth of AL1 in absence of the SACE_4237
expression. Western blot analysis of biotinylated proteins from cell-free extracts of S. erythraea NRRL23338 and the AL1 mutant strains grown +/− Th, and supplemented with or without oleic
acid, confirmed that SACE_4237 was not expressed in the absence of the inducer Th (Fig. 5B y 5C).
To prove that the absence of SACE_4237 expression in the mutant strain had a reduced ACC activity in vivo leading to lower levels of malonyl-CoA, we measured the incorporation of 14C-acetate
into the lipids in S. erythraea and the AL1 strains growing +/− Th and +/− oleic acid. As shown in Fig. 6 no incorporation of 14C-acetate into de novo synthesized fatty acids was observed
in strain AL1 confirming the absence of ACC activity. Altogether these results demonstrate that SACE_4237 is the essential ACC in S. erythraea responsible for the production of malonyl-CoA,
the elongation unit used for de novo fatty acids biosynthesis.
Figure 6Thin-layer chromatography (TLC) of unlabelled and 14C-labelled lipids from S. erythraea NRRL23338 and AL1 conditional mutant strains. Cultures were grown by duplicate in R5 medium to
exponential phase (OD600nm = 2) and supplemented with 14C-acetate and grown for one additional hour when indicated (lanes 2, 4, 6 and 8). Total lipids extracted from 3 mg of lyophilized cell
material with organic solvent and separated on silica gel TLC were developed with hexane-diethylether-acetic acid (50:50:1, v/v/v). (A) De novo synthesized lipids visualized using a Typhoon
FLA 7000 Phosphorimager. Image was overexposed for signal identification. (B) Total lipids detected by chemical staining with Cu-phosphoric stain, lipid bands were identified by their
co-migration with standards. SACE, S. erythraea NRRL23338 strain; SACE OA, S. erythraea NRRL23338 strain supplemented with oleic acid; AL1 Th, S. erythraea AL1 strain supplemented with Th;
AL1 OA, S. erythraea AL1 strain supplemented with oleic acid.
Full size imageLastly, since SACE_4237 also has PCC activity, we studied the influence of SACE_4237 on erythromycin production under different growth conditions. The parental and AL1 strains were grown for
four days in R5 medium in the presence or absence of Th or +/− oleic acid. As shown in Fig. 5D, the parental strain produced more erythromycin when grown in R5 supplemented with oleic acid.
On the contrary, the AL1 mutant strain produced more erythromycin in R5 with Th compared to the parental strain; however, reduced antibiotic production was observed in R5 medium
supplemented with oleic acid. Our results suggest that SACE_4237 presents PCC activity in vivo and provides methylmalonyl-CoA for polyketide production.
DiscussionACCs are enzymatic complexes distributed in all domains of life. In bacteria and eukaryotes, ACCs play an essential role in lipid metabolism1. Over the years, a vast variety of domain
arrangements of ACCs complexes have been established. Up to now, essential ACCs in bacteria were described as heteromeric complexes formed by different subunits. In all bacteria, except for
Thermotogales, Spirochaetes and Actinobacteria, ACCs are composed of four subunits26. In actinomycetes, such as Streptomyces or the mycolic-acid containing bacteria, ACCs are formed by two
large subunits, containing all catalytic domains, together with a small non-catalytic Ɛ subunit11,13. In other actinobacteria, like the erythromycin producer S. erythraea, no orthologue of
an essential ACC has been previously characterised. In this work, we identified SACE_4237, an ortholog of the multidomain eukaryotic ACC, and showed that its homodimeric complex behaves as
an essential ACC in S. erythraea.
S. erythraea also has a smaller putative multidomain acyl-CoA carboxylase, SACE_3154. This protein is an orthologue of other carboxylases already described in different groups of bacteria
like Pseudomonas and Mycobacterium22. Remarkably, biochemical and structural differences were found between SACE_4237 and the previously described single-chain multidomain carboxylases from
Pseudomonas and Mycobacterium22. First, the Pseudomonas like multidomain acyl-CoA carboxylase is considered as a long-chain acyl-CoA carboxylase (LCC) showing activity against acyl-CoAs with
chain lengths from C2 to C16. The enzyme shows comparable kcat values for all substrates, but the Km for palmitoyl-CoA is 350-fold lower than that for acetyl-CoA22. Instead, SACE_4237 only
shows activity towards short-chain acyl-CoAs (acetyl-CoA and propionyl-CoA) with similar catalytic efficiency. The LCCs are homo hexameric complexes and contain a BC-BCCP domain in the
carboxy-terminal region and a CT domain in the amino-terminal region (Fig. 1). On the other hand, active SACE_4237 enzyme forms a homodimer complex, that, in addition to the catalytic
domains, also contains a central region only found in eukaryotic ACCs6,27. Furthermore, sequence analysis of SACE_4237 central region revealed secondary structure elements that match
components present in the crystallographic structure of the S. cerevisiae ACC28 (see Supplementary Fig. S4). In addition, the phylogenetic analyses of the CT and BC domains (Figs 2 and S3)
of SACE_4237 and SACE_3154 show a divergent evolution of these complexes; suggesting that these two enzymes come from two independent fusion events of their domains.
Two isoforms of eukaryotic ACCs have been identified in mammalian cells, an essential cytoplasmic form that provides malonyl-CoA for fatty acid biosynthesis, and a mitochondrial form
involved in the regulation of fatty acid oxidation29. Both mammalian cytoplasmic and mitochondrial carboxylases form homodimer active complexes that are regulated transcriptionally and
post-translationally by different factors29. For example, the oligomeric state and kinetic activation of the enzyme activity are modulated by citrate and inactivated by phosphorylation29,30.
SACE_4237 is also active in a dimeric state; however, our in vitro experiments demonstrated that citrate is not required neither for oligomerization nor enzymatic activity. Proteomic
analyses carried out by Licona-Casani et al.31 showed that SACE_4237 is phosphorylated during the metabolic switch, a brief period of growth arrest preceding biosynthesis of secondary
metabolites. Thus, we hypothesise that SACE_4237 activity could be modulated by phosphorylation. In addition, the enzyme complex also showed a relaxed substrate specificity, being able to
carboxylate acetyl-CoA and propionyl-CoA with similar catalytic efficiency. This characteristic is shared by other heteromeric acyl-CoA carboxylases from actinomycetes11. In S. erythraea,
the PCC activity of SACE_4237 could be involved in providing methylmalonyl-CoA for erythromycin biosynthesis. In this regard, the sace_4237 conditional mutant strain AL1 grown in the
presence of oleic acid showed reduced erythromycin production compared to the parental strain (Fig. 5), suggesting that SACE_4237 also supplies methylmalonyl-CoA for erythromycin
biosynthesis under the condition tested. In previous studies, a sace_3398 deficient mutant showed reduced PCC activity, but the mutation did not have detrimental effects on erythromycin
production17. The overlapping activities of two PCC complexes make it difficult to predict their physiological effect on secondary metabolisms. Therefore, each of them should be carefully
evaluated to confirm their effect on erythromycin production.
Orthologues of SACE_4237 are present in other actinobacteria, including important secondary metabolites producers, like Micromonosporaceae, Nocardioidaceae, Pseudonocardiaceae,
Nocardiopsaceae, Streptosporangiaceae, Thermomonosporaceae, Cryptosporangiaceae, Kineosporiaceae. Thus, our results highlight the importance of characterizing this new family of homodimeric
multidomain acyl-CoA carboxylases for biotechnological purposes.
Evolution of multifunctional polypeptides has been proposed to occur by duplication, fusion and/or recombination of small monofunctional units32. Regarding eukaryotic ACCs, Lombard and
Moreira26 proposed that these complexes evolved by the fusion of two unit polypeptides bearing the BC-BCCP domains and the CT domain. Phylogenetic studies conducted in this work provide new
insights into the evolutionary origins of homodimeric eukaryotic ACCs. Our studies indicate that the CT domains of homodimeric acyl-CoA carboxylases from actinobacteria and eukaryotes shared
a common ancestor not previously reported (Fig. 2). In addition, the phylogenetic analysis of the BC domains of these enzyme complexes indicates the same distribution observed for the
corresponding CT domains (see Supplementary Fig. S4). The presence of a homologous central domain in all multidomain ACCs complexes, their active homodimeric form, and the phylogenetic
inferences here described, lead us to propose a new evolutionary theory where the occurrence of a gene fusion event resulted in an “eukaryotic-type” ACC among actinobacteria or in an
immediate actinobacterial ancestor. Two alternative scenarios can be foreseen, an actinobacterial organism bearing such fusion evolved into the precursor of all eukaryotes33 or, the genes
encoding for the multidomain ACC could have been laterally transferred from this prokaryotic organism to the last common ancestor of the eukaryotes. Regardless of the mechanism that leads to
the ACCs of present-day eukaryotes, we propose that the term “actinobacterial homodimeric ACCs” should replace the current “eukaryotic-like ACCs” designation.
MethodsBacterial Strains,Culture, and Transformation Conditions
S. erythraea strains (see Supplementary Table S1) were grown either on solid or liquid R5 medium (without sucrose) at 30 °C and supplemented when needed with the followings antibiotics: 150
µg ml−1 apramycin (Am), 10 µg ml−1 thiostrepton (Th) and/or 50 µg ml−1 kanamycin (Km)34. For conjugation experiments, E. coli strain ET12567 harbouring pUZ800235 and a movilisable plasmid
were mixed with S. erythraea spores and plated on ISP4 medium. After incubation at 30 °C for 16 hours, the plates were then overlaid with Th (300 µg per plate), Am (4.5 mg per plate) or Km
(1.5 mg per plate). Fatty acid supplementation studies were performed on R5 liquid medium supplemented with oleic acid 0.01% (v/v), Brij 58 0.075% (v/v) and appropriated antibiotics when
needed. All E. coli strains (see Supplementary Table S1) were grown in LB media and transformed following standard protocols. Transformants were selected on media supplemented with the
appropriate antibiotics at the following concentrations: 100 µg ml−1 ampicillin (Ap); 50 µg ml−1 Am; 25 µg ml−1 chloramphenicol (Cm); and 50 µg ml−1Km.
Gene cloning and plasmidconstruction
The synthetic oligonucleotides SACE4237Fw (5′TTCGCTCTAGAAGGAGATATACACATATGTTCAGTCGTGTTGCCATCGT3′) and SACE4237Rev (5′ TTACTAGTCACTGCGGGAGCATGCCCTTCTC 3′) were used to amplify sace_4237 gene.
The reaction mixture contained Q5 Pol reaction buffer, 0.02 U µl−1 Q5 DNA polymerase (New England Biolabs), 20 pmol of each primer, and 50 ng of S. erythraea chromosomal DNA in a final
volume of 25 µl. Samples were subjected to 30 cycles of denaturation (95 °C, 30 s), annealing (65 °C, 30 s), and extension (72 °C, 5 min). The utilization of SACE4237FW and SACE4237Rev
oligonucleotides allowed the introduction of an XbaI restriction site, a rbs site upstream of the start codon, an NdeI site at the translational start codon and a SpeI site downstream of the
stop codon of the sace_4237 gene. pSK-SACE4237: the PCR fragment of sace_4237 gene amplified with SACE_4237Fw and SACE_4237Rev oligonucleotides was digested with XbaI-SpeI and then cloned
in XbaI-SpeI cleaved pBluescript SK. Correct sequence and orientation of the cloned fragment was checked by DNA sequencing. pBAD-SACE4237: this plasmid was constructed by cloning the
NdeI-SpeI fragment from pSK-SACE4237 in a NdeI-SpeI-claeved pET28-BAD. pTL1 plasmid was constructed by cloning the NdeI-SphI fragment of sace_4237 from pSK-SACE4237 in a NdeI-SphI digested
pIJ860036, obtaining the first 4100pb of sace_4237 under the ptipA promoter and disrupting the integrase gene and the att site of pIJ8600. The plasmid pTL1 was transferred into S. erythraea
by conjugation37. Integration of pLT1 by homologous recombination through the sace_4237 5′ end, resulted in sace_4237 under the control of an inducible promoter ptipA (see Supplementary Fig.
S1). pERM-SACE4237: NdeI-SpeI fragment from pSK-SACE4237 was cloned into NdeI-SpeI digested pTR285 plasmid38. This plasmid contained an oriT and the BT integrase system which allowed its
conjugation and integration into the chromosome of AL1. pERM-SACE4237 was transferred into S. erythraea by conjugation37.
Bioinformatic AnalysisFor construction of the phylogenetic tree, several CT domains of different biotin-dependent carboxylases (many already characterized) were aligned by Clustal W and refined by hand. The
primary sequence of bacterial CT domain formed by two peptides (i.e. E. coli) were fused in silico as a single sequence, as found in all the biotin-dependent carboxylases of actinobacteria.
The phylogenetic tree was constructed by maximum likelihood using MEGA X software with 1000 bootstrap replicates23.
Protein MethodsCell-free extracts and purified proteins were analysed by SDS-PAGE using a Bio-Rad minigel apparatus. Coomassie Brilliant Blue was used to stain protein bands. For detection of biotinylated
proteins, proteins were electro-blotted after electrophoretic separation onto a nitrocellulose membrane (Bio-Rad) and probed with alkaline phosphatase-streptavidin conjugate (AP-streptavidin
diluted 1:10.000) (Roche) following the procedure provided for the supplier. Protein contents were determined by Bradford protein assay using bovine serum albumin as standard.
ProteinPurification
SACE_4237 was purified from cultures of E. coli BL21λ(DE3) carrying pBAD-SACE4237 and pCY216 plasmids and induced at 15 °C overnight after addition of 0.5% arabinose to cultures grown to
OD600 0.6. Cells were pelleted and resuspended in 50 mM Hepes pH 7.5, 150 mM NaCl, 20% glycerol and 0.1 mM PMSF, and disrupted by sonication. The lysates were clarified by centrifugation at
15,000 g at 4 °C for 30 min. Purification of SACE_4237 was performed by two column steps. First, a nickel-sepharose column was used since the protein was expressed as a His6 tag fusion at
the N-terminal end. Then, a molecular exclusion column Superdex-200 (GE) using AKTA basic (GE) allowed getting monomer and dimers of SACE_4237. The column was equilibrated in 50 mM Hepes pH
7.5, 150 mM NaCl, 20% glycerol and eluted with the same buffer. The molecular exclusion column was calibrated with the Gel Filtration Markers Kit for Protein Molecular Weights 29000–700000
Da (Sigma-Aldrich).
Enzymatic AssaysACC and PCC in vitro activity were measured following the incorporation of radioactive HCO3− into acid non-volatile material17. The reaction mixture contained 100 mM Hepes pH 7.5, 3 mM ATP,
5 mM MgCl2, 50 mM NaH14CO3 [specific activity 200 µCimmol−1 (740 kBq mmol−1)], 7.8 µM to 0.5 mM acetyl-CoA or propionyl-CoA, and 10 µg of pure SACE_4237 in a total reaction volume of 100 µl.
The reaction was initiated by the addition of NaH14CO3, allowed to proceed at 30 °C for 15 min, and stopped with 200 µl of 6 M HCl. The contents of the tubes were then evaporated to dryness
at 95 °C. The residue was resuspended in 100 µl of water, 1 ml of Optiphase liquid scintillation medium (Wallac Oy) was added, and 14C radioactivity was determined in a Beckman
scintillation liquid counter. Nonspecific CO2 fixation by crude extracts or pure protein was assayed in the absence of substrate. One unit of enzyme activity catalysed the incorporation of 1
µmol of 14C into acid-stable products/min.
Lipid AnalysisFor de novo TAG and fatty acids biosynthesis, S. erythraea NRRL23338 and AL1 strains were grown in R5 medium until exponential phase (OD600nm 1) and labelled for 1 h with 5 µCi [14C]-acetate
(50.5 mCi/mmol; Perkin-Elmer)38. Total lipids from S. erythraea NRRL23338 and AL1 strains were extracted twice from 3 mg of lyophilized cell material with chloroform:methanol (2:1, v/v).
The combined extracts were evaporated and analysed by thin-layer chromatography (TLC) on Silica Gel 60 F254 plate (Merck), using the solvent hexane-diethylether-acetic acid (50:50:1, v/v/v)
for TAG and fatty acids analysis. The radioactivity incorporated into each lipid fraction was quantified using Typhoon FLA 7000 Phosphorimager (GE). Lipid fractions were visualised by
Cu-phosphoric staining. Oleic acid was used as the fatty acid reference substance. Metabolite identity was based on the mobility of known standards.
Erythromycin A QuantificationBioassays were used to quantify the production of erythromycin A in steady-state cultures of S. erythraea. For this, 3 ml of Micrococcus luteus grown to OD600 0.6 were added per 125 ml of
Mueller-Hinton agar. 25 μl of supernatant of S. erythraea culture broth were added to 5 mm diameter holes at Mueller-Hinton agar plates and grown at 37 °C overnight. To calculate
erythromycin production the halos made by non-growing bacteria were measured and compared with the halos produced by a standard curve of erythromycin A39.
References Tong, L. Structure and function of biotin-dependent carboxylases. Cellular and Molecular Life Sciences 70, 863–891, https://doi.org/10.1007/s00018-012-1096-0 (2013).
Article CAS PubMed Google Scholar
Gago, G., Diacovich, L., Arabolaza, A., Tsai, S. C. & Gramajo, H. Fatty acid biosynthesis in actinomycetes. FEMS Microbiology Reviews 35, 475–497,
https://doi.org/10.1111/j.1574-6976.2010.00259.x (2011).
Article CAS PubMed PubMed Central Google Scholar
Tong, L. Acetyl-coenzyme A carboxylase: crucial metabolic enzyme and attractive target for drug discovery. Cellular and Molecular Life Sciences 62, 1784–1803,
https://doi.org/10.1007/s00018-005-5121-4 (2005).
Article CAS PubMed Google Scholar
Wang, C., Rajput, S., Watabe, K., Liao, D. F. & Cao, D. Acetyl-CoA carboxylase-a as a novel target for cancer therapy. Front Biosci (Schol Ed) 2, 515–526 (2010).
Google Scholar
Berg, I. A. et al. Autotrophic carbon fixation in archaea. Nature Reviews. Microbiology 8, 447–460, https://doi.org/10.1038/nrmicro2365 (2010).
Article CAS PubMed Google Scholar
Huang, C. S. et al. Crystal structure of the alpha(6)beta(6) holoenzyme of propionyl-coenzyme A carboxylase. Nature 466, 1001–1005, https://doi.org/10.1038/nature09302 (2010).
Article ADS CAS PubMed PubMed Central Google Scholar
Tong, L. Striking Diversity in Holoenzyme Architecture and Extensive Conformational Variability in Biotin-Dependent Carboxylases. Advances in Protein Chemistry and Structural Biology 109,
161–194, https://doi.org/10.1016/bs.apcsb.2017.04.006 (2017).
Article PubMed Google Scholar
Harder, M. E. et al. Temperature-sensitive mutants of Escherichia coli requiring saturated and unsaturated fatty acids for growth: isolation and properties. Proc Natl Acad Sci USA 69,
3105–3109 (1972).
Article ADS CAS PubMed Google Scholar
Silbert, D. F., Pohlman, T. & Chapman, A. Partial characterization of a temperature-sensitive mutation affecting acetyl coenzyme A carboxylase in Escherichia coli K-12. J Bacteriol 126,
1351–1354 (1976).
CAS PubMed PubMed Central Google Scholar
Li, S. J. & Cronan, J. E. Jr. The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. J Biol Chem 267, 855–863 (1992).
CAS PubMed Google Scholar
Rodriguez, E., Banchio, C., Diacovich, L., Bibb, M. J. & Gramajo, H. Role of an essential acyl coenzyme A carboxylase in the primary and secondary metabolism of Streptomyces coelicolor
A3(2). Appl Environ Microbiol 67, 4166–4176 (2001).
Article CAS PubMed PubMed Central Google Scholar
Kurth, D. G. et al. ACCase 6 is the essential acetyl-CoA carboxylase involved in fatty acid and mycolic acid biosynthesis in mycobacteria. Microbiology 155, 2664–2675,
https://doi.org/10.1099/mic.0.027714-0 (2009).
Article CAS PubMed PubMed Central Google Scholar
Gago, G., Kurth, D., Diacovich, L., Tsai, S. C. & Gramajo, H. Biochemical and structural characterization of an essential acyl coenzyme A carboxylase from Mycobacterium tuberculosis. J
Bacteriol 188, 477–486, https://doi.org/10.1128/JB.188.2.477-486.2006 (2006).
Article CAS PubMed PubMed Central Google Scholar
Oh, T. J., Daniel, J., Kim, H. J., Sirakova, T. D. & Kolattukudy, P. E. Identification and characterization of Rv3281 as a novel subunit of a biotin-dependent acyl-CoA Carboxylase in
Mycobacterium tuberculosis H37Rv. J Biol Chem 281, 3899–3908, https://doi.org/10.1074/jbc.M511761200 (2006).
Article CAS PubMed Google Scholar
Bazet Lyonnet, B. et al. Pleiotropic effect of AccD5 and AccE5 depletion in acyl-coenzyme A carboxylase activity and in lipid biosynthesis in mycobacteria. PloS One 9, e99853,
https://doi.org/10.1371/journal.pone.0099853 (2014).
Article ADS PubMed PubMed Central Google Scholar
Oliynyk, M. et al. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol 25, 447–453, https://doi.org/10.1038/nbt1297 (2007).
Article CAS PubMed Google Scholar
Donadio, S., Staver, M. J. & Katz, L. Erythromycin production in Saccharopolyspora erythraea does not require a functional propionyl-CoA carboxylase. Molecular Microbiology 19, 977–984
(1996).
Article CAS PubMed Google Scholar
Hunaiti, A. R. & Kolattukudy, P. E. Isolation and characterization of an acyl-coenzyme A carboxylase from an erythromycin-producing Streptomyces erythreus. Arch Biochem Biophys 216, 362–371
(1982).
Article CAS PubMed Google Scholar
Diacovich, L. et al. Crystal structure of the beta-subunit of acyl-CoA carboxylase: structure-based engineering of substrate specificity. Biochemistry 43, 14027–14036,
https://doi.org/10.1021/bi049065v (2004).
Article CAS PubMed Google Scholar
Li, S. J. & Cronan, J. E. Jr. The genes encoding the two carboxyltransferase subunits of Escherichia coli acetyl-CoA carboxylase. J Biol Chem 267, 16841–16847 (1992).
CAS PubMed Google Scholar
Tomassetti, M. et al. 3-methylcrotonyl Coenzyme A (CoA) carboxylase complex is involved in the Xanthomonas citri subsp. citri lifestyle during citrus infection. PloS One 13, e0198414,
https://doi.org/10.1371/journal.pone.0198414 (2018).
Article CAS PubMed PubMed Central Google Scholar
Tran, T. H. et al. Structure and function of a single-chain, multi-domain long-chain acyl-CoA carboxylase. Nature 518, 120–124, https://doi.org/10.1038/nature13912 (2015).
Article ADS CAS PubMed Google Scholar
Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Molecular Biology and Evolution 35, 1547–1549,
https://doi.org/10.1093/molbev/msy096 (2018).
Article CAS PubMed Google Scholar
An, J. H. & Kim, Y. S. A gene cluster encoding malonyl-CoA decarboxylase (MatA), malonyl-CoA synthetase (MatB) and a putative dicarboxylate carrier protein (MatC) in Rhizobium
trifolii-cloning, sequencing, and expression of the enzymes in Escherichia coli. FEBS J. 257, 395–402 (1998).
CAS Google Scholar
Chapman-Smith, A., Turner, D. L., Cronan, J. E. Jr., Morris, T. W. & Wallace, J. C. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier
protein of Escherichia coli acetyl-CoA carboxylase. Biochemical J. 302(Pt 3), 881–887 (1994).
Article CAS Google Scholar
Lombard, J. & Moreira, D. Early evolution of the biotin-dependent carboxylase family. BMC Evolutionary Biology 11, 232, https://doi.org/10.1186/1471-2148-11-232 (2011).
Article CAS PubMed PubMed Central Google Scholar
Hunkeler, M., Stuttfeld, E., Hagmann, A., Imseng, S. & Maier, T. The dynamic organization of fungal acetyl-CoA carboxylase. Nature. Communications 7, 11196,
https://doi.org/10.1038/ncomms11196 (2016).
Article CAS Google Scholar
Wei, J. & Tong, L. Crystal structure of the 500-kDa yeast acetyl-CoA carboxylase holoenzyme dimer. Nature 526, 723–727, https://doi.org/10.1038/nature15375 (2015).
Article ADS CAS PubMed PubMed Central Google Scholar
Brownsey, R. W., Boone, A. N., Elliott, J. E., Kulpa, J. E. & Lee, W. M. Regulation of acetyl-CoA carboxylase. Biochemical Society Transactions 34, 223–227,
https://doi.org/10.1042/BST20060223 (2006).
Article CAS PubMed Google Scholar
Beaty, N. B. & Lane, M. D. Kinetics of citrate-induced activation and polymerization of chick liver acetyl-CoA carboxylase. Annals of the New York Academy of Sciences 447, 23–37 (1985).
Article ADS CAS PubMed Google Scholar
Licona-Cassani, C., Lim, S., Marcellin, E. & Nielsen, L. K. Temporal dynamics of the Saccharopolyspora erythraea phosphoproteome. Molecular & Cellular Proteomics: MCP 13, 1219–1230,
https://doi.org/10.1074/mcp.M113.033951 (2014).
Article CAS Google Scholar
Jitrapakdee, S. & Wallace, J. C. The biotin enzyme family: conserved structural motifs and domain rearrangements. Current protein & Peptide Science 4, 217–229 (2003).
Article CAS Google Scholar
Cavalier-Smith, T. The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life. Cold Spring Harbor
Perspectives in. Biology 6, a016006, https://doi.org/10.1101/cshperspect.a016006 (2014).
Article CAS Google Scholar
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics. 2000 edn, (2000).
Bierman, M. et al. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116, 43–49 (1992).
Article CAS PubMed Google Scholar
Sun, J., Kelemen, G. H., Fernandez-Abalos, J. M. & Bibb, M. J. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology
145(Pt 9), 2221–2227, https://doi.org/10.1099/00221287-145-9-2221 (1999).
Article CAS PubMed Google Scholar
Labeda, D. P. Transfer of the Type Strain of Streptomyces erythraeus (Waksman 1923) Waksman and Henrici 1948 to the Genus Saccharopolyspora Lacey and Goodfellow 1975 as Saccharopolyspora
erythraea sp. nov., and Designation of a Neotype Strain for Streptomyces erythraeus. International Journal of Systematic and Evolutionary Microbiology 37, 19–22,
https://doi.org/10.1099/00207713-37-1-19 (1987).
Article Google Scholar
Arabolaza, A., Rodriguez, E., Altabe, S., Alvarez, H. & Gramajo, H. Multiple pathways for triacylglycerol biosynthesis in Streptomyces coelicolor. Appl Environ Microbiol 74, 2573–2582,
https://doi.org/10.1128/AEM.02638-07 (2008).
Article CAS PubMed PubMed Central Google Scholar
Peiru, S., Rodriguez, E., Menzella, H., Carney, J. & Gramajo, H. Metabolically engineered Escherichia coli for efficient production of glycosylated natural products. Microbial. Biotechnology
1, 476–486 (2008).
CAS Google Scholar
Download references
AcknowledgementsThis study was supported by PICT2015-2022 grant to H.G. and PICT2017-2421 grant to E.R. from ANPCyT. E. R. and H. G. are members of the Research Career, and A.L.L. is a doctoral fellow of
CONICET. We kindly thank Paula Casati, Gabriela Gago and Alejandro Viale for helpful comments. We kindly thank Marina Avecilla for technical assistance.
Author informationAuthor notes Laura Navone
Present address: Molecular Biology & Industrial Biotechnology, Science and Engineering Faculty, Queensland University of Technology, Brisbane, Queensland, Australia
Authors and Affiliations Instituto de Biología Molecular y Celular de Rosario, Facultad de Ciencias Bioquímicas y Farmacéuticas, Universidad Nacional de Rosario, Rosario, Argentina
Andrea L. Livieri, Hugo Gramajo & Eduardo Rodriguez
Australian Institute for Bioengineering and Nanotechnology, The University of Queensland, Brisbane, Queensland, Australia
Laura Navone & Esteban Marcellin
AuthorsAndrea L. LivieriView author publications You can also search for this author inPubMed Google Scholar
Laura NavoneView author publications You can also search for this author inPubMed Google Scholar
Esteban MarcellinView author publications You can also search for this author inPubMed Google Scholar
Hugo GramajoView author publications You can also search for this author inPubMed Google Scholar
Eduardo RodriguezView author publications You can also search for this author inPubMed Google Scholar
ContributionsA.L.L. and E.R. designed research; A.L.L. and L.N. performed research; A.L.L., L.N., H.G. and E.R. analyzed data; A.L.L., E.M., H.G. and E.R. wrote the paper. A.L.L. prepared all figures.
All authors reviewed the manuscript.
Corresponding authors Correspondence to Hugo Gramajo or Eduardo Rodriguez.
Ethics declarations Competing InterestsThe authors declare no competing interests.
Additional informationPublisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary informationSupplementaryInformationRights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or
format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or
other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in
the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the
copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and permissions
About this articleCite this article Livieri, A.L., Navone, L., Marcellin, E. et al. A novel multidomain acyl-CoA carboxylase in Saccharopolyspora erythraea provides malonyl-CoA for de novo
fatty acid biosynthesis. Sci Rep 9, 6725 (2019). https://doi.org/10.1038/s41598-019-43223-5
Download citation
Received: 06 December 2018
Accepted: 17 April 2019
Published: 30 April 2019
DOI: https://doi.org/10.1038/s41598-019-43223-5
Share this article Anyone you share the following link with will be able to read this content:
Get shareable link Sorry, a shareable link is not currently available for this article.
Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative




