- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
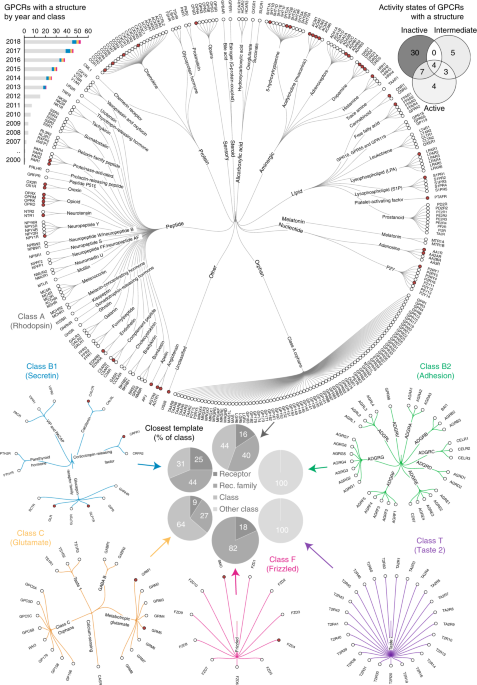
ABSTRACT G-protein-coupled receptors (GPCRs) transduce physiological and sensory stimuli into appropriate cellular responses and mediate the actions of one-third of drugs. GPCR structural
studies have revealed the general bases of receptor activation, signaling, drug action and allosteric modulation, but so far cover only 13% of nonolfactory receptors. We broadly surveyed the
receptor modifications/engineering and methods used to produce all available GPCR crystal and cryo-electron microscopy (cryo-EM) structures, and present an interactive resource integrated
in GPCRdb (http://www.gpcrdb.org) to assist users in designing constructs and browsing appropriate experimental conditions for structure studies. Access through your institution Buy or
subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get
Nature+, our best-value online-access subscription $32.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $259.00 per year only
$21.58 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout
ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS AN ONLINE GPCR STRUCTURE
ANALYSIS PLATFORM Article 10 November 2021 ALPHAFOLD3 VERSUS EXPERIMENTAL STRUCTURES: ASSESSMENT OF THE ACCURACY IN LIGAND-BOUND G PROTEIN-COUPLED RECEPTORS Article 06 December 2024
ALPHAFOLD2 VERSUS EXPERIMENTAL STRUCTURES: EVALUATION ON G PROTEIN-COUPLED RECEPTORS Article 01 July 2022 REFERENCES * Hauser, A. S., Attwood, M. M., Rask-Andersen, M., Schiöth, H. B. &
Gloriam, D. E. Trends in GPCR drug discovery: new agents, targets and indications. _Nat. Rev. Drug Discov._ 16, 829–842 (2017). LANDMARK REFERENCE FOR GPCR DRUGS, TARGETS AND INDICATIONS,
DESCRIBING RECENT DRUG DISCOVERY SUCCESSES AND NEW STRATEGIES IN CLINICAL TRIALS. Article CAS Google Scholar * Anonymous. Structure statistics. _GPCRdb_
http://gpcrdb.org/structure/statistics (2018). * Rasmussen, S. G. et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. _Nature_ 477, 549–555 (2011). Article CAS
Google Scholar * Liang, Y. L. et al. Phase-plate cryo-EM structure of a class B GPCR–G-protein complex. _Nature_ 546, 118–123 (2017). Article CAS Google Scholar * Koehl, A. et al.
Structure of the µ-opioid receptor–Gi protein complex. _Nature_ 558, 547–552 (2018). Article CAS Google Scholar * Kang, Y. et al. Cryo-EM structure of human rhodopsin bound to an
inhibitory G protein. _Nature_ 558, 553–558 (2018). Article CAS Google Scholar * Kang, Y. et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. _Nature_ 523,
561–567 (2015). Article CAS Google Scholar * Tautermann, C. S. & Gloriam, D. E. Editorial overview: New technologies: GPCR drug design and function—exploiting the current (of)
structures. _Curr. Opin. Pharmacol._ 30, vii–x (2016). Article CAS Google Scholar * Manglik, A. et al. Structural insights into the dynamic process of β2-adrenergic receptor signaling.
_Cell_ 161, 1101–1111 (2015). Article CAS Google Scholar * Van Eps, N. et al. Conformational equilibria of light-activated rhodopsin in nanodiscs. _Proc. Natl Acad. Sci. USA_ 114,
E3268–E3275 (2017). Article Google Scholar * Staus, D. P. et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. _Nature_
535, 448–452 (2016). Article CAS Google Scholar * Ye, L., Van Eps, N., Zimmer, M., Ernst, O. P. & Prosser, R. S. Activation of the A2A adenosine G-protein-coupled receptor by
conformational selection. _Nature_ 533, 265–268 (2016). Article CAS Google Scholar * Van Eps, N. et al. Gi- and Gs-coupled GPCRs show different modes of G-protein binding. _Proc. Natl
Acad. Sci. USA_ 115, 2383–2388 (2018). Article Google Scholar * Violin, J. D., Crombie, A. L., Soergel, D. G. & Lark, M. W. Biased ligands at G-protein-coupled receptors: promise and
progress. _Trends Pharmacol. Sci._ 35, 308–316 (2014). Article CAS Google Scholar * Congreve, M., Oswald, C. & Marshall, F. H. Applying structure-based drug design approaches to
allosteric modulators of GPCRs. _Trends Pharmacol. Sci._ 38, 837–847 (2017). Article CAS Google Scholar * Munk, C., Harpsøe, K., Hauser, A. S., Isberg, V. & Gloriam, D. E. Integrating
structural and mutagenesis data to elucidate GPCR ligand binding. _Curr. Opin. Pharmacol._ 30, 51–58 (2016). Article CAS Google Scholar * Isberg, V. et al. GPCRdb: an information system
for G protein-coupled receptors. _Nucleic Acids Res._ 44, D356–D364 (2016). Article CAS Google Scholar * Isberg, V. et al. GPCRDB: an information system for G protein-coupled receptors.
_Nucleic Acids Res._ 42, D422–D425 (2014). Article CAS Google Scholar * Flock, T. et al. Selectivity determinants of GPCR–G-protein binding. _Nature_ 545, 317–322 (2017). Article CAS
Google Scholar * Pándy-Szekeres, G. et al. GPCRdb in 2018: adding GPCR structure models and ligands. _Nucleic Acids Res._ 46, D440–D446 (2018). Article Google Scholar * Munk, C. et al.
GPCRdb: the G protein-coupled receptor database—an introduction. _Br. J. Pharmacol._ 173, 2195–2207 (2016). Article CAS Google Scholar * Rose, P. W. et al. The RCSB Protein Data Bank:
integrative view of protein, gene and 3D structural information. _Nucleic Acids Res._ 45, D271–D281 (2017). Article CAS Google Scholar * Velankar, S. et al. SIFTS: Structure Integration
with Function, Taxonomy and Sequences resource. _Nucleic Acids Res._ 41, D483–D489 (2013). RESOURCE FOR RESIDUE-LEVEL MAPPING OF UNIPROT (PROTEIN) AND PDB (STRUCTURE) ENTRIES THAT ALSO
INTEGRATES ANNOTATIONS FROM MANY MORE MAJOR DATABASES. Article CAS Google Scholar * Anonymous. Construct alignments. _GPCRdb_ http://gpcrdb.org/construct/ (2018). * Hutchings, C. J.,
Koglin, M., Olson, W. C. & Marshall, F. H. Opportunities for therapeutic antibodies directed at G-protein-coupled receptors. _Nat. Rev. Drug Discov._ 16, 787–810 (2017). Article CAS
Google Scholar * Anonymous. Structure. _GPCRdb_ http://gpcrdb.org/structure (2018). * Renaud, J. P. et al. Cryo-EM in drug discovery: achievements, limitations and prospects. _Nat. Rev.
Drug Discov._ 17, 471–492 (2018). REVIEWS THE RECENT ADVANCES IN CRYO-EM AND PROVIDES AN OUTLOOK OF WHAT TO EXPECT IN THE NEAR FUTURE. Article CAS Google Scholar * García-Nafría, J., Lee,
Y., Bai, X., Carpenter, B. & Tate, C. G. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. _eLife_ 7, e35946 (2018). Article Google
Scholar * Su, X. et al. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. _Science_ 357, 815–820 (2017). Article CAS Google Scholar * Liang, Y. L. et al.
Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor–Gs complex. _Nature_ 555, 121–125 (2018). Article CAS Google Scholar * Khoshouei, M., Radjainia, M.,
Baumeister, W. & Danev, R. Cryo-EM structure of haemoglobin at 3.2 Å determined with the Volta phase plate. _Nat. Commun._ 8, 16099 (2017). Article CAS Google Scholar * Zhang, Y. et
al. Cryo-EM structure of the activated GLP-1 receptor in complex with a G protein. _Nature_ 546, 248–253 (2017). Article CAS Google Scholar * Draper-Joyce, C. J. et al. Structure of the
adenosine-bound human adenosine A1 receptor–Gi complex. _Nature_ 558, 559–563 (2018). Article CAS Google Scholar * Tsai, C. J. et al. Crystal structure of rhodopsin in complex with a
mini-Go sheds light on the principles of G protein selectivity. _Sci. Adv._ 4, eaat7052 (2018). Article Google Scholar * García-Nafría, J., Nehmé, R., Edwards, P. C. & Tate, C. G.
Cryo-EM structure of the serotonin 5-HT1B receptor coupled to heterotrimeric Go. _Nature_ 558, 620–623 (2018). Article Google Scholar * Chun, E. et al. Fusion partner toolchest for the
stabilization and crystallization of G protein-coupled receptors. _Structure_ 20, 967–976 (2012). Article CAS Google Scholar * Anonymous. Fusion construct analysis. _GPCRdb_
http://gpcrdb.org/construct/analysis#fusions (2018). * Isberg, V. et al. Generic GPCR residue numbers—aligning topology maps while minding the gaps. _Trends Pharmacol. Sci._ 36, 22–31
(2015). GENERIC NUMBERING OF RECEPTOR RESIDUES CRUCIAL FOR ALL GPCR STRUCTURE–FUNCTION STUDIES. Article CAS Google Scholar * Anonymous. Mutation construct analysis. _GPCRdb_
http://gpcrdb.org/construct/analysis#mutations (2018). * Pace, C. N. & Scholtz, J. M. A helix propensity scale based on experimental studies of peptides and proteins. _Biophys. J._ 75,
422–427 (1998). Article CAS Google Scholar * Jazayeri, A. et al. Crystal structure of the GLP-1 receptor bound to a peptide agonist. _Nature_ 546, 254–258 (2017). Article CAS Google
Scholar * Venkatakrishnan, A. J. et al. Molecular signatures of G-protein-coupled receptors. _Nature_ 494, 185–194 (2013). PIONEERING GPCR STRUCTURE ANALYSIS UNCOVERING COMMON CONTACT
NETWORKS STABILIZING THE RECEPTOR FOLD, AND CHARACTERISTIC FEATURES OF LIGAND BINDING AND RECEPTOR ACTIVATION. Article CAS Google Scholar * Anonymous. Stabilising Mutation Analyser.
_GPCRdb_ http://gpcrdb.org/construct/stabilisation (2018). * Roth, C. B., Hanson, M. A. & Stevens, R. C. Stabilization of the human β2-adrenergic receptor TM4-TM3-TM5 helix interface by
mutagenesis of Glu122(3.41), a critical residue in GPCR structure. _J. Mol. Biol._ 376, 1305–1319 (2008). Article CAS Google Scholar * White, K. L. et al. Structural connection between
activation microswitch and allosteric sodium site in GPCR signaling. _Structure_ 26, 259–269 (2018). Article CAS Google Scholar * Nygaard, R., Frimurer, T. M., Holst, B., Rosenkilde, M.
M. & Schwartz, T. W. Ligand binding and micro-switches in 7TM receptor structures. _Trends Pharmacol. Sci._ 30, 249–259 (2009). Article CAS Google Scholar * Katritch, V., Cherezov, V.
& Stevens, R. C. Structure-function of the G protein-coupled receptor superfamily. _Annu. Rev. Pharmacol. Toxicol._ 53, 531–556 (2013). Article CAS Google Scholar * Carpenter, B.
& Tate, C. G. Active state structures of G protein-coupled receptors highlight the similarities and differences in the G protein and arrestin coupling interfaces. _Curr. Opin. Struct.
Biol._ 45, 124–132 (2017). Article CAS Google Scholar * Anonymous. Truncation analysis. _GPCRdb_ http://gpcrdb.org/construct/analysis#truncations (2018). * Cilia, E., Pancsa, R., Tompa,
P., Lenaerts, T. & Vranken, W. F. The DynaMine webserver: predicting protein dynamics from sequence. _Nucleic Acids Res._ 42, W264–W270 (2014). Article CAS Google Scholar * Petersen,
T. N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. _Nat. Methods_ 8, 785–786 (2011). Article CAS Google Scholar *
Anonymous. Mutation rules. _GPCRdb_ http://files.gpcrdb.org/mutation_rules.html (2018). * Anonymous. Local installation. _GPCRdb_ http://docs.gpcrdb.org/local_installation.html (2018). *
Peng, Y. et al. 5-HT2C receptor structures reveal the structural basis of GPCR polypharmacology. _Cell_ 172, 719–730 (2018). Article CAS Google Scholar * Magnani, F., Shibata, Y.,
Serrano-Vega, M. J. & Tate, C. G. Co-evolving stability and conformational homogeneity of the human adenosine A2a receptor. _Proc. Natl Acad. Sci. USA_ 105, 10744–10749 (2008). Article
CAS Google Scholar * Anonymous. Experiment Browser. _GPCRdb_ http://gpcrdb.org/construct/experiments (2018). * Zhang, X., Stevens, R. C. & Xu, F. The importance of ligands for G
protein-coupled receptor stability. _Trends Biochem. Sci._ 40, 79–87 (2015). Article Google Scholar * Milić, D. & Veprintsev, D. B. Large-scale production and protein engineering of G
protein-coupled receptors for structural studies. _Front. Pharmacol._ 6, 66 (2015). PubMed PubMed Central Google Scholar * Lv, X. et al. In vitro expression and analysis of the 826 human
G protein-coupled receptors. _Protein Cell_ 7, 325–337 (2016). Article CAS Google Scholar * Luo, P. & Baldwin, R. L. Origin of the different strengths of the (i,i+4) and (i,i+3)
leucine pair interactions in helices. _Biophys. Chem._ 96, 103–108 (2002). Article CAS Google Scholar * Anonymous. Mutations. _GPCRdb_ http://gpcrdb.org/construct/mutations (2018).
Download references ACKNOWLEDGEMENTS We acknowledge A. Tsolakou, D. Milic and K.S. Harpsøe for help with data annotation; I. Carson for development of the preliminary version of the
Stabilising Mutation Analyser; and C.-J. Tsai for input on the description of cryo-EM construct engineering and experiments. This work was supported in part by the ERC (Starting Grant 639125
to D.E.G.), the Lundbeck Foundation (grants R163-2013-16327 and R218-2016-1266 to D.E.G.), the Swiss National Science Foundation (grant CRSII2_160805 to X.D.), the European Commisions
Seventh Framework Program (FP7/2007-2013; grant 290605 (COFUND: PSI-FELLOW) to E.M.) and the COST Action CM1207 (‘GLISTEN’). AUTHOR INFORMATION Author notes * Vignir Isberg Present address:
Novozymes A/S, Copenhagen, Denmark * These authors contributed equally: Christian Munk, Eshita Mutt. AUTHORS AND AFFILIATIONS * Department of Drug Design and Pharmacology, University of
Copenhagen, Copenhagen, Denmark Christian Munk, Vignir Isberg, Louise F. Nikolajsen, Janne M. Bibbe & David E. Gloriam * Paul Scherrer Institute, Villigen, Switzerland Eshita Mutt,
Tilman Flock & Xavier Deupi * GPCR Consortium, San Marcos, CA, USA Michael A. Hanson * Departments of Biological Sciences and Chemistry, Bridge Institute, USC Michelson Center for
Convergent Bioscience, University of Southern California, Los Angeles, CA, USA Raymond C. Stevens * iHuman Institute, ShanghaiTech University, Shanghai, China Raymond C. Stevens Authors *
Christian Munk View author publications You can also search for this author inPubMed Google Scholar * Eshita Mutt View author publications You can also search for this author inPubMed Google
Scholar * Vignir Isberg View author publications You can also search for this author inPubMed Google Scholar * Louise F. Nikolajsen View author publications You can also search for this
author inPubMed Google Scholar * Janne M. Bibbe View author publications You can also search for this author inPubMed Google Scholar * Tilman Flock View author publications You can also
search for this author inPubMed Google Scholar * Michael A. Hanson View author publications You can also search for this author inPubMed Google Scholar * Raymond C. Stevens View author
publications You can also search for this author inPubMed Google Scholar * Xavier Deupi View author publications You can also search for this author inPubMed Google Scholar * David E.
Gloriam View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS C.M., D.E.G., J.M.B. and T.F. made the construct analyses and figures; C.M. and
V.I. developed the online resources; E.M. and C.M. annotated and analyzed published experimental data; L.F.N. conducted the mutagenesis experiments; M.A.H. and R.C.S. provided critical input
on the project, manuscript writing and data analysis; X.D. and D.E.G. drafted the paper; all authors commented on the drafted manuscript; D.E.G. and X.D. designed the project; and D.E.G.
managed the project. CORRESPONDING AUTHORS Correspondence to Christian Munk or David E. Gloriam. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests.
ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION
SUPPLEMENTARY TEXT AND FIGURES Supplementary Note 1 and Supplementary Tables 1–5 RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Munk, C., Mutt, E.,
Isberg, V. _et al._ An online resource for GPCR structure determination and analysis. _Nat Methods_ 16, 151–162 (2019). https://doi.org/10.1038/s41592-018-0302-x Download citation *
Received: 18 April 2018 * Accepted: 14 December 2018 * Published: 21 January 2019 * Issue Date: February 2019 * DOI: https://doi.org/10.1038/s41592-018-0302-x SHARE THIS ARTICLE Anyone you
share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the
Springer Nature SharedIt content-sharing initiative