- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT The oral cavity harbours a complex microbiome that is linked to dental diseases and serves as a route to other parts of the body. Here, the aims were to characterize the oral
microbiota by deep sequencing in a low-caries population with regular dental care since childhood and search for association with caries prevalence and incidence. Saliva and tooth biofilm
from 17-year-olds and mock bacteria communities were analysed using 16S rDNA Illumina MiSeq (v3-v4) and PacBio SMRT (v1-v8) sequencing including validity and reliability estimates. Caries
was scored at 17 and 19 years of age. Both sequencing platforms revealed that _Firmicutes_ dominated in the saliva, whereas _Firmicutes_ and _Actinobacteria_ abundances were similar in tooth
biofilm. Saliva microbiota discriminated caries-affected from caries-free adolescents, with enumeration of _Scardovia wiggsiae_, _Streptococcus mutans_, _Bifidobacterium longum_,
_Leptotrichia sp_. HOT498, and _Selenomonas spp_. in caries-affected participants. Adolescents with _B_. _longum_ in saliva had significantly higher 2-year caries increment. PacBio SMRT
revealed _Corynebacterium matruchotii_ as the most prevalent species in tooth biofilm. In conclusion, both sequencing methods were reliable and valid for oral samples, and saliva microbiota
was associated with cross-sectional caries prevalence, especially _S. wiggsiae, S. mutans_, and _B. longum_; the latter also with the 2-year caries incidence. SIMILAR CONTENT BEING VIEWED BY
OTHERS HIGH ABUNDANCE OF SUGAR METABOLISERS IN SALIVA OF CHILDREN WITH CARIES Article Open access 24 February 2021 ORAL MICROBIOTA AND DENTAL CARIES DATA FROM MONOZYGOTIC AND DIZYGOTIC TWIN
CHILDREN Article Open access 13 October 2020 RELATIONSHIP BETWEEN DENTAL AND PERIODONTAL HEALTH STATUS AND THE SALIVARY MICROBIOME: BACTERIAL DIVERSITY, CO-OCCURRENCE NETWORKS AND
PREDICTIVE MODELS Article Open access 13 January 2021 INTRODUCTION The most prevalent oral diseases, dental caries and periodontal diseases, are complex infectious diseases where life style
and host factors, including behavioural, socioeconomic factors, medical status and genetics, interact1, 2. The oral cavity is reported to harbour more than 700 species/phylotypes, among
which some species have been cultured3, 4. The bacteria are organized in site-specific biofilm communities5, with dysbiosis in the tooth surface or gingival pocket communities preceding
caries and periodontal diseases6. For dental caries, which result from the demineralization of tooth tissues by acids produced by bacterial fermentation of dietary carbohydrates, the
aciduric and acidophilic mutans streptococci (_Streptococcus mutans_ and _Streptococcus sobrinus_) have been specifically linked to disease development6. However, any acidogenic species,
including the mutans streptococci, aciduric non-mutans streptococci, _Bifidobacterium_, _Lactobacillus, Actinomyces_, and _Scardovia_ 7,8,9,10, may contribute to disease development. The
relative impact of a species from these genera may vary between populations and within a population over time. For example, it was recently reported that in Romanian adolescents, in whom
dental care is limited and disease activity is high, the frequency of both _S. mutans_ and _S. sobrinus_ was very high compared to in Swedish adolescents, who are exposed to life-long
disease prevention and treatment programmes11. In 1973, _S. mutans_ infection was as prevalent among Swedish adolescents (96%)12 as that reported for Romanian adolescents in 201311. Eleven
years later (1984), the prevalence had decreased to 77% in Swedish adolescents12, and it had decreased to 50% in 201311. Non-targeted methods are warranted to characterize a
disease-associated bacterial community, i.e., methods not limited by present knowledge or expectations. Over recent decades, non-targeted multiplex DNA sequencing of the 16S rRNA gene and
taxonomic determination from gene databases have commonly been used13. To date, most studies searching for caries-associated microbial patterns have been conducted in young children with
severe caries. Only a limited number of studies have targeted adolescents or adults14, 15. Currently, Illumina technology is most commonly used for the microbiome characterization of
clinical samples. This platform generates many sequences, but taxonomic resolution is limited. Recently, multiplex amplicon sequencing was launched for the Single-Molecule Real-Time (PacBio
SMRT, Pacific Biosciences of California, USA) platform, which produces long reads, i.e., up to nine variable regions of the 16S rDNA, and improved taxonomic resolution16. The above described
results indicate systematic differences in the oral microbiota between populations by socio-economic status and that shifts occur over time not only at the individual but also at the
population level. Therefore, studies targeting various well-characterized populations are required to understand determinants of health and disease under given conditions. The aims of the
present study were to (i) characterize and compare saliva and tooth biofilm microbiota via deep sequencing using the Illumina MiSeq and the PacBio SMRT platforms and the Human Oral
Microbiome Database (HOMD, www.HOMD.org) for taxa resolution17 in a population with long-term caries prevention and low disease activity, (ii) compare saliva and tooth biofilm microbiota in
caries-affected and caries-free adolescents cross-sectionally and longitudinally, and (iii) evaluate the relative validity between and within the two sequencing methods. The study will add
knowledge concerning saliva and tooth biofilm microbiota in adolescents, representing the most typical situation in Western countries18. The study will also be the first to employ the PacBio
SMRT amplicon technique for determination of oral microbiota. RESULTS ILLUMINA MISEQ GLOBAL SEQUENCING RESULTS Summary data from saliva (n = 64) and tooth biofilm (n = 49) sequencing using
the Illumina MiSeq platform are presented in Table 1. The average read length of these sequences was 427 bp (v3-v4), and the average number of cleaned sequences per sample was 71,047 for
saliva and 20,056 for tooth biofilm samples. Taxa were found in 10 phyla in both sample types and in 98 and 86 genera in saliva and tooth biofilm, respectively (Table 1). ProbeSeq
(http://homings.forsyth.org/index2.html) and QIIME19 processing of raw sequences produced similar results (Table 1). In the following sections, data are presented for ProbeSeq processing
unless stated otherwise. DETERMINATION OF PHYLA IN SALIVA AND TOOTH BIOFILM SAMPLES USING ILLUMINA MISEQ _Firmicutes_ dominated in saliva (48% abundance, % of all sequences) followed by
_Actinobacteria_ (20%), whereas the abundances of _Firmicutes_ and _Actinobacteria_ were more similar (32% and 24%, respectively) in tooth biofilms. _Bacteroidetes, Fusobacteria_, and
_Proteobacteria_, which were also represented in all adolescents and both sample types, together constituted 32% and 41% in saliva and tooth biofilm, respectively (Supplementary Tables S1
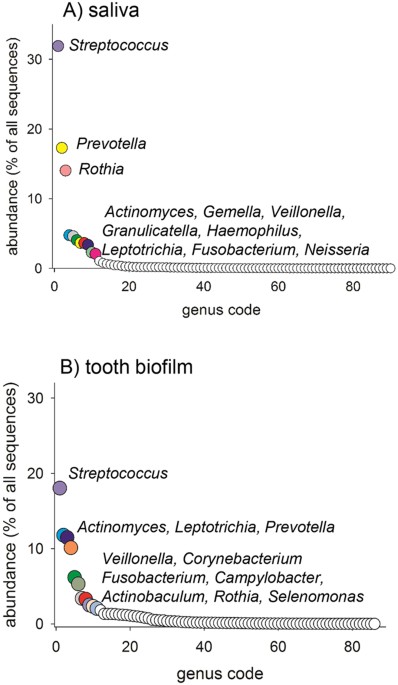
and S2 ). DETERMINATION OF GENERA IN SALIVA AND TOOTH BIOFILM USING ILLUMINA MISEQ In total, 11 genera in saliva and 11 genera in tooth biofilm each represented 2% or more of all sequences
(Fig. 1A,B, Supplementary Tables S1 and S2). Of these, 7 genera were detected in both sample types (i.e., _Actinomyces, Fusobacterium, Leptotrichia, Prevotella, Rothia, Streptococcus_, and
_Veillonella_) (Fig. 1A,B). Although these genera were represented in all adolescents, the individual abundance was highly varied, as exemplified for saliva in Fig. 2. In total, 16 genera
were found in both sample types and all adolescents. At the genus level, _Streptococcus_ dominated (32% abundance) in saliva, followed by _Prevotella_ (17%), _Rothia_ (14%), and 8 additional
genera (5-2%) (Fig. 1A). In tooth biofilm, _Streptococcus_, _Actinomyces, Leptotrichia_, and _Prevotella_ prevalence rates were more similar (18%, 12%, 11%, and 10%, respectively) as well
as 7 additional genera (6-2%) (Fig. 1B). The abundances of all identified genera are presented in Supplementary Tables S1 and S2. SPECIES/PHYLOTYPES/GENUS PROBES IN SALIVA AND TOOTH BIOFILM
USING ILLUMINA MISEQ In saliva, 30% of all sequences were recognized by the _Streptococcus_ Genus probe 4 (recognizing 32 different _Streptococcus_ species/phylotypes). Other prevalent
species/phylotypes/Genus probes (>2% abundance) in saliva included, in descending order, _Rothia mucilaginosa, Prevotella melaninogenica, Haemophilus parainfluenzae_, species recognized
by the _Granulicatella_ Genus probe, and _Gemella sanguinis_ (Supplementary Table S3). A full list of abundances is shown in Supplementary Table S3, and the species recognized by the Genus
probes are listed at http://homings.forsyth.org/Genus%20probe%20list%20for%20website_v2.0.pdf. In tooth biofilm, 11% of all sequences were recognized by the _Streptococcus_ Genus probe 4.
Other prevalent species/phylotypes/Genus probes (>2% abundance) in tooth biofilm included _Corynebacterium matruchotii_ (4%)_, Actinobaculum sp_. HOT183_, Actinomyces gerencseriae,
Actinomyces sp_. HOT448_, Campylobacter gracilis_, species recognized by _Fusobacterium_ Genus probe 4_, Leptotrichia wadei, Prevotella melaninogenica, Prevotella nigrescens, Veillonella
dispar_, and species recognized by _Veillonella_ Genus probe 2 (all between 2 and 3%). A full list of abundances is given in Supplementary Table S4. PACBIO SMRT SEQUENCING OF TOOTH BIOFILM
SAMPLES The average read length of the PacBio SMRT sequences was 1,360 bp (v1-v8), and the average number of cleaned sequences per sample was 3,367 (Table 1). Thus, the PacBio SMRT read
length was significantly longer, but the sequencing depth was significantly lower than that of Illumina MiSeq sequencing. In total, 345 species/phylotypes in 10 phyla and 77 genera were
detected. Similar to the Illumina MiSeq sequencing of tooth biofilm, the abundances of sequences in the _Firmicutes_ and _Actinobacteria_ phyla were similar (23 and 26%, respectively),
followed by _Bacteroidetes, Fusobacteria_ and _Proteobacteria_ (in total 39% of all sequences). _Actinomyces_ was the most abundant genus (19%), followed by _Streptococcus_ (10%),
_Prevotella_ (9%), _Leptotrichia_ (8%), and _Fusobacterium_, _Corynebacterium_, and _Veillonella_ (all 6%). The most frequently detected species was _Corynebacterium matruchotii_ (5%
abundance). The full lists of abundances are given in Supplementary Tables S5 and S6. CORE MICROBIOME IN SALIVA AND TOOTH BIOFILM SAMPLES Using Illumina MiSeq, 50 species/phylotypes/Genus
probes were detected in all saliva samples, and 23 were detected in all tooth biofilm samples (100% prevalence). Nineteen of these “core species” were shared by saliva and tooth biofilm
(Supplementary Tables S3 and S4). The PacBio SMRT analyses confirmed that _Campylobacter gracilis, Corynebacterium matruchotii, Streptococcus mitis, Veillonella dispar_, and _Veillonella
parvula_ were present in all of the analysed tooth biofilm samples (Supplementary Table S6). TOOTH BIOFILM MICROBIOTA IN ADOLESCENTS WITH OR WITHOUT CARIES STUDY GROUP CHARACTERISTICS The
characteristics of the 17-year-old adolescents in the caries-affected versus caries-free group are presented in Table 2. In addition to higher caries scores, the former group had a higher
BMI (p = 0.033), more colony forming units (CFUs) of mutans streptococci in saliva (cultivation) (p < 0.001), and a higher _S. mutans_ prevalence by PCR (p = 0.001). Additionally, they
brushed their teeth less frequently (p = 0.012). Caries, caries-associated variables, and detected taxa were not systematically different between boys and girls. MICROBIOME PROFILE BY CARIES
STATUS Microbiota richness. The species richness in saliva and tooth biofilm samples did not differ between subjects with or without caries (Fig. 3A). Microbiota diversity. PCA modelling
with species/phylotypes/Genus probes in saliva (Illumina MiSeq ProbeSeq processing), tooth brushing, BMI, sugar intake, and tobacco use included as potential confounders separated
caries-affected adolescents from those who were caries free (Fig. 3B). Similarly, PCoA modelling by QIIME identified Operational Taxonomic Units (OTUs) tended to separate most caries
affected subjects from caries free subjects (Supplementary Fig. 1). A linear discriminant analysis (LDA) effect size-based cladogram, in which successive circles represent a phylogenetic
level (phylum, class, family, genus) indicated that species in the _Synergistetes_ phylum, _Synergistia_ class_, Clostridiales_ [F-1] and _Synergistaceae_ families, and _Dialister_,
_Scardovia, Clostridiales_ [F-1][G-1], _Fretibacterium, Shuttleworthia, Peptostreptococcaceae_ [11] [G-6] and [11][G-9], and _Veillonellaceae_ [G-1] genera were enriched in subjects with
caries, whereas taxa in the _Fusobacteria_ phylum, _Fusobacteriia class, Actinomycetaceae_ and _Ruminococcaceae_ families, and _Actinomyces, Ruminococcaceae_ [G-1], and [G-2] genera were
enriched in caries-free subjects (Fig. 4A). These results are also illustrated in an LDA bar graph (Fig. 4B). Partial least squares (PLS) modelling with caries status (DFS yes/no) at 17
years of age as the dependent variable and an independent block of species/phylotypes/Genus probes (yes/no) in saliva along with the same set of confounders as in PCA yielded a model with
four significant components and a cross-validated predictive capacity (Q2) of 37% of the two first components. The strongest associations (PLS correlation coefficients >0.1) with the
presence of caries were found for _Scardovia wiggsia_e, _Streptococcus mutans, Selenomonas_ Genus probe 1, _Bifidobacterium longum_, and _Leptotrichia sp_. HOT498, whereas _Mycoplasma orale_
and _Porphyromonas sp_. HOT278 were most strongly (PLS correlation coefficients >0.1) associated with being caries free. In addition, BMI was significantly associated with having caries,
and tooth brushing was significantly associated with being caries free (Fig. 5). The findings of the multivariate PLS model were consistent with univariate comparisons in which _S.
wiggsiae_ was found in the saliva of 100% of individuals with caries compared to 65.4% of caries-free adolescents (p < 0.0001, which was significant after false discovery correction)
(Supplementary Table S3). Other PLS identified caries-associated species prevalence rates were also higher in univariate comparisons (p ≤ 0.008) but did not reach significance after false
discovery correction, i.e., _S. mutans, Selenomonas_ Genus probe 1, _B. longum_, and _Leptotrichia sp_. HOT498. In addition, the mean percent of all sequences (abundance) of S. _mutans was_
significantly higher (p < 0.0001) in caries-affected than in caries-free adolescents and tended to be higher (p ≤ 0.008) for _Bifidobacterium longum, Fusobacterium nucleatum subsp.
nucleatum_, and _Selenomonas_ Genus probe 1, whereas the opposite was found for _Mycoplasma orale_ and _Porphyromonas sp_. HOT278 (both p = 0.009) (Supplementary Table S3). For tooth biofilm
microbiota (Illumina MiSeq, ProbeSeq processing), PCA did not indicate any clustering of subjects, and PLS modelling was not applied. Similarly, neither the prevalence nor abundance
displayed a significant difference between the two caries groups, although the prevalence of the _Capnocytophaga_ Genus probe 1 and _Veillonella rogosae_ tended to be higher for
caries-affected than caries-free adolescents _(_p ≤ 0.008) as did the abundance of _S. mutans_ (Supplementary Table 4). Similarly, PCA modelling of PacBio SMRT sequences did not indicate the
clustering of subjects by tooth biofilm taxa, and no significant differences were found in univariate analyses between the two groups. However, caries-diseased adolescents tended to have a
higher prevalence of _Actinobaculum sp_. HOT183, _Actinomyces sp_. HOT171_, Granulicatella adiascens_, _Rothia aeria_, and _Veillonellaceae_ [G-1] _sp_. HOT150 (p ≤ 0.008). _Actinobaculum
sp_. HOT183 and _Veillonellaceae_ [G-1] _sp_. HOT150 also tended to be more abundant in caries-affected than caries-free adolescents, whereas _Fusobacterium nucleatum subsp. polymorphum_
tended to be more abundant in the caries-free group (p ≤ 0.008) (Supplementary Table 6). SALIVA MICROBIOTA AND THE 2-YEAR CARIES INCREMENT FROM 17 TO 19 YEARS OF AGE PCA did not reveal any
systematic clustering of subjects by 2-year caries increment but a higher proportion of adolescents who had developed caries over the 2-year follow-up period were smokers and had received
additional fluoride treatments (Table 2). However, when 2-year mean caries increments were compared in adolescents with or without _B. longum_ in saliva (means adjusted for sex, oral
hygiene, sugar intake, BMI, and tobacco use by general linear modelling (glm)), the former group had a significantly higher increment (2.1 versus 0.5 new caries lesions, p = 0.005).
Adolescents with _S. mutans_ or _S. wiggsiae_ had a numerically higher 2-year mean caries increment (both 0.8 versus 0.3), but the differences did not reach significance (p > 0.05).
METHOD VALIDATIONS RELIABILITY OF SEQUENCING ANALYSIS Nine tooth biofilm DNA samples in triplicate and saliva samples from 5 subjects collected one day apart during one week were analysed
using the Illumina MiSeq platform. Triplicate samples clustered close together, and samples were distinctly separated from each other (Fig. 6A). The repeated saliva samples clustered
together for three subjects, whereas the sample from one day deviated for one subject and for all three samples in one subject (Fig. 6B). VALIDITY Illumina MiSeq sequencing detected all 10
species in the _Streptococcus_ mock correctly among OTUs with ≥10 sequences but also suggested 5 OTUs as unnamed _Streptococcus_ phylotypes. Of the 20 _Lactobacillus_ species in that mock,
16 were correctly identified among OTUs with ≥10 sequences, and 4 could not be detected because they were not included in the HOMD database (_L. curvatus, L. colehominis, L. gallinarium_,
and _L. graminis_). Similarly, with ≥10 sequences/OTU as the cut-off, 25 (of 32) species from the mixed genera mock were correctly identified. For the 8 non-identified species, 1 was
represented by 2 sequences only, and 7 species of different genera were not detected at all. The specificity was low for OTUs with fewer than 10 sequences. The number of sequences for the
detected species varied greatly, i.e., 38,862 sequences for _S. mutans_ and 3,721 for _S. sanguinis_ in the streptococcus mock. The sensitivity and specificity for _S. mutans_ detection by
both Illumina MiSeq and PCR were calculated. The sensitivity values for both saliva and tooth biofilm were high (0.98 and 0.90, respectively). ROC curves were calculated (Supplementary Fig.
2), and the area under the curve was 0.94 for saliva and 0.74 for tooth biofilm. PacBio SMRT sequences of the _Streptococcus_ mock identified 9 of 10 species with >1 sequence. The 10th
was present with 1 sequence. In the _Lactobacillus_ mock, 16 of 20 species were detected, with the same undetected species described above, i.e., 4 species not in the HOMD database.
Similarly, a more than 10-fold variation in the number of detected sequences per species was observed, i.e., for streptococci, from 553 for _S. sobrinus_ to 49 for _S. sanguinis_ (in
addition to _S. mitis_ with 1 sequence); for lactobacilli, from 343 for _L. paracasei_ to 10 for _L. crispatus_. In addition, a negative control sample (ultra-pure water) subjected to
Illumina MiSeq sequencing yielded 620 reads, which was far below the acceptance criterion for this method, i.e., ≥5,000 reads per sample. CORRELATION BETWEEN SALIVA AND TOOTH BIOFILM TAXA
Spearman correlation coefficients between abundances in the saliva and tooth biofilm by Illumina MiSeq sequencing varied from 0.9 to −0.1 for various genera, reflecting the similarities and
differences between the hard- and soft-tissue niches. The Spearman correlation coefficients between saliva and tooth biofilm abundances for the caries-associated taxa were as follows: _S.
wiggsiae_ and _Leptotrichia sp_. HOT498 r = 0.6 (p < 0.001), _S. mutans_ r = 0.7 (p < 0.001), and _B. longum_ and _Selenomonas_ Genus probe 1 r = 0.3 (p < 0.05). DISCUSSION In the
present study, saliva and tooth biofilm samples were analysed using Illumina MiSeq, and tooth biofilm samples were also analysed using PacBio SMRT sequencing. This study is the first to
report the use of the latter technology on oral samples. PacBio SMRT sequencing confirmed the results from Illumina MiSeq sequencing at the phylum and genus levels and partially at the
species level, including the predominance of _C. matruchotii_ in tooth biofilms. _C. matruchotii_ was overlooked in many previous studies using PCR-based or metagenomic sequencing but was
recognized as a prominent species in metatranscriptome20, metaproteome21, and spectral imaging fluorescence hybridization5 analyses. Saliva microbiota, but not tooth biofilm microbiota,
distinguished adolescents with caries from caries-free adolescents, with _S. wiggsiae, S. mutans, B. longum, Leptotrichia sp_. HOT498 and species recognized by _Selenomonas_ Genus probe 1
(_S. noxia_ and _Selenomonas sp_. HOT140) as the most influential taxa for group distinction. The dominant taxa in saliva and tooth biofilms largely coincided with those reported in previous
studies, especially at the phylum and genus levels14, 20,21,22,23. Thus, according to the Illumina MiSeq results, phylum _Firmicutes_ and genus _Streptococcus_ dominated in saliva, whereas
abundances of the phyla _Firmicutes_ and _Actinobacteria_ were similar in tooth biofilm samples, and the _Streptococcus_ genus was only slightly more abundant than _Actinomyces_. PacBio SMRT
sequencing also identified similar levels of _Firmicutes_ and _Actinobacteria_ but a considerably higher abundance of _Actinomyces_ than _Streptococcus_. Generally, studies using PCR-based
454 FLX+ pyrosequencing or Illumina MiSeq sequencing report a higher abundance of _Streptococcus_ than _Actinomyces_, but the opposite has been indicated in newer metatranscriptome20 and
metaproteome21 studies. The reason for the discrepancy between the two sequencing methods observed in the present study cannot be determined but likely relates to method-specific
biases/strengths and possibly the difference in sequencing depth between the two methods. Five samples were placed per PacBio SMRT cell, but fewer would have been preferential making cost of
PacBio SMRT sequencing compared to Illumina MiSeq a hindrance. However, comparisons with other studies must be performed with caution because the sequencing methods, age of the study
subjects, dentition (primary versus permanent teeth), and socio-economic conditions of the target populations differed between most studies. Although the PacBio SMRT method has been used for
samples from non-oral sites and mock communities24, 25, the fact that the present study was the first to report tooth biofilm microbiota limits comparisons with other studies. Thus, in
accordance with results from metaproteomic profiling21, PacBio SMRT identified _Actinomyces_, _Streptococcus_, _Corynebacterium_, _Leptotrichia_, and _Veillonella_ as the five most abundant
genera, whereas _Rothia_ was less abundant by PacBio SMRT than by both metaproteomic profiling21 and Illumina MiSeq sequencing in the present study. The structure of tooth biofilm has
recently been elegantly illustrated by spectral imaging fluorescence _in situ_ hybridization5. The arrangement was referred to as a “hedgehog structure” with clustering of species in
_Corynebacterium, Streptococcus, Porphyromonas, Haemophilus/Aggregatibacter, Neisseriaceae, Fusobacterium, Leptotrichia, Capnocytophaga, and Actinomyces_, with dominance of the 3 first
genera and a central role of _C. matruchotii_. In the present study, most of these (_Corynebacterium, Streptococcus, Porphyromonas, Fusobacterium, Leptotrichia, Capnocytophaga, and
Actinomyces)_ were found in all or nearly all of the adolescents regardless of the sequencing method, and _Neisseria_ and _Haemophilus/Aggregatibacter_ were found by the more extensive
Illumina MiSeq sequencing_. C. matruchotii_, which was also detected in all adolescents by both sequencing methods, was the most abundant species by PacBio SMRT and ranked second by Illumina
MiSeq, reflecting its central position in the hedgehog or so-called “corn-cob formation”5. Overall, saliva sequences represented more genera and species than tooth biofilm sequences. This
may reflect that saliva is in contact with various surfaces in the mouth, i.e., mucosal surfaces, tongue epithelium, teeth, and gingival margins, rather than transient bacteria or the effect
of a higher sequencing depth in the saliva than in tooth biofilm samples. By contrast, bacteria on teeth are selectively recruited by discriminatory attachment to tooth tissue adhering
saliva proteins/peptides and interactions with neighbouring bacteria4, 5, 22, 26. Among the earliest saliva proteins/peptides that adhere to a clean tooth surface are the acidic proline-rich
proteins, statherin and cystatins27, which unfold their binding epitopes for _Streptococcus_ and _Actinomyces_ upon tooth attachment22. Yet, other explanations cannot be ruled out, such as
an influence of total amount of DNA captured in saliva versus tooth biofilm samples and systematic species differences in DNA retrieval. Notably, a mixture of enzymes were used in the
present study to break the resistant cell walls of Gram positive bacteria, and Gram positive species in the mock with mixed genera, i.e. species in _Bifidobacteria, Scardovia_ and _Rothia_,
were detected in numbers that were similar to Gram negative species. Some validations were performed in the present study, including negative and positive controls and repeated analyses. It
was concluded that false positives due to contamination were negligible, and reproducibility was high for Illumina MiSeq sequencing; however, the sample composition may vary daily in some
individuals. The latter is consistent with results reported by David _et al_.28. The detection specificity at the species level was acceptable for both the Illumina MiSeq and PacBio SMRT
methods when the appropriate cut-offs were applied for taxa allocation. Unexpectedly, the numbers of sequences per species in the mock varied largely, even though equal volumes of bacterial
suspensions of the same OD were added; therefore, this finding should be considered in the data interpretation and modelling. We chose to use a more conservative dichotomous variable
(yes/no) in statistical models of taxa in the independent data block. The present study population was characterized by a significant caries reduction over the last five decades29, regular
high-standard dental care, including compulsory preventive strategies, from early childhood18, and generally slow development of caries symptoms. The selected conservative approach better
addressed time fluctuations in microbiota abundances. Use of the ProbeSeq or QIIME approach for the bioinformatics of Illumina sequences did not confer any substantial difference in the
interpretation of results. Because the ProbeSeq approach has the advantage of clustering species with very high 16S rDNA identity into a Genus probe group, this approach was preferred. Thus,
for the present clinical research question, we found the ProbeSeq processing of oral samples and dichotomous measures most suitable and that our results supported the previously reported
validity of ProbeSeq-processed Illumina MiSeq data30. In the present study group, saliva microbiota separated caries-affected from caries-free subjects, whereas microbiota from tooth biofilm
did not separate these groups. There are several plausible explanations for this finding, including that tooth biofilm samples only represent sampled surfaces, whereas saliva is the pool
from which tooth colonizers are recruited and reflects most surfaces in the mouth. In contrast to our findings, it has been claimed that saliva does not reflect tooth biofilm microbiota and
variations among single tooth surfaces31. In the present study, the caries-associated bacterial species in saliva were consistent with those found in previous studies that used various
techniques (cultivation, PCR, DNA hybridization chips) on tooth biofilm samples, and the abundances of the caries-associated taxa in saliva were significantly correlated with the
corresponding abundance in tooth biofilms. This result supports the use of saliva; however, as recommended32, a combination with more traditional techniques, such as culturing and PCR,
should be used for optimal identification. Thus, based on the present findings, we suggest that chewing-stimulated saliva is valid for the characterization of caries-associated microbiota in
large-scale epidemiological studies, but this finding has been contradicted by others32, 33. The species that were most strongly associated with caries were _S. wiggsiae, S. mutans, B.
longum, Leptotrichia sp_. HOT498, and species detected by _Selenomonas_ Genus probe 1_. Selenomonas_ Genus probe 1 targets _S. noxia_ and _Selenomonas sp_. HOT140, of which _S. noxia_ has
been found to be increased in patients with gingivitis34. Thus, a higher prevalence of _Selenomonas_ Genus probe 1 likely reflected less frequent tooth brushing and gingivitis in
caries-affected adolescents rather than an aetiological role in the caries process. Consistent with the present finding of _Leptotrichia sp_. HOT498 being more prevalent in the saliva of
caries-affected than caries-free adolescents, it was recently reported that this species was more prevalent in severely caries-affected Romanian adolescents than in Swedish adolescents with
a low caries prevalence11. This difference was found in tooth biofilms and for sequences obtained by the LibL 454 FLX+ pyrosequencing option covering the first four variable regions of the
16S rRNA gene. However, _S. wiggsiae, S. mutans_, and _B. longum_ are well-documented aciduric and acidophilic species that have been associated with caries in many earlier
studies6,7,8,9,10, 35, 36. Among these three species, _B. longum_ was significantly associated with the 2-year caries incidence; however, because the number of subjects carrying _B. longum_
was low, the association must be evaluated in a larger sample. In addition, smoking and having topical fluoride treatment besides a fluoride toothpaste were associated with 2-year caries
increment. Smoking is well documented to be associated with caries prevalence and even in this small study sample the association was evident. In contrast, fluoride is a well documented
caries protective compound. It may therefore seem contradictory to find such treatment more prevalent among adolescents with disease development. However, in the study population persons
with a high risk to develop caries and/or high caries activity have annual dental visits and preventive measures are required. For this reason fluoride treatment in this population reflects
caries treatment in high caries subjects and not the effect of fluoride per se. The strengths of the present study include that (i) the study groups were larger than those of previous
studies, and the study extended knowledge beyond early childhood caries and caries in deciduous teeth, (ii) the criteria for sequence inclusion were based on results from simultaneously
analysed mock samples, (iii) the sparsely used PacBio SMRT method with improved taxa resolution was applied, (iv) the study population represented populations with a substantial caries
decline in the 20th century, and (v) prospective caries incidence was evaluated. The weaknesses include that (i) tooth biofilm could not be analysed in all participants due to limitations in
DNA accessibility, (ii) it cannot be assured that every single tooth surface was sampled, (iii) the sequencing depth was low for PacBio SMRT, and (iv) saliva was not analysed using PacBio
SMRT. CONCLUSIONS Based on the present findings, we conclude that saliva microbiota can separate adolescents with caries from caries-free adolescents in a low-caries population with daily
tooth brushing using fluoridated tooth paste18. The three most influential species were _S. wiggsiae, S. mutans_, and _B. longum_. We also showed that _C. matruchotii_ is enriched in tooth
biofilms relative to saliva and that overall PacBio SMRT sequencing covering most of the 16S rDNA supported the findings of Illumina sequencing. However, some discrepancies need to be
followed up in future studies. METHODS STUDY SUBJECTS Seventeen-year-old adolescents, who were caries-free (no present or previous caries) or had present caries activity and a high caries
risk37, were invited to participate. Subjects were consecutively recruited from three Public Dental Health Care Clinics in the city of Umeå, Sweden. The exclusion criteria were that the
adolescent (or caregiver) did not consent, had a chronic disease or was on medication, had taken antibiotics during the latest 6 months, or was unable to communicate in Swedish or English.
The caries status was evaluated at 17 and 19 years of age. Baseline data were collected in 2013. Twenty-six caries-free and 37 caries-affected adolescents were recruited, and 55 of these
subjects were re-evaluated at 19 years of ages. The population from which the participants were recruited has an overall low caries prevalence, as well as annual dental care, including
preventive measures, provided since 2–3 years of age (http://www.socialstyrelsen.se/nationalguidelines). SAMPLING FOR MICROBIOTA ANALYSES Whole saliva (5 ml) was collected into ice-chilled
sterile test tubes while chewing on a 1-g piece of paraffin wax. Next, 100 μl of fresh saliva was transferred to a transport medium for cultivation, the remaining saliva centrifuged at 4 °C
(13,000 rpm for 5 min), and the pellets stored at −80 °C until DNA extraction and amplicon sequencing by Illumina MiSeq (http://www.illumina.com). Supragingival tooth biofilm was collected
using sterile wooden toothpicks, pooled by subject in 100 µl TE-buffer (10 mM Tris, 1 mM EDTA, pH 7.6), and stored at −80 °C until DNA extraction and 16S rDNA amplicon sequencing by Illumina
MiSeq and PacBio RS II SMRT (Pacific Biosciences, Menlo Park, CA, USA) was conducted. CARIES SCORING AND LIFESTYLE INFORMATION The number of teeth, cavitated carious lesions (D), fillings
(F), and missing (M) tooth surfaces were scored from visual and radiograph examinations in dental clinics. The sum of the decayed and filled tooth surfaces (DFS) was calculated (caries
prevalence). The M-component was not included in the caries index because losses of teeth were due to orthodontic or tooth hypomineralization reasons. The caries incidence (increment) was
calculated as the DFS increase from 17 to 19 years of age. Information on the general health status, medication, oral hygiene, dietary habits, and tobacco use (smoking and snuff) was
obtained by a questionnaire when the adolescent was 17 years old. MOCK COMMUNITIES Three mock communities of oral species were created. These included (_i_) 20 _Lactobacillus_ species (_L.
acidophilus, L. brevis, L. buchneri, L. casei, L. coleohominis, L. crispatus, L. curvatus, L. fermentum, L. gallinarium, L. gasseri, L. graminis, L. jensenii, L. johnsonii, L. panis, L.
paracasei, L. pentosus, L. reuteri, L. rhamnosus, L. salivarius, and L. vaginalis_); (_ii_) 10 _Streptococcus_ species, (_S. gordonii, S. intermedius, S. mitis, S. mitis bv 2, S. mutans, S.
oralis, S. parasanguinis I, S. salivarius, S. sanguinis, and S. sobrinus_); and (_iii_) 25 species of mixed genera (_Actinomyces gerencseriae, Actinomyces meyeri, Actinomyces odontolyticus,
Bifidobacterium longum, Escherichia coli, Fusobacterium nucleatum, L. acidophilus, L. casei, L. coleohominis, Leptotrichia buccalis, Porphyromonas gingivalis, Prevotella denticola,
Prevotella oris, Rothia dentocariosa, Scardovia wiggsiae, S. gordoni, S. intermedius, S. mitis, S. mutans, S. oralis, S. parasanguinis I, S. salivarius, S. sanguinis, S. sobrinus, and
Veillonella parvula_). Equal aliquots were obtained from each bacterial suspension (OD600 = 2.0), resulting in a total volume of 1 ml. All 3 mocks were used for Illumina MiSeq sequencing,
and the _Streptococcus_ and _Lactobacillus_ mocks for PacBio SMRT sequencing. DNA EXTRACTION Genomic DNA was extracted from the saliva pellets, tooth biofilm samples, 3 mock communities, and
a sample of ultra-pure water (negative control) with a Gene elute™ Bacterial Genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA) as described previously11. Briefly, samples were (_i_)
centrifuged for 5 min at 13,000 rpm, (_ii_) lysed in buffer with lysozyme and mutanolysin for 30 min at 37 °C, (_iii_) treated with RNase for 2 min at room temperature followed by Proteinase
K for 10 min at 55 °C, (_iv_) mixed with ethanol and transferred to the binding column, and (_v_) washed and eluted in 100 µl of elution buffer. The DNA quality and quantity were evaluated
using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) to meet the standard set by the sequencing facilities (OD 260/280 ratio ≥1.8). DNA was extracted from
undivided samples, and after the DNA content had been determined, the volumes at concentrations requested by the sequencing facilities were subjected to further preparation. SEQUENCING
Multiplex 16S rDNA amplicon sequencing was performed using two different platforms, Illumina MiSeq (http://www.illumina.com) and PacBio RS II SMRT
(http://www.pacb.com/smrt-science/smrt-sequencing/). The former analyses were performed at Forsyth Research Institute (Cambridge, MA, USA), and the latter at GATC Biotech AG (Konstanze,
Germany). For Illumina MiSeq sequencing, the HOMINGS protocol was applied as described previously30. Briefly, the V3-V4 hypervariable regions of the 16S rDNA were PCR amplified using the
forward 341F (AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGT_CCT_ ACGGGAGGCAGCAG) and reverse 806R (CAAGCAGAAGACGGCATACGAGATNNNNNNNNNNNNAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT) primers (primer sequences
are in bold, and sample-specific sequence tags (barcodes) are indicated by NNNNNNNNNNNN). Amplicons were purified (AMPure beads; Beckman Coulter Genomics, Danvers, MA); 100 ng of each
library was pooled, gel purified, and quantified; and 20% PhiX was added to 12 pmol of the library mixture and run on the MiSeq system. Pair-end reads were fused; barcodes, primers, and
ambiguous and chimeric sequences were removed; and taxa were identified by the ProbeSeq customized BLAST programme (HOMINGS, http://homings.forsyth.org/index2.html) for the recognition of
538 species by 638 probes of 17 to 40 bases followed by the detection of 129 genus-level probes for closely related species. Species/phylotypes targeted by the Genus probes are described at
http://homings.forsyth.org/index2.html. Taxa represented by <10 sequences were excluded. In parallel, the QIIME pipeline (version 1.8.0) was applied to raw sequences for comparisons,
i.e., sequences were quality filtered according to QIIME default values and binned into clusters (OTUs) using UCLUST at 97.0% similarity. Potential chimaeras were removed (USEARCH), and
taxonomic determination was performed by BLAST against the HOMD database for oral bacteria (www.HOMD.org). The named or unnamed species or phylotypes were identified by their HOMD Human Oral
Taxon (HOT) identity. OTUs with <10 sequences were excluded. For PacBio SMRT amplicon sequencing, hairpin adaptors were ligated to V1-V8 hypervariable regions of the 16S rDNA, sample
barcodes were added to the end of the amplicons, and standard PacBio SMRT bell adapters were ligated to the barcoded amplicon24. The obtained sequences were quality filtered as described
above, and potential chimaera sequences were removed. These steps were performed at GATC. Chimaera-free OTU clusters were taxonomically identified by BLAST against the HOMD database at 98.5%
similarity. The original sequencing data are available from the Figshare (accession link: 10.6084/m9.figshare.4552849). COMPLEMENTARY PCR AND CULTIVATION The presence of _S. mutans_ in
saliva and tooth biofilm was evaluated by PCR using the KAPA2G Robust HotStart PCR Ready Mix (2´) kit (Kapa Biosystems, Boston, MA, USA) as described previously11. In addition, colony
forming units (CFUs) per ml of saliva for mutans streptococci (_S. mutans_ and _S. sobrinus_) were assessed in fresh saliva by cultivation on mitis salivarius sucrose agar supplemented with
0.2 U of bacitracin (Becton, Dickinson and Company, Stockholm, Sweden) and incubated at 37 °C in 5% CO2 for 48 h. STATISTICAL ANALYSES Normally distributed variables are presented as means
with 95% confidence intervals, and group differences were analysed using ANOVA or unpaired _t_-tests. The bacterial taxa mean percentage of all sequences (abundance) and the proportion of
adolescents in whom the taxa were identified (prevalence) were calculated and tested with non-parametric tests (Mann-Whitney U and Chi-squared/Fisher’s exact test, respectively).
Correlations were evaluated by Spearman correlation coefficients. The mean caries increment was adjusted by sex, BMI, sugar intake, and oral hygiene in general linear modelling (glm). These
analyses were performed using SPSS version 23 (IBM Corporation, Armonk, NY, USA). P-values of taxa prevalence comparisons were evaluated with Benjamini-Hochberg corrections for multiple
testing, i.e., p-values ≤ 0.001 were considered significant. For other comparisons, a p-value <0.05 was considered significant. Tests were two-sided. Rarefaction curves plotting the
number of observed species as a function of the number of sequences per sample were established to compare the microbial richness (α diversity) among the samples. Principal component
analysis (PCA) was used to explore the clustering of subjects by the ProbeSeq taxa, and principal coordinate analysis (PCoA) was used to define clustering by QIIME identified OTUs. The
Linear Discriminant Analysis Effect Size (LEfSe) algorithm was used to identify taxa (genus level or higher) that differed in relative abundance between the two groups38. The online Galaxy
Version 1.0 interface (http://huttenhower.sph.harvard.edu) was used, and the threshold for the logarithmic LDA score was set at 1.0. The results are displayed in a cladogram and a bar graph.
Partial least square (PLS) regression was used to identify taxa associated with the caries status. The PLS models included taxa prevalence and potential lifestyle confounders, i.e., sex,
BMI, oral hygiene, sugar intake, and tobacco use. The results are presented as PLS correlation coefficients from a column loading plot. The variables for which the 95% CI did not include
zero were considered significant. SIMCA P+ version 12.0 (Umetrics AB, Umeå, Sweden) was used for PCA and PLS. ETHICAL APPROVAL The study was approved by the Regional Ethical Review Board in
Umeå, Sweden (Dnr 2012-111-31 M) with an addendum for the 2-year follow up (Dnr 2015-389-32 M). All experiments and data collections were performed in accordance with relevant guidelines and
regulations, including that all adolescents and their caregivers provided informed consent and data collection and handling followed the Helsinki declaration and the Swedish Law on personal
data act (PuL). REFERENCES * Selwitz, R. H., Ismail, A. I. & Pitts, N. B. Dental caries. _Lancet_ 369, 51–59 (2007). Article CAS PubMed Google Scholar * Chapple, I. L. _et al_.
Interaction of lifestyle, behaviour or systemic diseases with dental caries and periodontal diseases: consensus report of group 2 of the joint EFP/ORCA workshop on the boundaries between
caries and periodontal diseases. _J. Clin. Periodontol._ 44(Suppl 18), S39–S51 (2017). Article PubMed Google Scholar * Dewhirst, F. E. _et al_. The human oral microbiome. _J. Bacteriol._
192, 5002–5017 (2010). Article CAS PubMed PubMed Central Google Scholar * Kilian, M. _et al_. The oral microbiome - an update for oral healthcare professionals. _Br. Dent. J._ 221,
657–666 (2016). Article CAS PubMed Google Scholar * Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E. & Borisy, G. G. Biogeography of a human oral microbiome at the
micron scale. _Proc. Natl. Acad. Sci. USA_ 113, E791–800 (2016). Article ADS CAS PubMed PubMed Central Google Scholar * Takahashi, N. & Nyvad, B. The role of bacteria in the caries
process: ecological perspectives. _J. Dent. Res._ 90, 294–303 (2011). Article CAS PubMed Google Scholar * Ruoff, K. L. Nutritionally variant streptococci. _Clin. Microbiol. Rev._ 4,
184–190 (1991). Article CAS PubMed PubMed Central Google Scholar * Ma, C. _et al_. Comparison of Oral Microbial Profiles between Children with Severe Early Childhood Caries and
Caries-Free Children Using the Human Oral Microbe Identification Microarray. _PLoS One_ 10, e0122075 (2015). Article PubMed PubMed Central Google Scholar * Tanner, A. C. _et al_.
Microbiota of severe early childhood caries before and after therapy. _J. Dent. Res._ 90, 1298–1305 (2011). Article CAS PubMed PubMed Central Google Scholar * Mantzourani, M. _et al_.
The isolation of bifidobacteria from occlusal carious lesions in children and adults. _Caries Res._ 43, 308–313 (2009). Article CAS PubMed Google Scholar * Johansson, I., Witkowska, E.,
Kaveh, B., Lif Holgerson, P. & Tanner, A. C. The Microbiome in Populations with a Low and High Prevalence of Caries. _J. Dent. Res._ 95, 80–86 (2016). Article CAS PubMed PubMed
Central Google Scholar * Klock, B. & Krasse, B. Caries status and microbial conditions in children in 1973 and 1984. _Scand. J. Dent. Res._ 95, 13–17 (1987). CAS PubMed Google
Scholar * Yang, F. _et al_. Saliva microbiomes distinguish caries-active from healthy human populations. _ISME J_ 6, 1–10 (2012). Article PubMed Google Scholar * Belstrom, D., Paster, B.
J., Fiehn, N. E., Bardow, A. & Holmstrup, P. Salivary bacterial fingerprints of established oral disease revealed by the Human Oral Microbe Identification using Next Generation
Sequencing (HOMINGS) technique. _J. Oral Microbiol_ 8, 30170 (2016). Article PubMed Google Scholar * Zhou, J. _et al_. Influences of pH and iron on the salivary microbiome in individuals
with and without caries. _Appl. Environ. Microbiol_. Epub ahead of print (2016). * Franzen, O. _et al_. Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic
hierarchical clustering. _Microbiome_ 3, 43 (2015). Article PubMed PubMed Central Google Scholar * Chen, T. _et al_. The Human Oral Microbiome Database: a web accessible resource for
investigating oral microbe taxonomic and genomic information. _Database_ (_Oxford_), baq013 (2010). * Splieth, C. H., Christiansen, J. & Foster Page, L. A. Caries Epidemiology and
Community Dentistry: Chances for Future Improvements in Caries Risk Groups. Outcomes of the ORCA Saturday Afternoon Symposium, Greifswald, 2014. Part 1. _Caries Res._ 50, 9–16 (2016).
Article PubMed Google Scholar * Caporaso, J. G. _et al_. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. _Proc. Natl. Acad. Sci. USA._ 108(Suppl 1),
4516–4522 (2011). Article ADS CAS PubMed Google Scholar * Benitez-Paez, A., Belda-Ferre, P., Simon-Soro, A. & Mira, A. Microbiota diversity and gene expression dynamics in human
oral biofilms. _BMC Genomics_ 15, 311 (2014). Article PubMed PubMed Central Google Scholar * Belda-Ferre, P. _et al_. The human oral metaproteome reveals potential biomarkers for caries
disease. _Proteomics_ 15, 3497–3507 (2015). Article CAS PubMed Google Scholar * Wade, W. G. The oral microbiome in health and disease. _Pharmacol. Res._ 69, 137–143 (2013). Article CAS
PubMed Google Scholar * Zhou, J. _et al_. Exploration of Human Salivary Microbiomes–Insights into the Novel Characteristics of Microbial Community Structure in Caries and Caries-Free
Subjects. _PLoS One_ 11, e0147039 (2016). Article PubMed PubMed Central Google Scholar * Schloss, P. D., Jenior, M. L., Koumpouras, C. C., Westcott, S. L. & Highlander, S. K.
Sequencing 16S rRNA gene fragments using the PacBio SMRT DNA sequencing system. _PeerJ_ 4, e1869 (2016). Article PubMed PubMed Central Google Scholar * Wagner, J. _et al_. Evaluation of
PacBio sequencing for full-length bacterial 16S rRNA gene classification. _BMC Microbiol._ 16, 274 (2016). Article PubMed PubMed Central Google Scholar * Kolenbrander, P. E. Intergeneric
coaggregation among human oral bacteria and ecology of dental plaque. _Annu. Rev. Microbiol._ 42, 627–656 (1988). Article CAS PubMed Google Scholar * Heller, D., Helmerhorst, E. J.
& Oppenheim, F. G. Saliva and Serum Protein Exchange at the Tooth Enamel Surface. _J. Dent. Res_. Epub ahead of print (2016). * David, L. A. _et al_. Host lifestyle affects human
microbiota on daily timescales. _Genome Biol._ 15, R89 (2014). Article PubMed PubMed Central Google Scholar * Norderyd, O. _et al_. Oral health of individuals aged 3–80 years in
Jonkoping, Sweden during 40 years (1973–2013). II. Review of clinical and radiographic findings. _Swed. Dent. J._ 39, 69–86 (2015). PubMed Google Scholar * Gomes, B. P., Berber, V. B.,
Kokaras, A. S., Chen, T. & Paster, B. J. Microbiomes of Endodontic-Periodontal Lesions before and after Chemomechanical Preparation. _J. Endod._ 41, 1975–1984 (2015). Article PubMed
Google Scholar * Simon-Soro, A. _et al_. Microbial geography of the oral cavity. _J. Dent. Res._ 92, 616–621 (2013). Article CAS PubMed Google Scholar * Nyvad, B., Crielaard, W., Mira,
A., Takahashi, N. & Beighton, D. Dental caries from a molecular microbiological perspective. _Caries Res._ 47, 89–102 (2013). Article CAS PubMed Google Scholar * Foxman, B. _et al_.
The effects of family, dentition, and dental caries on the salivary microbiome. _Ann. Epidemiol._ 26, 348–354 (2016). Article PubMed PubMed Central Google Scholar * Tanner, A. C.
Anaerobic culture to detect periodontal and caries pathogens. _J. Oral Biosci._ 57, 18–26 (2015). Article PubMed Google Scholar * Henne, K., Rheinberg, A., Melzer-Krick, B. & Conrads,
G. Aciduric microbial taxa including Scardovia wiggsiae and Bifidobacterium spp. in caries and caries free subjects. _Anaerobe_ 35, 60–65 (2015). Article PubMed Google Scholar * Valdez,
R. M. _et al_. Comparative _in vitro_ investigation of the cariogenic potential of bifidobacteria. _Arch. Oral Biol._ 71, 97–103 (2016). Article CAS PubMed Google Scholar * Soderstrom,
U., Johansson, I. & Sunnegardh-Gronberg, K. A retrospective analysis of caries treatment and development in relation to assessed caries risk in an adult population in Sweden. _BMC Oral
Health_ 14, 126 (2014). Article PubMed PubMed Central Google Scholar * Segata, N. _et al_. Metagenomic biomarker discovery and explanation. _Genome Biol._ 12, R60 (2011). Article PubMed
PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS The present study was supported by grants from The Swedish Patent Revenue Foundation and TUA, the County Council of
Västerbotten, Sweden. None of the funding bodies had any influence on the design, data collection, analysis, interpretation or writing of the manuscript. Carina Öhman and Agnetha Rönnlund
are acknowledged for skilful laboratory work. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Odontology, section of Cariology, Umeå University, Umeå, Sweden Linda Eriksson &
Ingegerd Johansson * Department of Odontology,section of Pedodontics, Umeå University, Umeå, Sweden Linda Eriksson & Pernilla Lif Holgerson Authors * Linda Eriksson View author
publications You can also search for this author inPubMed Google Scholar * Pernilla Lif Holgerson View author publications You can also search for this author inPubMed Google Scholar *
Ingegerd Johansson View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS I.J. and P.L.H. designed the study, granted various permissions, and
provided laboratory supplies, L.E. and I.J. compiled and analysed data, L.E. and I.J. drafted the manuscript and all authors contributed to the final article. CORRESPONDING AUTHOR
Correspondence to Ingegerd Johansson. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare that they have no competing interests. ADDITIONAL INFORMATION PUBLISHER'S NOTE:
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. ELECTRONIC SUPPLEMENTARY MATERIAL SUPPLEMENTARY FIGURES S1 AND S2
DATASET 1 RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and
reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if
changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the
material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to
obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Eriksson, L., Lif Holgerson, P. & Johansson, I. Saliva and tooth biofilm bacterial microbiota in adolescents in a low caries community. _Sci Rep_ 7, 5861 (2017).
https://doi.org/10.1038/s41598-017-06221-z Download citation * Received: 30 January 2017 * Accepted: 08 June 2017 * Published: 19 July 2017 * DOI: https://doi.org/10.1038/s41598-017-06221-z
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative



:max_bytes(150000):strip_icc():focal(749x0:751x2)/Travis-Kelce-Brock-Perdy-Patrick-Mahomes-super-bowl-2024-021124-1b558979da1e49e6b1583d5a0773eadd.jpg)