- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
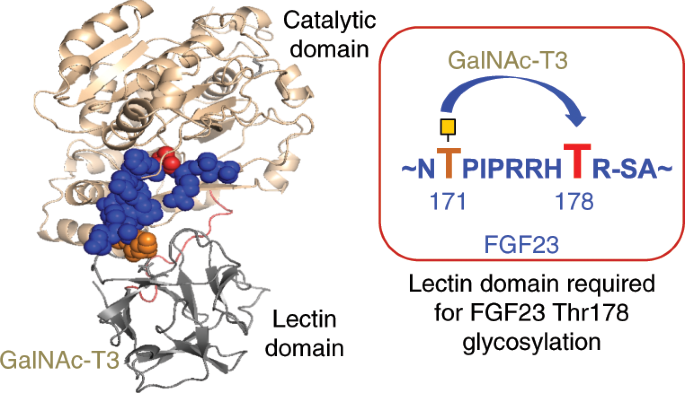
ABSTRACT Polypeptide GalNAc-transferase T3 (GalNAc-T3) regulates fibroblast growth factor 23 (FGF23) by _O_-glycosylating Thr178 in a furin proprotein processing motif RHT178R↓S. FGF23
regulates phosphate homeostasis and deficiency in _GALNT3_ or _FGF23_ results in hyperphosphatemia and familial tumoral calcinosis. We explored the molecular mechanism for GalNAc-T3
glycosylation of FGF23 using engineered cell models and biophysical studies including kinetics, molecular dynamics and X-ray crystallography of GalNAc-T3 complexed to glycopeptide
substrates. GalNAc-T3 uses a lectin domain mediated mechanism to glycosylate Thr178 requiring previous glycosylation at Thr171. Notably, Thr178 is a poor substrate site with limiting
glycosylation due to substrate clashes leading to destabilization of the catalytic domain flexible loop. We suggest GalNAc-T3 specificity for FGF23 and its ability to control circulating
levels of intact FGF23 is achieved by FGF23 being a poor substrate. GalNAc-T3’s structure further reveals the molecular bases for reported disease-causing mutations. Our findings provide an
insight into how GalNAc-T isoenzymes achieve isoenzyme-specific nonredundant functions. Access through your institution Buy or subscribe This is a preview of subscription content, access via
your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days
cancel any time Learn more Subscribe to this journal Receive 12 print issues and online access $259.00 per year only $21.58 per issue Learn more Buy this article * Purchase on SpringerLink *
Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional
subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS CRYSTAL STRUCTURE OF LRG1 AND THE FUNCTIONAL SIGNIFICANCE OF LRG1 GLYCAN FOR LPHN2 ACTIVATION
Article Open access 01 May 2023 MOLECULAR STRUCTURE AND ENZYMATIC MECHANISM OF THE HUMAN COLLAGEN HYDROXYLYSINE GALACTOSYLTRANSFERASE GLT25D1/COLGALT1 Article Open access 16 April 2025
GLYCOSYLTRANSFERASE 8 DOMAIN-CONTAINING PROTEIN 1 (GLT8D1) IS A UDP-DEPENDENT GALACTOSYLTRANSFERASE Article Open access 07 December 2023 DATA AVAILABILITY The crystal structures of
_Tg_GalNAc-T3-UDP-P3 and _Tg_GalNAc-T3-UDP–FGF23c complexes were deposited at the RCSB PDB with accession codes 6S24 and 6S22, respectively. REFERENCES * Hurtado-Guerrero, R. Recent
structural and mechanistic insights into protein _O_-GalNAc glycosylation. _Biochem Soc. Trans._ 44, 61–67 (2016). CAS PubMed Google Scholar * Bennett, E. P. et al. Control of mucin-type
_O_-glycosylation: a classification of the polypeptide GalNAc-transferase gene family. _Glycobiology_ 22, 736–756 (2012). CAS PubMed Google Scholar * de Las Rivas, M., Lira-Navarrete, E.,
Gerken, T. A. & Hurtado-Guerrero, R. Polypeptide GalNAc-Ts: from redundancy to specificity. _Curr. Opin. Struct. Biol._ 56, 87–96 (2019). PubMed Google Scholar * Lira-Navarrete, E. et
al. Substrate-guided front-face reaction revealed by combined structural snapshots and metadynamics for the polypeptide _N_-acetylgalactosaminyltransferase 2. _Angew. Chem. Int. Ed. Engl._
53, 8206–8210 (2014). CAS PubMed Google Scholar * Gerken, T. A. et al. Emerging paradigms for the initiation of mucin-type protein _O_-glycosylation by the polypeptide GalNAc transferase
family of glycosyltransferases. _J. Biol. Chem._ 286, 14493–14507 (2011). CAS PubMed PubMed Central Google Scholar * Gerken, T. A. et al. The lectin domain of the polypeptide GalNAc
transferase family of glycosyltransferases (ppGalNAc Ts) acts as a switch directing glycopeptide substrate glycosylation in an N- or C-terminal direction, further controlling mucin type
_O_-glycosylation. _J. Biol. Chem._ 288, 19900–19914 (2013). CAS PubMed PubMed Central Google Scholar * Revoredo, L. et al. Mucin-type _O_-glycosylation is controlled by short- and
long-range glycopeptide substrate recognition that varies among members of the polypeptide GalNAc transferase family. _Glycobiology_ 26, 360–376 (2016). CAS PubMed Google Scholar *
Lira-Navarrete, E. et al. Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein _O_-glycosylation. _Nat. Commun._ 6, 6937 (2015). CAS PubMed
Google Scholar * Rivas, M. L. et al. The interdomain flexible linker of the polypeptide GalNAc transferases dictates their long-range glycosylation preferences. _Nat. Commun._ 8, 1959
(2017). PubMed Central Google Scholar * de Las Rivas, M. et al. Structural analysis of a GalNAc-T2 mutant reveals an induced-fit catalytic mechanism for GalNAc-Ts. _Chemistry_ 24,
8382–8392 (2018). PubMed Google Scholar * Topaz, O. et al. Mutations in GALNT3, encoding a protein involved in _O_-linked glycosylation, cause familial tumoral calcinosis. _Nat. Genet._
36, 579–581 (2004). CAS PubMed Google Scholar * Kato, K. et al. Polypeptide GalNAc-transferase T3 and familial tumoral calcinosis. Secretion of fibroblast growth factor 23 requires
_O_-glycosylation. _J. Biol. Chem._ 281, 18370–18377 (2006). CAS PubMed Google Scholar * White, K. E. et al. Autosomal dominant hypophosphataemic rickets is associated with mutations in
FGF23. _Nat. Genet_ 26, 345–348 (2000). CAS Google Scholar * Benet-Pages, A., Orlik, P., Strom, T. M. & Lorenz-Depiereux, B. An FGF23 missense mutation causes familial tumoral
calcinosis with hyperphosphatemia. _Hum. Mol. Genet._ 14, 385–390 (2005). CAS PubMed Google Scholar * Simpson, M. A. et al. Mutations in FAM20C also identified in non-lethal
osteosclerotic bone dysplasia. _Clin. Genet._ 75, 271–276 (2009). CAS PubMed Google Scholar * Tagliabracci, V. S. et al. Dynamic regulation of FGF23 by Fam20C phosphorylation, GalNAc-T3
glycosylation, and furin proteolysis. _Proc. Natl Acad. Sci. USA_ 111, 5520–5525 (2014). CAS PubMed PubMed Central Google Scholar * Urakawa, I. et al. Klotho converts canonical FGF
receptor into a specific receptor for FGF23. _Nature_ 444, 770–774 (2006). CAS PubMed Google Scholar * Bennett, E. P., Hassan, H. & Clausen, H. cDNA cloning and expression of a novel
human UDP-N-acetyl-alpha-d-galactosamine. Polypeptide _N_-acetylgalactosaminyltransferase, GalNAc-t3. _J. Biol. Chem._ 271, 17006–17012 (1996). CAS PubMed Google Scholar * Kong, Y. et al.
Probing polypeptide GalNAc-transferase isoform substrate specificities by in vitro analysis. _Glycobiology_ 25, 55–65 (2015). CAS PubMed Google Scholar * Schjoldager, K. T. et al.
Deconstruction of _O_-glycosylation–GalNAc-T isoforms direct distinct subsets of the _O_-glycoproteome. _EMBO Rep._ 16, 1713–1722 (2015). CAS PubMed PubMed Central Google Scholar *
Khetarpal, S. A. et al. Loss of function of GALNT2 lowers high-density lipoproteins in humans, nonhuman primates, and rodents. _Cell Metab._ 24, 234–245 (2016). CAS PubMed PubMed Central
Google Scholar * Wang, S. et al. Site-specific _O_-glycosylation of members of the low-density lipoprotein receptor superfamily enhances ligand interactions. _J. Biol. Chem._ 293, 7408–7422
(2018). CAS PubMed PubMed Central Google Scholar * Yoshimura, Y. et al. Elucidation of the sugar recognition ability of the lectin domain of UDP-GalNAc:polypeptide
_N_-acetylgalactosaminyltransferase 3 by using unnatural glycopeptide substrates. _Glycobiology_ 22, 429–438 (2012). CAS PubMed Google Scholar * Frishberg, Y. et al.
Hyperostosis-hyperphosphatemia syndrome: a congenital disorder of _O_-glycosylation associated with augmented processing of fibroblast growth factor 23. _J. Bone Min. Res_ 22, 235–242
(2007). CAS Google Scholar * Chen, G. et al. alpha-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. _Nature_ 553, 461–466 (2018). CAS PubMed PubMed Central
Google Scholar * Hagen, F. K., Hazes, B., Raffo, R., deSa, D. & Tabak, L. A. Structure-function analysis of the UDP-_N_-acetyl-d-galactosamine:polypeptide
_N_-acetylgalactosaminyltransferase. Essential residues lie in a predicted active site cleft resembling a lactose repressor fold. _J. Biol. Chem._ 274, 6797–6803 (1999). CAS PubMed Google
Scholar * Kozarsky, K., Kingsley, D. & Krieger, M. Use of a mutant cell line to study the kinetics and function of _O_-linked glycosylation of low density lipoprotein receptors. _Proc.
Natl Acad. Sci. USA_ 85, 4335–4339 (1988). CAS PubMed PubMed Central Google Scholar * Bar-Even, A. et al. The moderately efficient enzyme: evolutionary and physicochemical trends shaping
enzyme parameters. _Biochemistry_ 50, 4402–4410 (2011). CAS PubMed Google Scholar * de Las Rivas, M. et al. Structural and mechanistic insights into the catalytic-domain-mediated
short-range glycosylation preferences of GalNAc-T4. _ACS Cent. Sci._ 4, 1274–1290 (2018). PubMed Google Scholar * Lira-Navarrete, E. et al. Structural insights into the mechanism of
protein _O_-fucosylation. _PLoS ONE_ 6, e25365 (2011). CAS PubMed PubMed Central Google Scholar * Ghirardello, M. et al. Glycomimetics targeting glycosyltransferases: synthetic,
computational and structural studies of less-polar conjugates. _Chemistry_ 22, 7215–7224 (2016). CAS PubMed Google Scholar * Yamamoto, H. et al. Posttranslational processing of FGF23 in
osteocytes during the osteoblast to osteocyte transition. _Bone_ 84, 120–130 (2016). CAS PubMed Google Scholar * Bennett, E. P. et al. Cloning and characterization of a close homologue of
human UDP-_N_-acetyl-alpha-d-galactosamine: polypeptide _N_-acetylgalactosaminyltransferase-T3, designated GalNAc-T6. Evidence for genetic but not functional redundancy. _J. Biol. Chem._
274, 25362–25370 (1999). CAS PubMed Google Scholar * Joshi, H. J. et al. Glycosyltransferase genes that cause monogenic congenital disorders of glycosylation are distinct from
glycosyltransferase genes associated with complex diseases. _Glycobiology_ 28, 284–294 (2018). CAS PubMed PubMed Central Google Scholar * Minisola, S. et al. Tumour-induced osteomalacia.
_Nat. Rev. Dis. Prim._ 3, 17044 (2017). PubMed Google Scholar * Rafaelsen, S., Johansson, S., Raeder, H. & Bjerknes, R. Long-term clinical outcome and phenotypic variability in
hyperphosphatemic familial tumoral calcinosis and hyperphosphatemic hyperostosis syndrome caused by a novel GALNT3 mutation; case report and review of the literature. _BMC Genet._ 15, 98
(2014). PubMed PubMed Central Google Scholar * Ramnitz, M. S., Gafni, R. I. & Collins, M. T. Hyperphosphatemic familial tumoral calcinosis. in _GeneReviews (R)_ (eds Adams, M. P. et
al.) https://www.ncbi.nlm.nih.gov/books/NBK476672/ (Univ. Wash., Seattle, 2018). * Takashi, Y. et al. Activation of unliganded FGF receptor by extracellular phosphate potentiates proteolytic
protection of FGF23 by its _O_-glycosylation. _Proc. Natl Acad. Sci. USA_ 116, 11418–11427 (2019). CAS PubMed PubMed Central Google Scholar * Hintze, J. et al. Probing the contribution
of individual polypeptide GalNAc-transferase isoforms to the _O_-glycoproteome by inducible expression in isogenic cell lines. _J. Biol. Chem._ 293, 19064–19077 (2018). CAS PubMed PubMed
Central Google Scholar * Wandall, H. H. et al. The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc: lectin binding to
GalNAc-glycopeptide substrates is required for high density GalNAc-_O_-glycosylation. _Glycobiology_ 17, 374–387 (2007). CAS PubMed Google Scholar * Steentoft, C. et al. A validated
collection of mouse monoclonal antibodies to human glycosyltransferases functioning in mucin-type _O_-glycosylation. _Glycobiology_ 29, 645–656 (2019). PubMed PubMed Central Google Scholar
* Yang, Z. et al. Engineered CHO cells for production of diverse, homogeneous glycoproteins. _Nat. Biotechnol._ 33, 842–844 (2015). CAS PubMed Google Scholar * Lonowski, L. A. et al.
Genome editing using FACS enrichment of nuclease-expressing cells and indel detection by amplicon analysis. _Nat. Protoc._ 12, 581–603 (2017). CAS PubMed PubMed Central Google Scholar *
Wandall, H. H. et al. Substrate specificities of three members of the human UDP-N-acetyl-alpha-d-galactosamine: polypeptide _N_-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and
-T3. _J. Biol. Chem._ 272, 23503–23514 (1997). CAS PubMed Google Scholar * Kabsch, W. Xds. _Acta Crystallogr. D._ 66, 125–132 (2010). CAS PubMed PubMed Central Google Scholar * Winn,
M. D. et al. Overview of the CCP4 suite and current developments. _Acta Crystallogr. D._ 67, 235–242 (2011). CAS PubMed PubMed Central Google Scholar * Collaborative Computational
Project, Number 4. The CCP4 suite: programs for protein crystallography. _Acta Crystallogr. D._ 50, 760–763 (1994). * Emsley, P. & Cowtan, K. Coot: model-building tools for molecular
graphics. _Acta Crystallogr. D._ 60, 2126–2132 (2004). PubMed Google Scholar * Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. _Acta Crystallogr.
D._ 67, 355–367 (2011). CAS PubMed PubMed Central Google Scholar * Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray
crystallography using ARP/wARP version 7. _Nat. Protoc._ 3, 1171–1179 (2008). CAS PubMed PubMed Central Google Scholar * Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein
structures and complexes. _Nucleic Acids Res._ 46, W296–W303 (2018). CAS PubMed PubMed Central Google Scholar * Maier, J. A. et al. ff14SB: Improving the accuracy of protein side chain
and backbone parameters from ff99SB. _J. Chem. Theory Comput._ 11, 3696–3713 (2015). CAS PubMed PubMed Central Google Scholar * Plattner, C., Hofener, M. & Sewald, N. One-pot
azidochlorination of glycals. _Org. Lett._ 13, 545–547 (2011). CAS PubMed Google Scholar * Horcas, I. et al. WSXM: a software for scanning probe microscopy and a tool for nanotechnology.
_Rev. Sci. Instrum._ 78, 013705 (2007). CAS PubMed Google Scholar * Lostao, A., Peleato, M. L., Gomez-Moreno, C. & Fillat, M. F. Oligomerization properties of FurA from the
cyanobacterium _Anabaena_ sp. PCC 7120: direct visualization by in situ atomic force microscopy under different redox conditions. _Biochim. Biophys. Acta_ 1804, 1723–1729 (2010). CAS PubMed
Google Scholar * Sun, T., Lin, F. H., Campbell, R. L., Allingham, J. S. & Davies, P. L. An antifreeze protein folds with an interior network of more than 400 semi-clathrate waters.
_Science_ 343, 795–798 (2014). CAS PubMed Google Scholar * Franke, D. et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. _J.
Appl. Crystallogr._ 50, 1212–1225 (2017). CAS PubMed PubMed Central Google Scholar * Tria, G., Mertens, H. D., Kachala, M. & Svergun, D. I. Advanced ensemble modelling of flexible
macromolecules using X-ray solution scattering. _IUCr. J._ 2, 207–217 (2015). CAS Google Scholar * Sali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of spatial
restraints. _J. Mol. Biol._ 234, 779–815 (1993). CAS PubMed Google Scholar * Bernado, P., Mylonas, E., Petoukhov, M. V., Blackledge, M. & Svergun, D. I. Structural characterization of
flexible proteins using small-angle X-ray scattering. _J. Am. Chem. Soc._ 129, 5656–5664 (2007). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank the Diamond Light
Source (Oxford) synchrotron beamline I24 (experiment nos. MX14739-6 and MX14739-11) and the SOLEIL synchrotron (Gif-sur-Yvette) SWING beamline (experiment nos. 99170088). We thank ARAID,
MEC (grant no. CTQ2013-44367-C2-2-P, BFU2016-75633-P and RTI2018-099592-B-C21), the National Institutes of Health (grant no. GM113534 and instrument grant no. GM113534-01S), the Danish
National Research Foundation (grant no. DNRF107), the FCT-Portugal (grant no. UID/Multi/04378/2013) and Gobierno de Aragón (grant nos. E34_R17, E35_17R and LMP58_18) with FEDER (grant no.
2014-2020) funds for ‘Building Europe from Aragón’ for financial support. I.C. thanks the Universidad de La Rioja for the FPI grant. F.M. and H.C. thank FCT-Portugal for IF Investigator
(IF/00780/2015), PTDC/BIA-MIB/31028/2017 and UID/Multi/04378/2019 projects, and PTNMR (grant no. ROTEIRO/0031/2013 and PINFRA/22161/2016). P.B. acknowledges support from the Labex EpiGenMed,
an ‘Investissements d’avenir’ program (grant no. ANR-10-LABX-12-01). The CBS (Montpellier) is a member of France-BioImaging (FBI, ANR-10-INBS-04-01) and the French Infrastructure for
Integrated Structural Biology (FRISBI, ANR-10-INBS-05). The research leading to these results has also received funding from the FP7 (2007–2013) under BioStruct-X (grant agreement nos.
283570 and BIOSTRUCTX_5186). We also thank I. Echániz for technical support and K. Moremen from the University of Georgia, Complex Carbohydrate Research Center, for supplying the
pGEn2-_HsGalNAc-T6_ and pGEn2-_HsGalNAc-T12_ plasmids. AUTHOR INFORMATION Author notes * These authors contributed equally: Matilde de las Rivas, Earnest James Paul Daniel, Yoshiki
Narimatsu, Ismael Compañón. AUTHORS AND AFFILIATIONS * BIFI, University of Zaragoza, Mariano Esquillor s/n, Campus Rio Ebro, Edificio I+D, Zaragoza, Spain Matilde de las Rivas, Laura
Ceballos-Laita & Ramon Hurtado-Guerrero * Department of Biochemistry, Case Western Reserve University, Cleveland, OH, USA Earnest James Paul Daniel & Thomas A. Gerken * Copenhagen
Center for Glycomics, Department of Cellular and Molecular Medicine, School of Dentistry, University of Copenhagen, Copenhagen, Denmark Yoshiki Narimatsu, Kentaro Kato, Lars Hansen, Henrik
Clausen & Ramon Hurtado-Guerrero * Departamento de Química, Universidad de La Rioja, Centro de Investigación en Síntesis Química, Logroño, Spain Ismael Compañón & Francisco Corzana *
Department of Eco-epidemiology, Institute of Tropical Medicine Nagasaki University, Nagasaki, Japan Kentaro Kato * Laboratorio de Microscopías Avanzadas, Instituto de Nanociencia de Aragón,
Universidad de Zaragoza, Zaragoza, Spain Pablo Hermosilla & Anabel Lostao * Swing Beamline, Synchrotron SOLEIL, Gif sur Yvette, France Aurélien Thureau * UCIBIO, REQUIMTE, Departamento
de Química, Faculdade de Ciências e Tecnologia, Universidade de Nova de Lisboa, Caparica, Portugal Helena Coelho & Filipa Marcelo * CIC bioGUNE, Bizkaia Technology Park, Derio, Spain
Helena Coelho * Centre de Biochimie Structurale. INSERM, CNRS, Université de Montpellier, Montpellier, France Pau Bernadó * Department of Hematology, Graduate School of Medicine, Kyoto
University, Kyoto, Japan Ryota Maeda * Fundación ARAID, Zaragoza, Spain Anabel Lostao & Ramon Hurtado-Guerrero * Instituto de Ciencia de Materiales de Aragón, Universidad de
Zaragoza-CSIC, Zaragoza, Spain Anabel Lostao Authors * Matilde de las Rivas View author publications You can also search for this author inPubMed Google Scholar * Earnest James Paul Daniel
View author publications You can also search for this author inPubMed Google Scholar * Yoshiki Narimatsu View author publications You can also search for this author inPubMed Google Scholar
* Ismael Compañón View author publications You can also search for this author inPubMed Google Scholar * Kentaro Kato View author publications You can also search for this author inPubMed
Google Scholar * Pablo Hermosilla View author publications You can also search for this author inPubMed Google Scholar * Aurélien Thureau View author publications You can also search for
this author inPubMed Google Scholar * Laura Ceballos-Laita View author publications You can also search for this author inPubMed Google Scholar * Helena Coelho View author publications You
can also search for this author inPubMed Google Scholar * Pau Bernadó View author publications You can also search for this author inPubMed Google Scholar * Filipa Marcelo View author
publications You can also search for this author inPubMed Google Scholar * Lars Hansen View author publications You can also search for this author inPubMed Google Scholar * Ryota Maeda View
author publications You can also search for this author inPubMed Google Scholar * Anabel Lostao View author publications You can also search for this author inPubMed Google Scholar *
Francisco Corzana View author publications You can also search for this author inPubMed Google Scholar * Henrik Clausen View author publications You can also search for this author inPubMed
Google Scholar * Thomas A. Gerken View author publications You can also search for this author inPubMed Google Scholar * Ramon Hurtado-Guerrero View author publications You can also search
for this author inPubMed Google Scholar CONTRIBUTIONS R.H.-G. designed the crystallization construct and solved the crystal structures. M.R. and R.H.-G. purified the enzymes, crystallized
the complexes and refined the crystal structures. I.C. and F.C. synthesized the glycopeptides. F.C. performed the MD simulations. H.Coelho and F.M. performed and analyzed the NMR
experiments. T.A.G. and E.J.P.D. performed the kinetic studies together with the Edman amino acid sequencing. Y.N., K.K. and R.M. performed the experiments in cells and did the MALDI–TOF MS
mass spectrometry experiments. P.H. and A.L. performed the AFM studies. A.T. and P.B. performed the SAXS experiments. L.C.-L. conducted the expression and purification of GalNAc-T6 and T12
in HEK293 cells. L.H. identified the GalNAc-T3 mutations associated to disease. R.H.-G., T.A.G. and H.Clausen wrote the article with the other authors’ contributions. All authors read and
approved the final manuscript. CORRESPONDING AUTHOR Correspondence to Ramon Hurtado-Guerrero. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL
INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
INFORMATION Supplementary Figs 1–17 and Tables 1–6. REPORTING SUMMARY SUPPLEMENTARY VIDEO 1 A 500 ns MD simulation of TgGalNAc-T3 complexed to UDP-Mn+2 and FGF23c in explicit water
SUPPLEMENTARY VIDEO 2 A 500 ns MD simulation of HsGalNAc-T4 complexed to UDP-Mn+2 and FGF23c in explicit water SUPPLEMENTARY VIDEO 3 A 500 ns MD simulation of HsGalNAc-T6 complexed to
UDP-Mn+2 and FGF23c in explicit water SUPPLEMENTARY VIDEO 4 A 500 ns MD simulation of HsGalNAc-T12 complexed to UDP-Mn+2 and FGF23c in explicit water RIGHTS AND PERMISSIONS Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE de las Rivas, M., Paul Daniel, E.J., Narimatsu, Y. _et al._ Molecular basis for fibroblast growth factor 23 _O_-glycosylation by GalNAc-T3.
_Nat Chem Biol_ 16, 351–360 (2020). https://doi.org/10.1038/s41589-019-0444-x Download citation * Received: 25 June 2019 * Revised: 14 November 2019 * Accepted: 25 November 2019 * Published:
13 January 2020 * Issue Date: March 2020 * DOI: https://doi.org/10.1038/s41589-019-0444-x SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get
shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative