- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Citizen science video games are designed primarily for users already inclined to contribute to science, which severely limits their accessibility for an estimated community of 3
billion gamers worldwide. We created _Borderlands Science_ (_BLS_), a citizen science activity that is seamlessly integrated within a popular commercial video game played by tens of millions
of gamers. This integration is facilitated by a novel game-first design of citizen science games, in which the game design aspect has the highest priority, and a suitable task is then
mapped to the game design. _BLS_ crowdsources a multiple alignment task of 1 million 16S ribosomal RNA sequences obtained from human microbiome studies. Since its initial release on 7 April
2020, over 4 million players have solved more than 135 million science puzzles, a task unsolvable by a single individual. Leveraging these results, we show that our multiple sequence
alignment simultaneously improves microbial phylogeny estimations and UniFrac effect sizes compared to state-of-the-art computational methods. This achievement demonstrates that
hyper-gamified scientific tasks attract massive crowds of contributors and offers invaluable resources to the scientific community. SIMILAR CONTENT BEING VIEWED BY OTHERS THE AUSTRONESIAN
GAME TAXONOMY: A CROSS-CULTURAL DATASET OF HISTORICAL GAMES Article Open access 12 May 2021 CROSS-CULTURAL PATTERNS IN MOBILE PLAYTIME: AN ANALYSIS OF 118 BILLION HOURS OF HUMAN DATA Article
Open access 07 January 2023 VOLUNTEER CONTRIBUTIONS TO WIKIPEDIA INCREASED DURING COVID-19 MOBILITY RESTRICTIONS Article Open access 02 November 2021 MAIN In 2022, more than 3 billion
people played video games worldwide1. The growth of this community is expected to continue in upcoming years. The size and diversity of the gamer community show that video games are a
universal entertainment activity that transcends generations, gender and cultures2. The ubiquity of video games created new opportunities to communicate and teach scientific
concepts3,4,5,6,7, and one of the most remarkable applications of this media is the development of citizen science games (CSGs) that engage participants in the analysis of real scientific
data. Introduced with _Foldit_ in 2008, which recruited online citizen scientists to help with the prediction of the structure of proteins8, CSGs have been subsequently applied to
comparative genomics9, RNA folding10, neuron segmentation11 and quantum physics12,13. In the last decade, CSGs had a dramatic impact on the practice of citizen science, lowering the barrier
to entry of scientific activities and bringing hundreds of thousands of new participants into the community. However, successful implementation of this concept also comes with its own
challenges, such as reaching potential participants and maintaining player engagement14,15. The classical approach to design CSGs relies on gamification—that is, the introduction of game
design elements that facilitate the acquisition of the expertise needed to complete the activity and increase engagement16,17,18. The design of these CSGs is strongly rooted in the canonical
presentation of the scientific task, which may hinder its entertainment value and, therefore, participation of the general public. Hence, their target audience focuses on users with prior
interest in science15, which eventually amplifies the demographic skew observed in citizen science projects19. To answer to the challenge of user acquisition and engagement in citizen
science, in 2015, Massively Multiplayer Online Science (MMOS) proposed to embed citizen science activities into commercial video games played by massive established gamer communities20,
relying on the leadership of game designers to ensure a seamless integration of the citizen science activity in the host video game user experience. The first incarnation of this concept was
_Project Discovery_ in the massively multiplayer online game _EVE Online_ in 2016 (ref. 21). _EVE Online_ has a user base with a welcoming stance toward scientific narratives, and it had
not yet been shown that citizen science can work in any type of video game, specifically gamer communities not familiar with scientific narratives. In the present work, we reinforce the
primacy of game design to overcome the challenges of integration of citizen science in commercial games. We also leverage our previous experience with _Phylo_9,22,23 to develop a CSG for
microbiome data analysis that follows ‘game-first design’ principles. This approach aims to create a pure game that integrates scientific computation mechanisms in its core gameplay rather
than adapting existing scientific research modus operandi (Supplementary Information, section 6). Eventually, this methodology can lead to fundamental alterations in the canonical
presentation of the data that will prevent the complexity of the scientific task from obfuscating the accessibility or altering the user experience. We found that this approach of
redesigning the original task with a fresh ‘untrained’ eye can bring valuable improvements from these other disciplines of game or narrative design. This approach results in massive gains in
public participation without sacrificing the relevance of the collected answers. More importantly, it provides access to new human computation resources capable of executing complex
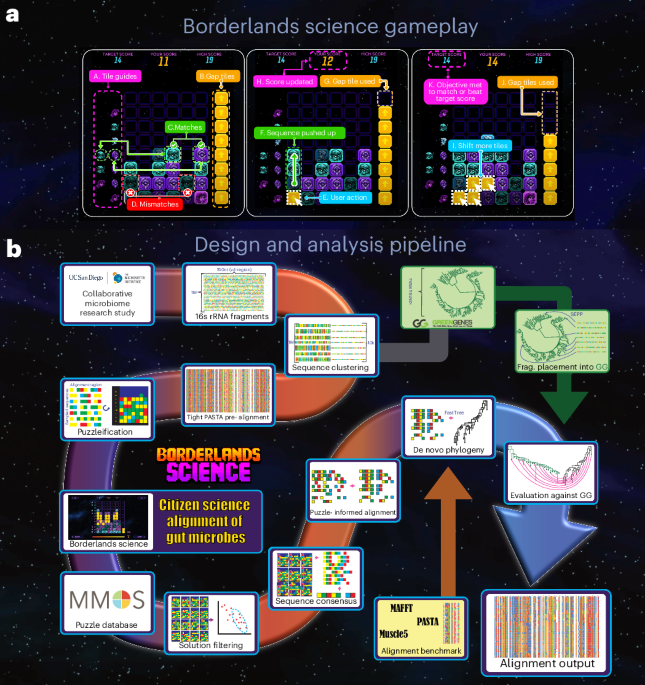
computational tasks on large scientific datasets, which would otherwise be unfeasible with classical CSGs. We showcase this concept with _Borderlands Science_ (_BLS_; Fig. 1)24, a
tile-matching mini game released on 7 April 2020 as a free downloadable content for _Borderlands 3_ (the most recent release of a video game franchise that sold over 77 million copies
worldwide). _BLS_ is designed to improve a reference multiple alignment of 1 million 16S ribosomal RNA (rRNA) sequences from The Microsetta Initiative’s (TMI) American Gut Project (AGP)25, a
citizen science initiative collecting gut microbial genomic data. Since its launch, more than 4 million gamers have played _BLS_ and solved more than 135 million puzzles. Our results show
that the multiple sequence alignment (MSA) obtained from _BLS_ (Fig. 1b and Methods) improves microbial phylogeny inference compared to other alignment methods and enables improved UniFrac26
effect size estimation for many variables known to be important to human digestive health. Beyond these results, _BLS_ demonstrates that commercial video games can provide massive human
processing resources for the manual curation and analysis of large-scale genomic datasets that could not be completed otherwise. RESULTS TASK AND DATA The data featured in this article come
from a set of 953,000 rRNA fragments sequenced from stool samples submitted by participants in the AGP25, powered by TMI (https://microsetta.ucsd.edu/). The fragments are 150 nucleotides
long and come from the V4 region of the 16S rRNA gene. In the _BLS_ mini game (Fig. 1a), the player sees 7–20 sequences of 4–10 nucleotides. Each sequence is displayed as a vertical pile of
bricks, each color representing a nucleotide (through a random mapping between colors and nucleotides). The bricks are collapsed (all gaps are removed), and the player is asked to insert a
finite number of gaps to improve a score determined by the number of bricks correctly aligned to the guides. These targets, located on the left, display the most common nucleotides in the
corresponding alignment column. By inserting gap tokens, the player is using their natural knack for pattern matching to realign a region of the scaffold alignment. The limited number of
tokens, set by a naive greedy artificial intelligence (AI) player based on how easily it could improve the alignment, forces the player to make tough choices. This greedy player also sets a
target score that the player must beat to progress to the next puzzle, enforcing a minimal effort. During the first year of the initiative, we collected approximately 75 million puzzle
solutions (mean, 43 per puzzle). The solutions were used as ‘votes’ by players on potential errors in the scaffold alignment, and a corrected alignment was generated. Here we report results
for our _BLS_ alignment and benchmark alignments produced by several state-of-the-art de novo multiple alignment programs (PASTA, MUSCLE and MAFFT) as well as an alignment produced by a
greedy algorithm (Methods and Supplementary Information, sections 12–15). COMPLEX CITIZEN SCIENCE TASKS CAN BE EMBEDDED IN VIDEO GAMES Integrating a scientific task in a commercial video
game from a franchise that sells millions of copies constitutes a risk. The scientific mini game must not look out of place in the broader game as it could break immersion, an important
component of role-playing games. This is especially true when integrating a puzzle game into a shooter–looter role-playing game such as _Borderlands 3_, in which the player expects to face
fast-paced action and humor. _BLS_ was specifically designed to address this potential issue by integrating dialogues with characters from the _Borderlands_ universe. The virtual arcade
booth was located in a virtual laboratory within the game universe and is presented as belonging to the resident scientist. The gameplay was simplified to make sure that the pace matched a
shooter–looter game, and all the visuals were specifically designed to make sense in-universe. As of May 2023, 3 years after launch, 4.45 million players had visited the arcade booth. Of
these players, over 4 million completed the 10-min tutorial and at least one real task. This represents an engagement rate of 90%, which is a substantial improvement over _Phylo_, the
previous sequence alignment CSG, which has reported an engagement rate of around 10%. This level of player engagement demonstrates that the ultra-gamification of the task worked and that its
integration into the commercial game was seamless. _BLS_ OUTPUTS A HIGH-QUALITY MSA The evaluation of the _BLS_ alignment output is complicated by the absence of a universal ground truth to
benchmark against. Nevertheless, there are several qualities typically expected from a high-quality alignment that we can investigate. Indeed, a high-quality alignment of homologous genomic
sequences tends to: * a. Have a high sum-of-pairs score * b. Have a gap frequency compatible with that RNA family’s indel frequency * c. Allow the inference of phylogenetic trees that
resemble the state of the art for that family * d. Appropriately separate taxa associated with different illnesses, behaviors and profiles in the host * e. Be compatible with the structural
signature of the family We assert that the _BLS_ alignment satisfies all these criteria and, thus, constitutes a high-quality MSA. In Table 1 (top), we show that _BLS_ improves the
sum-of-pairs score compared to all benchmarks when excluding heavily gapped columns from the scoring, thus satisfying A. In Fig. 2b, we show the gap frequency sampled from sub-alignments (we
sample sub-alignments to account for differences in number of sequences among Greengenes, Rfam and _BLS_) for _BLS_ and benchmarks. These benchmarks include PASTA27, MUSCLE28 and MAFFT29,
the pyNAST30 and SSU-ALIGN31 alignments from the Greengenes32 database and the structural Rfam33 alignment. We observed that all alignments had a gap frequency in the same order of magnitude
as PASTA, Rfam and pyNAST. This low gap frequency is consistent with the strongly structured nature of the V4 region, which varies in length by only a few base pairs between microbes. Thus,
the _BLS_ alignment shows a gap frequency that is compatible with our state-of-the-art knowledge of the V4 region, and criterion B is satisfied. Our results in Table 1 and Fig. 2 indicate
that the additional gaps inserted by _BLS_ compared to PASTA, MAFFT, MUSCLE and the greedy algorithm led to a improvement of the tree structure. We present our investigation of criteria C, D
and E in the following subsections. THE _BLS_ ALIGNMENT IMPROVES DE NOVO PHYLOGENY A central objective of improving alignments of microbial genomic sequences was to better understand their
phylogeny. To assess whether this goal was achieved, we inferred phylogenetic trees from our alignments with FastTree34 and then assessed their similarity to a reference tree built by
placing our sequences into the Greengenes 13.5 (ref. 32) phylogeny with SEPP35. Greengenes has been previously benchmarked for fragment insertion, and SEPP with Greengenes has been shown to
outperform de novo phylogeny inference, when a high-quality tree and alignment are already available36. Our phylogenetic similarity results, shown in Table 1 for two distance metrics,
Kendall–Colijn37 and Triplet38 distance, indicate that the _BLS_ phylogenies are closer to the reference than standard MSA approaches. This result shows that the _BLS_ alignment outperforms
alternatives for the task of inferring a reliable phylogeny, thus satisfying criterion C. THE _BLS_ PHYLOGENY LEADS TO IMPROVED UNIFRAC EFFECT SIZES We also formulated the hypothesis that
improved alignments (and, by extension, de novo phylogenies) would lead to some improvement in the separation of taxa associated with different behaviors, profiles and diseases. To confirm
this, we measured effect sizes on UniFrac26 distances over 74 non-technical variables available in the AGP metadata associated with the samples used for sequencing. These variables relate to
the host’s lifestyle, health condition, food or general profile. The strongest effect sizes that we observed were for teeth brushing frequency and prior _Clostridium difficile_ infection
(full list in Fig. 3 and Supplementary Information, section 14). We report the average pairwise effect sizes between _BLS_ and Greengenes + SEPP in Fig. 3. We observed that _BLS_ outperforms
SEPP on many variables, including several that have been linked to gut microbe diversity and human health25,39,40,41,42. The top five variables with highest delta are, respectively, teeth
brushing frequency, diabetes, number of types of plants, antibiotic history and alcohol frequency. We backed up this analysis by assessing whether the effect sizes obtained were
significantly different than what could be observed in a random assignment of categories to samples. We annotated Fig. 3 with the _P_ values associated with the null hypothesis that our
results are compatible with a random outcome. We observed an enrichment of significant outcomes in the variables with high effect sizes and, in particular, 13 variables for which _BLS_
achieves a higher significance category than SEPP, whereas the opposite occurs 10 times, generally on variables with lower effect sizes. These improvements observed on most variables confirm
that the previously reported improvements to phylogeny can be perceived in meta-analyses. Although overall improvement over SEPP is limited as SEPP still outperforms _BLS_ on important
variables, such as age category, _BLS_ does outperform SEPP, and the two approaches lead to distinct solutions that are complementary in improving understanding of gut microbe phylogeny and
its impact on human health, thus satisfying criterion D. THE _BLS_ ALIGNMENT IMPROVES SUPPORT FOR 16S RRNA STRUCTURE As stated previously, the structure is an important component of a
high-quality MSA of a strongly structured RNA region. Given that the _BLS_ alignment contains about 99% of bacterial sequences, it is possible to map its columns to the bases of the
state-of-the-art structural model of the 16S bacterial rRNA defined on the Comparative RNA Web43. To estimate the quality of such mapping, we report the proportion of non-gap nucleotides
that cannot be mapped to the structure (see Table 1, bottom). It turns out that alignments such as _BLS_ and PASTA map rather easily, whereas MUSCLE and MAFFT lose substantially more
information. To deepen our evaluation of the structural quality of the two alignments that map well, we investigated column by column whether the differences between _BLS_ and PASTA
alignments tend to agree or disagree with the model. In Fig. 4, we show that the changes added by human players tend to agree with the 16S structural model, especially in regions linked to
important functional sites, such as S8 and S15 binding sites, and a region that undergoes conformational readjustment during 30S assembly44,45,46,47,48,49. This agreement is a strong
argument in favor of the _BLS_ alignment as the CRW model considers long-range base pairs, a context that is not available to the players solving small puzzles. This is important because, as
shown in Fig. 2a, the region of the alignment with the most gaps and the most variation between methods is the top of the V4 stem, a region with considerable mapping improvements for _BLS_
in this analysis. This demonstrates that the modifications to the PASTA scaffold added by _BLS_ led to an improvement of the structural and functional signal of the alignment, thus
satisfying criterion E. Additionally, in Fig. 2a, we show the gap frequency per column (excluding highly gapped columns) of different alignment methods, including _BLS_ and our benchmarks
PASTA27, MUSCLE28 and MAFFT29. These gap frequencies help reveal the differences between _BLS_ alignments and alternatives, on top of satisfying criteria A–E. DISCUSSION Since the release of
_Foldit_ in 2008, many high-impact CSGs have been released, such as _EteRNA_10, _Project Discovery_, _EyeWire_, _Phylo_ and _Sea Hero Quest_ (Fig. 5). To this day, a CSG that would reach
500,000 players would be considered a massive success. This evaluation is made in context of the relative difficulty for a scientific team to reach people who are likely to be interested in
playing a game for science. _Galaxy Zoo_50 was the first initiative of this type to break 1 million participants after 10 years in activity. _Sea Hero Quest_ was the first to achieve the
feat in less than 1 year, by working with Deutsche Telecom, a company with 300 million customers51. _BLS_ built on this idea of improving reach through collaboration with non-scientific
companies and went to the next level by reaching gamers directly where they are, within the virtual universe of high-profile video games. Now that we have shown that puzzle-style CSGs can be
seamlessly integrated into a high-profile game and engage players, we expect other projects to take advantage of this opportunity to enter an era where million-players CSGs are no longer an
exception. _Phylo_ struggled to retain players for more than a couple puzzles, with an average of 5.7 tasks completed by users, and a median of two. Thus, _Phylo_ relies heavily on expert
players. Indeed, in _Phylo_, the experts can collectively outperform casual gamers by up to 40% depending on the alignment, despite being a small group completing a tiny fraction of the
tasks solved by casual players. On the other hand, _BLS_ players complete an average of 35 tasks, with a median of 12. Unlike in _Phylo_, where experts achieving high scores form the
backbone of the scientific contribution, _BLS_ filters tasks based on consensus between players and provides rewards for just reaching a target score. Therefore, _BLS_ does not rely on
experts to the same degree. Indeed, removing all contributions of the 10% most active players represents only a 14% drop in evaluation metrics compared to removing the same amount of data at
random. This lack of reliance on experts marks a change of paradigm from lightly gamified CSGs that rely on experts to a new category of CSGs to which everyone can contribute. Two core
objectives guide the design of CSGs: maximizing player engagement and maximizing the scientific relevance of the game. Unfortunately, these two objectives often conflict with one another; a
game with optimal scientific relevance would be a pure crowdsourced scientific task with no gaming elements, such as image annotation in Stardust@home. Gamification of the task, by design,
will make mapping player solutions to scientific contributions more difficult. However, the more gamified the task, the higher the potential for it to play like a real game and be fun enough
to stimulate player engagement and retention. In the present study, we pushed the envelope on gamification, and our observations match the conundrum: _BLS_ achieved a very high level of
player engagement, but the average scientific contribution of one task was lower than in _Phylo_, where a few hundreds of puzzles are sufficient to improve a small alignment. Here, millions
of tasks are combined to improve alignments. Nevertheless, the ultra-gamification of _BLS_ led to a much higher player engagement and retention than in _Phylo_, which strongly alleviates
that downside. Another aspect that we think indicates potential for growth in CSGs is the feedback that we received from the players. Through our exchanges with gamers in the form of quality
assurance testing feedback, blog posts, email and social media (Supplementary Information, section 11.5), the most common response that we received was enthusiasm and curiosity about
science. Although quantifying this attitude would be its own study and, thus, left for future work, our experience in this project leads us to think that player engagement was driven by a
combination of game-first design and genuine interest in science from the public. Moreover, beyond the optimization of the tradeoff described above, the inherent benefits of participating in
a citizen science initiative, such as improvements in science literacy and increased connectedness of the public to the scientific world, further justifies the exploration of the extra
gamification region of that tradeoff: reaching more participants is a valid objective in its own right (Supplementary Information, section 11.4). In the present study, we focused on phase 1
of _BLS_, which includes data from the first year, and over two-thirds of the total amount of data. There are, thus, 2 years of data remaining to analyze. These data include different types
of puzzles aimed at studying how sequence alignment CSGs can be made more efficient. After phase 1, we started producing puzzles with alternative scaffolds, and, as such, the analysis
presented here does not apply to them, which is why they are not included in this study (Supplementary Information, section 5). Additionally, we included a guide offset in some of the
puzzles sent to users (Supplementary Information, section 5.2.6). Along with presenting more stimulating gameplay, these guide offsets allow the user to correct potential errors in the
scaffold alignments at a larger scale. As a result, they are more noisy than regular puzzles (Supplementary Information, section 8.5.4). _BLS_ alignments built with offset puzzles show
potential, as they outperform _BLS_ on some important effect size variables, such as age category. Nevertheless, because they require a different type of analysis, their investigation is
left for future work. Another consideration for future work is the integration of synthetic data in the game. Indeed, in the present study, we relied on the Greengenes phylogeny as a proxy
for a ground truth. Although this phylogeny is well understood and state of the art in the field, it is not perfect. In future projects of this type, we will endeavor to integrate synthetic
data into the design of the game, to complement our proxy for the ground truth. Nevertheless, it must be mentioned that synthetic data often embed assumptions that match the true
evolutionary process poorly, especially under heterogeneous substitution models in different parts of the tree and when RNA secondary structure is present, both of which we know are the case
in this situation. Finally, we have been working in parallel on strategies to extend the impact of player participation by training reinforcement learning agents to reproduce player
strategies, and, recently, we published two studies on the matter. In the first study52, we established that it is possible for reinforcement learning agents to learn player strategies for
solving _BLS_ puzzles, unlocking potential for propagating these player strategies to data outside of the game. In the second study53, we demonstrated that injecting human solutions into a
Q-learning algorithm outperforms pure AI. Combined with the aggregation techniques presented in the present study, these results suggest promising avenues to design an automated pipeline for
massive alignment of 16S rRNA sequences. CONCLUSION In the present study, we present _BLS_, a sequence alignment game aiming to open a path for gaming-focused CSGs targeting large-scale
problems with large-scale audiences. The project achieved player engagement unseen before for a game of its category and reached its goal of pooling gamers together to solve a large-scale
collective task while also serving as a large-scale science outreach initiative. We showed that, despite being gamified to a degree beyond that of previous efforts, the contributions of the
players led to clear improvements over state-of-the-art sequence alignment methods, unlocking the potential for a new generation of gaming-focused CSGs. METHODS The data featured in this
article come from a set of 953,000 rRNA fragments sequenced from stool samples submitted by participants in the AGP25, powered by TMI (https://microsetta.ucsd.edu/). The fragments are 150
nucleotides long and come from the V4 region of the 16S rRNA gene. The 953,000 150-nucleotide V4 fragments were first clustered with CD-HIT54 into 10,200 clusters. Very dissimilar clusters
representing a small minority of sequences (about 5,000) were removed, and a final set of 9,667 cluster representative sequences was obtained, which accounts for 946,740 sequences of the
initial dataset. These 9,667 sequences were then aligned with PASTA 1.9 (ref. 27), generating a very tight (183 columns) initial alignment, which served as a scaffold for the creation of CSG
puzzles. GAME DESIGN In the _BLS_ mini game, the player sees 7–20 sequences of 4–10 nucleotides (the size of the grid changes with the difficulty level). Each sequence is displayed as a
vertical pile of bricks, each color representing a random base. The bricks are collapsed (all gaps are removed), and the player is then asked to insert a finite number of gaps to improve a
score determined by the number of bricks correctly aligned to the guides. These targets, located on the left, display the most common nucleotides in the corresponding alignment column. By
inserting gap tokens, the player is using their natural knack for pattern matching to realign a region of the scaffold alignment. The limited number of tokens, set by a naive greedy AI
player based on how easily it could improve the alignment, forces the player to make tough choices. This greedy player also sets a par score that the player must beat to progress to the next
puzzle, enforcing a minimal effort. The game is displayed in Fig. 1, and more details on game design can be found in the Supplementary Information (section 6). BUILDING PUZZLES Puzzles were
added to the game on a regular basis. The results presented in this paper use the solutions of the first 1.4 million puzzles (a total of about 75 million player tasks), submitted to the
players between April 2020 and July 2021. To get a wide enough distribution of solutions, each puzzle was released in three different versions, each with a slightly different number of gap
tokens available. Each different version was aimed to be played by about 15 players, for a total of 45 per puzzle. For each alignment region (a sequence of alignment columns of variable
length), sequences are sampled from the PASTA alignment. To compensate for the bias that comes from the vertical orientation, puzzles are built both from left to right and from right to
left. Additionally, each sequence in each puzzle has a small probability (~0.15) to be shortened or elongated to show puzzle sequences of different lengths, allowing the player to simulate
deletions. Finally, an offset was sometimes introduced between the guides and the puzzle sequences. An offset of 1 would mean that puzzles using sequences from the alignment columns 10–14
would have the guides from columns 9–13, to enhance the user’s action space; they would then have to push up many bricks just to get back to the initial PASTA configuration. Through these
variations in the puzzles, the players are given opportunities to re-evaluate alignment decisions made by the PASTA algorithm. PUZZLE AND SOLUTION FILTERING Puzzles are discarded if they
offer little incentive to move bricks because the starting gravity effect already yields a local optimum. This led to some alignment regions being overrepresented in the puzzles due to a
higher acceptance rate, which we interpreted as these regions presenting more room for improvement via puzzles. For each puzzle, the 20–60 (objective, 45; real mean, 43.4) user solutions
were filtered by their similarity to the distribution of gap positions from all submitted solutions to that puzzle. The player solutions were filtered using two criteria: (1) how far from
optimality the solution was and (2) how far from the player consensus the solution was. The distance from optimality in (1) was defined as the score difference against the solution with the
highest score using the same number of gaps for that puzzle. The player consensus in (2) was defined as the centroid of the distribution of solutions. Solutions that were too far from this
combined objective were excluded, and the exclusion threshold was set to exclude about two-thirds of the solutions, to keep a subset that was most representative of the ensemble of
high-quality player solutions. This threshold was selected experimentally based on visual exploration of rejected solutions: we selected a threshold for which no obvious outliers were
included, and all excluded solutions were clear outliers. ALIGNMENT IMPROVEMENT The puzzle solutions are converted to positional sequence annotations, which are normalized to account for
variance in coverage, and unannotated positions are filled with PASTA and/or Rfam version 14 (ref. 33) alignment information. We consider each high-quality annotation from one puzzle solved
by one player as a vote. Our algorithm leverages dynamic programming through a compromise between Needleman–Wunsch55 and the voting system, in which we align a sequence to the array of
votes. VALIDATION Because the alignments produced by _BLS_ are very different from the ones produced by state-of-the-art software, such as MUSCLE version 5.1 (ref. 28) or MAFFT version 7.490
(ref. 29), it is difficult to fairly compare them as alignments. Thus, the validation process for our methods is centered around the comparison of phylogenetic trees estimated from the
alignment with FastTree 2.1 (ref. 34). SMALL ALIGNMENT BENCHMARK Along with the PASTA alignment that we computed as the scaffold for the puzzles, we ran MAFFT and MUSCLE on the same 9,667
sequences using both the standard and slowest settings. We then generated phylogenies for all with FastTree’s standard settings. These trees were then re-rooted using sequences classified by
naive Bayes with QIIME version 2’s q2-feature-classifier56 against Greengenes 13.8 (ref. 32) with Archaea as the outgroup. COMPARISON TO SEPP The state-of-the-art method for building
phylogenetic trees from alignments has been shown to be outperformed on gut microbiome data by the placement of the sequences into an existing tree with SEPP35,36, which, when sufficient
data are available, outperforms de novo phylogeny estimation from alignments36. To build a reference phylogeny, we placed 9,667 sequence cluster representatives into the Greengenes 13.5
phylogenetic tree with SEPP and then removed all the other tips and re-rooted the tree to Archaea. We refer to this tree as the Greengenes-SEPP reference tree. We compare the de novo
alignment methods to this Greengenes-SEPP reference tree by computing distances between randomly sampled subtrees of the de novo and reference trees, using two distance metrics:
Kendall–Colijn metric37, which compares the placement of the most recent common ancestor of each pair of tips, specifically designed for assessing similarity in evolutionary patterns and
specifically designed for situations where the Robinson–Foulds distance is not sensitive enough. Triplet distance38, a metric that measures the structural dissimilarity of two phylogenetic
trees by counting the number of rooted trees with exactly three leaves that occur in one but not both trees. When comparing large trees, because these distances are very computationally
expensive, we sample 400 nodes for Kendall–Colijn and 100 nodes for Triplet. This process is repeated, respectively, 100 and five times. We performed these evaluations with the R treeDist57
library, version 2.7.0. Trees were resized and sheared in Python 3.8 with scikit-bio version 0.5. APPLICATION TO META-ANALYSES To assess the impact of the _BLS_ alignment on meta-analyses
involving gut microbiome, the alignment results were propagated from cluster representatives to other sequences to build an alignment of 946,740 sequences, for which a phylogenetic tree was
estimated with FastTree. To assess effect sizes, a feature table representing the AGP data was obtained from redbiom version 0.3.5 (ref. 58), filtered for blooms and rarefied to 1,000
sequences per sample, and unweighted UniFrac26 distances were computed for each tree (for example, the _BLS_ trees, de novo trees and the SEPP fragment insertion tree). Pairwise effect sizes
were then calculated with Evident version 0.4.0 (ref. 59) on the distance matrix using Cohen’s _d_ distributions for each tree. The pairwise effect sizes of 74 non-technical variables were
investigated (all variables available on QIIME 2 meeting a minimum of two categories including 50 samples). We reproduced these experiments with shuffled metadata to assess significance. We
also examined two technical variables—the sample plates and the liquid handling robot—and observed no significant change. It should be noted that the aggregation of player solutions and the
production of an alignment were not optimized for performance on effect sizes. This choice aims to avoid overfitting by training to optimize for a phenotype, due to the relatively small size
of those data. Furthermore, it was established in previous work that there is a relationship between the quality of the tree and the observed phenotype (effect sizes)36. Thus, alignments
were optimized on tree metrics, and effect sizes were kept as an independent evaluation. As expected from previous work, the _BLS_ alignment with the best tree metrics (listed in Table 1)
also had the best effect sizes. ETHICS STATEMENT Participation in the project was done entirely virtually, through the _Borderlands 3_ game. It was fully optional. No participants were
recruited, and potential participants were informed of how and why the data would be used through a presentation video played before gameplay. Only gameplay data were collected. No
identifying or personal information about the participants was collected, and participants were fully anonymous to the research team. REPORTING SUMMARY Further information on research design
is available in the Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY All the data discussed in this paper are publicly available for scientists and individuals
at https://games.cs.mcgill.ca/bls/. The data are also publicly available on mirror sites at https://gitlab.com/borderlands-science/BLS1 and https://doi.org/10.6084/m9.figshare.24962349 (ref.
60). These data include all the puzzles that players solved, all the solutions submitted by the players and related data, such as the order in which they made their moves to solve the
puzzle. The players are identified by unique alphanumeric strings as it was not possible to obtain their informed consent to share their personal information, as these data were collected
through a video game. CODE AVAILABILITY All the code used to generate data, process data and compute results presented in this paper is freely available as a GitLab repository linked on the
project website: https://games.cs.mcgill.ca/bls/. The code is also publicly available on mirror sites at https://gitlab.com/borderlands-science/BLS1 (ref. 61) and
https://doi.org/10.6084/m9.figshare.24962349 (ref. 60). REFERENCES * _Newzoo Global Games Market Report 2022_ (newzoo, 2022);
https://newzoo.com/resources/trend-reports/newzoo-global-games-market-report-2022-free-version * Essential facts about the video game industry. _Entertainment Software Association_
https://www.theesa.com/resource/2022-essential-facts-about-the-video-game-industry/ (2022). * Nebel, S., Schneider, S. & Rey, G. D. Mining learning and crafting scientific experiments: a
literature review on the use of Minecraft in education and research. _J. Educ. Technol. Soc._ 19, 355–366 (2016). Google Scholar * Santos, D., Zagalo, N. & Morais, C. Players
perception of the chemistry in the video game No Man’s Sky. _Simul. Gaming_ 54, 375–394 (2023). Article Google Scholar * Cheng, M. T. et al. The use of serious games in science education:
a review of selected empirical research from 2002 to 2013. _J. Comput. Educ._ 2, 353–375 (2015). Article Google Scholar * Muehrer, R. et al. Challenges and opportunities: using a
science-based video game in secondary school settings. _Cult. Stud. Sci. Educ._ 7, 783–805 (2012). Article Google Scholar * Stegman, M. Immune Attack players perform better on a test of
cellular immunology and self confidence than their classmates who play a control video game. _Faraday Discuss._ 169, 403–423 (2014). Article CAS PubMed PubMed Central Google Scholar *
Cooper, S. et al. Predicting protein structures with a multiplayer online game. _Nature_ 466, 756–760 (2010). Article CAS PubMed PubMed Central Google Scholar * Kawrykow, A. et al.
Phylo: a citizen science approach for improving multiple sequence alignment. _PLoS ONE_ 7, e31362 (2012). Article CAS PubMed PubMed Central Google Scholar * Lee, J. et al. RNA design
rules from a massive open laboratory. _Proc. Natl Acad. Sci. USA_ 111, 2122–2127 (2014). Article PubMed PubMed Central Google Scholar * Kim, J. S. et al. Space-time wiring specificity
supports direction selectivity in the retina. _Nature_ 509, 331–336 (2014). Article CAS PubMed PubMed Central Google Scholar * Jensen, J. H. M. et al. Crowdsourcing human common sense
for quantum control. _Phys. Rev. Res._ 3, 013057 (2021). * Wootton, J. R. Getting the public involved in Quantum Error Correction. Preprint at https://arxiv.org/abs/1712.09649 (2017). *
Kreitmair, K. V. & Magnus, D. C. Citizen science and gamification. _Hastings Cent. Rep._ 49, 40–46 (2019). Article PubMed Google Scholar * Miller, J. A. et al. A survey of citizen
science gaming experiences. _Citiz. Sci._ 7, 34 (2022). Google Scholar * Miller, J. A. et al. Expertise and engagement: re-designing citizen science games with players’ minds in mind. In
_Proc. 14th International Conference on the Foundations of Digital Games_ https://doi.org/10.1145/3337722.3337735 (Association for Computing Machinery, 2019). * Miller, J. A. & Cooper,
S. Barriers to expertise in citizen science games. In _Proc._ _CHI Conference on Human Factors in Computing Systems_ https://doi.org/10.1145/3491102.3517541 (Association for Computing
Machinery, 2022). * Franzoni, C., Poetz, M. & Sauermann, H. Crowds, citizens, and science: a multi-dimensional framework and agenda for future research. _Ind. Innov._ 29, 251–284 (2022).
* Allf, B. C. et al. Citizen science as an ecosystem of engagement: implications for learning and broadening participation. _Bioscience_ 72, 651–663 (2022). Article PubMed PubMed Central
Google Scholar * Szantner, A. in _Levelling Up: The Cultural Impact of Contemporary Videogames_ (eds Kuhn, B. & Bhéreur-Lagounaris, A.) 103–110 (Brill, 2016). * Sullivan, D. P. et al.
Deep learning is combined with massive-scale citizen science to improve large-scale image classification. _Nat. Biotechnol._ 36, 820–828 (2018). Article CAS PubMed Google Scholar *
Singh, A. et al. Lessons from an online massive genomics computer game. In _Proc. AAAI Conference on Human Computation and Crowdsourcing_ https://doi.org/10.1609/hcomp.v5i1.13309 (PKP,
2017). * Kwak, D. et al. Open-Phylo: a customizable crowd-computing platform for multiple sequence alignment. _Genome Biol._ 14, R116 (2013). Article PubMed PubMed Central Google Scholar
* Waldispühl, J. et al. Leveling up citizen science. _Nat. Biotechnol._ 38, 1124–1126 (2020). Article PubMed Google Scholar * McDonald, D. et al. American Gut: an open platform for
citizen science microbiome research. _mSystems_ 3, e00031–18 (2018). Article CAS PubMed PubMed Central Google Scholar * Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method
for comparing microbial communities. _Appl. Environ. Microbiol._ 71, 8228–8235 (2005). Article CAS PubMed PubMed Central Google Scholar * Mirarab, S. et al. PASTA: ultra-large multiple
sequence alignment for nucleotide and amino-acid sequences. _J. Comput. Biol._ 22, 377–386 (2015). Article CAS PubMed PubMed Central Google Scholar * Edgar, R. C. MUSCLE: multiple
sequence alignment with high accuracy and high throughput. _Nucleic Acids Res._ 32, 1792–1797 (2004). Article CAS PubMed PubMed Central Google Scholar * Katoh, K. & Standley, D. M.
MAFFT multiple sequence alignment software version 7: improvements in performance and usability. _Mol. Biol. Evol._ 30, 772–780 (2013). Article CAS PubMed PubMed Central Google Scholar
* Caporaso, J. G. et al. PyNAST: a flexible tool for aligning sequences to a template alignment. _Bioinformatics_ 26, 266–267 (2010). Article CAS PubMed Google Scholar * Nawrocki, E. P.,
Kolbe, D. L. & Eddy, S. R. Infernal 1.0: inference of RNA alignments. _Bioinformatics_ 25,1335–1337 (2009). * DeSantis, T. Z. et al. Greengenes, a chimera-checked 16S rRNA gene database
and workbench compatible with ARB. _Appl. Environ. Microbiol._ 72, 5069–5072 (2006). Article CAS PubMed PubMed Central Google Scholar * Kalvari, I. et al. Rfam 14: expanded coverage of
metagenomic, viral and microRNA families. _Nucleic Acids Res._ 49, D192–D200 (2021). Article CAS PubMed Google Scholar * Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree
2—approximately maximum-likelihood trees for large alignments. _PLoS ONE_ 5, e9490 (2010). Article PubMed PubMed Central Google Scholar * Mirarab, S., Nguyen, N. & Warnow, T. SEPP:
SATé-enabled phylogenetic placement. _Pac. Symp. Biocomput_. 247–258 (2012). * Janssen, S. et al. Phylogenetic placement of exact amplicon sequences improves associations with clinical
information. _mSystems_ 3, e00021–18 (2018). Article CAS PubMed PubMed Central Google Scholar * Kendall, M. & Colijn, C. Mapping phylogenetic trees to reveal distinct patterns of
evolution. _Mol. Biol. Evol._ 33, 2735–2743 (2016). Article CAS PubMed PubMed Central Google Scholar * Dress, A. W. M., Huber, K. T. & Steel, M. ‘Lassoing’ a phylogenetic tree I:
basic properties, shellings, and covers. _J. Math. Biol._ 65, 77–105 (2012). Article PubMed Google Scholar * Singhal, S. et al. The role of oral hygiene in inflammatory bowel disease.
_Dig. Dis. Sci._ 56, 170–175 (2011). Article PubMed Google Scholar * Lau, W. L. et al. Diabetes and the gut microbiome. _Semin. Nephrol._ 41, 104–113 (2021). Article CAS PubMed Google
Scholar * Capurso, G. & Lahner, E. The interaction between smoking, alcohol and the gut microbiome. _Best Pract. Res. Clin. Gastroenterol._ 31, 579–588 (2017). Article CAS PubMed
Google Scholar * Mutlu, E. A. et al. Colonic microbiome is altered in alcoholism. _Am. J. Physiol. Gastrointest. Liver Physiol._ 302, G966–G978 (2012). Article CAS PubMed PubMed Central
Google Scholar * Gutell, R. R., Lee, J. C. & Cannone, J. J. The accuracy of ribosomal RNA comparative structure models. _Curr. Opin. Struct. Biol._ 12, 301–310 (2002). Article CAS
PubMed Google Scholar * Moine, H. et al. In vivo selection of functional ribosomes with variations in the rRNA-binding site of _Escherichia coli_ ribosomal protein S8: evolutionary
implications. _Proc. Natl Acad. Sci. USA_ 97, 605–610 (2000). Article CAS PubMed PubMed Central Google Scholar * Hori, N., Denesyuk, N. A. & Thirumalai, D. Shape changes and
cooperativity in the folding of the central domain of the 16S ribosomal RNA. _Proc. Natl Acad. Sci. USA_ 118, e2020837118 (2021). * Li, W., Ma, B. & Shapiro, B. A. Binding interactions
between the core central domain of 16S rRNA and the ribosomal protein S15 determined by molecular dynamics simulations. _Nucleic Acids Res._ 31, 629–638 (2003). Article CAS PubMed PubMed
Central Google Scholar * Nikulin, A. et al. Crystal structure of the S15-rRNA complex. _Nat. Struct. Biol._ 7, 273–277 (2000). Article CAS PubMed Google Scholar * Serganov, A. A. et
al. The 16S rRNA binding site of _Thermus thermophilus_ ribosomal protein S15: comparison with _Escherichia coli_ S15, minimum site and structure. _RNA_ 2, 1124–1138 (1996). CAS PubMed
PubMed Central Google Scholar * Yang, B., Wang, Y. & Qian, P.-Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. _BMC Bioinformatics_
17, 135 (2016). Article PubMed PubMed Central Google Scholar * Masters, K. L. Twelve years of Galaxy Zoo. _Proc. IAU_ 14, 205–212 (2019). * Spiers, H. J., Coutrot, A. & Hornberger,
M. Explaining world‐wide variation in navigation ability from millions of people: citizen science project Sea Hero Quest. _Top. Cogn. Sci._ 15, 120–138 (2023). Article PubMed Google
Scholar * Mutalova, R. et al. Playing the system: can puzzle players teach us how to solve hard problems? In _Proc. 2023 CHI Conference on Human Factors in Computing Systems_
https://doi.org/10.1145/3544548.3581375 (Association for Computing Machinery, 2023). * Mutalova, R. et al. Player-guided AI outperforms standard AI in sequence alignment puzzles. In _Proc.
ACM Collective Intelligence Conference_ https://doi.org/10.1145/3582269.3615597 (Association for Computing Machinery, 2023). * Fu, L. et al. CD-HIT: accelerated for clustering the
next-generation sequencing data. _Bioinformatics_ 28, 3150–3152 (2012). Article CAS PubMed PubMed Central Google Scholar * Needleman, S. B. & Wunsch, C. D. A general method
applicable to the search for similarities in the amino acid sequence of two proteins. _J. Mol. Biol._ 48, 443–453 (1970). Article CAS PubMed Google Scholar * Estaki, M. et al. QIIME 2
enables comprehensive end-to-end analysis of diverse microbiome data and comparative studies with publicly available data. _Curr. Protoc. Bioinformatics_ 70, e100 (2020). Article PubMed
PubMed Central Google Scholar * Smith, M. R. Robust analysis of phylogenetic tree space. _Syst. Biol._ 71, 1255–1270 (2022). Article PubMed Google Scholar * McDonald, D. et al. redbiom:
a rapid sample discovery and feature characterization system. _mSystems_ 4, e00215–e00219 (2019). Article PubMed PubMed Central Google Scholar * Rahman, G. et al. Determination of
effect sizes for power analysis for microbiome studies using large microbiome databases. _Genes_ 14, 1239 (2023). * Sarrazin-Gendron, R. & Waldispühl, J. Borderlands science Git
repository. _figshare_ https://doi.org/10.6084/m9.figshare.24962349 (2024). * Sarrazin-Gendron, R. et al. Improving microbial phylogeny with citizen science within a mass-market video game.
_GitHub_ https://gitlab.com/borderlands-science/BLS1 (2024). Download references ACKNOWLEDGEMENTS We thank all members of the Gearbox Entertainment Company who contributed directly and
indirectly to the making of _Borderlands Science_, as well as the millions of _Borderlands Science_ players, whom we are unfortunately not able to credit individually but without whom this
project could not have happened. We also thank Z. Bányai, a former principal collaborator of Massively Multiplayer Online Science (MMOS), who contributed to the early development of MMOS. We
consider all _Borderlands Science_ players to be essential contributors to this publication, but it is not possibe to name them individually, which is why they are included as a group in
the author lists. This work was supported by a Genome Canada and Génome Québec grant (Genomic Application Partnership Program) to J.W., A.Sz., M.B. and S.C. R.K. is partially supported by a
Director’s Pioneer Award from the National Institute of Health (DP1AT010885). AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * School of Computer Science, McGill University, Montréal, QC,
Canada Roman Sarrazin-Gendron, Parham Ghasemloo Gheidari, Alexander Butyaev, Timothy Keding, Eddie Cai, Jiayue Zheng, Renata Mutalova, Julien Mounthanyvong, Yuxue Zhu, Elena Nazarova,
Chrisostomos Drogaris, Mathieu Blanchette, Attila Szantner & Jérôme Waldispühl * Massively Multiplayer Online Science, Gryon, Switzerland Kornél Erhart & Attila Szantner * Gearbox
Studio Québec, Québec, QC, Canada David Bélanger, Michael Bouffard, Mathieu Falaise, Vincent Fiset, Steven Hebert, Jonathan Huot, Jonathan Moreau-Genest, Ludger Saintélien, Amélie
Brouillette, Gabriel Richard & Sébastien Caisse * Gearbox Entertainment Company, Frisco, TX, USA Joshua Davidson, Dan Hewitt, Seung Kim, David Najjab, Steve Prince & Randy Pitchford
* Department of Pediatrics, University of California, San Diego, La Jolla, CA, USA Daniel McDonald & Rob Knight * Department of Computer Science, University of California, San Diego, La
Jolla, CA, USA Rob Knight * Department of Bioengineering, University of California, San Diego, La Jolla, CA, USA Rob Knight * Center for Microbiome Innovation, University of California, San
Diego, La Jolla, CA, USA Rob Knight Authors * Roman Sarrazin-Gendron View author publications You can also search for this author inPubMed Google Scholar * Parham Ghasemloo Gheidari View
author publications You can also search for this author inPubMed Google Scholar * Alexander Butyaev View author publications You can also search for this author inPubMed Google Scholar *
Timothy Keding View author publications You can also search for this author inPubMed Google Scholar * Eddie Cai View author publications You can also search for this author inPubMed Google
Scholar * Jiayue Zheng View author publications You can also search for this author inPubMed Google Scholar * Renata Mutalova View author publications You can also search for this author
inPubMed Google Scholar * Julien Mounthanyvong View author publications You can also search for this author inPubMed Google Scholar * Yuxue Zhu View author publications You can also search
for this author inPubMed Google Scholar * Elena Nazarova View author publications You can also search for this author inPubMed Google Scholar * Chrisostomos Drogaris View author publications
You can also search for this author inPubMed Google Scholar * Kornél Erhart View author publications You can also search for this author inPubMed Google Scholar * Amélie Brouillette View
author publications You can also search for this author inPubMed Google Scholar * Gabriel Richard View author publications You can also search for this author inPubMed Google Scholar * Randy
Pitchford View author publications You can also search for this author inPubMed Google Scholar * Sébastien Caisse View author publications You can also search for this author inPubMed
Google Scholar * Mathieu Blanchette View author publications You can also search for this author inPubMed Google Scholar * Daniel McDonald View author publications You can also search for
this author inPubMed Google Scholar * Rob Knight View author publications You can also search for this author inPubMed Google Scholar * Attila Szantner View author publications You can also
search for this author inPubMed Google Scholar * Jérôme Waldispühl View author publications You can also search for this author inPubMed Google Scholar CONSORTIA BORDERLANDS SCIENCE
DEVELOPMENT TEAM * David Bélanger * , Michael Bouffard * , Joshua Davidson * , Mathieu Falaise * , Vincent Fiset * , Steven Hebert * , Dan Hewitt * , Jonathan Huot * , Seung Kim * , Jonathan
Moreau-Genest * , David Najjab * , Steve Prince * & Ludger Saintélien BORDERLANDS SCIENCE PLAYERS CONTRIBUTIONS R.S.-G. contributed to the design of the game, designed the puzzles used
in the game and designed and wrote the alpha version of the code related to the puzzle generation pipeline, the solution processing and analysis pipeline and the initial versions of the
solution filtering and alignment improvement pipelines. R.S.-G. also designed and implemented the validation process and computed most of the results, authored most of the figures and wrote
most of the paper. P.G.G. designed and implemented much of the final version of the alignment improvement pipeline. A.B. designed and implemented much of the database systems, managed the
data and hardware systems and prepared the data release. T.K. contributed to the puzzle generation and alignment improvement pipelines and improved the implementation of much of the project
codebase. E.C. designed and implemented much of the final version of the solution filtering pipeline. J.Z., R.M., J.M., Y.Z., E.N. and C.D. are the other members of the scientific team who
contributed to the design and/or the implementation of specific scripts and figures. This group performed this work under the supervision of J.W. and M.B. at the School of Computer Science
at McGill University. M.B. contributed to the analysis of the results and reviewed the paper. J.W. contributed to the design of the game and methods, analysis of the results, writing of the
manuscript and the acquisition of funding. The _Borderlands Science_ mini game was developed by the Gearbox Entertainment _Borderlands Science_ development team. The game design was
performed as a team under the supervision of G.R. The overall development and integration of _Borderlands Science_ into _Borderlands 3_ was coordinated by A.B., under the leadership of S.C.
and R.P. Data collection and sequencing were performed by The Microsetta Initiative, led by D.M. and R.K. They contributed to the scientific aspects of the project on aspects related to
data, phylogeny, effect size computations and validation and reviewed the paper. A.Sz. presented the initial idea of the _Borderlands Science_ concept and contributed to the writing of the
paper. A.Sz. and K.E. designed, developed and maintain the technological framework connecting the scientific and game development teams. CORRESPONDING AUTHOR Correspondence to Jérôme
Waldispühl. ETHICS DECLARATIONS COMPETING INTERESTS J.W. is supported by a Genome Canada grant (Genomic Applications Partnership Program), which is co-funded by Gearbox Studio Québec, Inc.,
and Massively Multiplayer Online Science (MMOS Sàrl). J.W. is an occasional scientific consultant for Takeda pharmaceuticals, for which he receives income. The agreement has been reviewed
and approved by McGill University in accordance with its conflict of interest policies. A.Sz. is the CEO and founder of MMOS, a Swiss innovator company that presented the initial idea and
set up the first major collaborations between citizen science and video games and provides the underlying technological services to connect these two entities. MMOS received operating fees
from Gearbox Studio Québec, Inc. S.C. is co-studio head at Gearbox Studio Québec, Inc., which develops the game _Borderlands 3_ and the citizen science game _Borderlands Science_. R.P. is
president of Gearbox Entertainment Company, Inc., which develops the game _Borderlands 3_ and the citizen science game _Borderlands Science._ _Borderlands Science_ is a free mini game
available within the _Borderlands 3_ game, but _Borderlands 3_ is sold as a premium game. R.K. is a scientific advisory board member and consultant for BiomeSense, Inc., has equity and
receives income. He is a scientific advisory board member and has equity in GenCirq. He is a consultant and scientific advisory board member for DayTwo and receives income. He has equity in
and acts as a consultant for Cybele. He is a co-founder of Biota, Inc., and has equity. He is a cofounder of Micronoma and has equity and is a scientific advisory board member. D.M. is a
consultant for, and has equity in, BiomeSense, Inc. The terms of these arrangements have been reviewed and approved by the University of California, San Diego, in accordance with its
conflict of interest policies. PEER REVIEW PEER REVIEW INFORMATION _Nature Biotechnology_ thanks Paul Gardner, Firas Khatib and the other, anonymous, reviewer(s) for their contribution to
the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
SUPPLEMENTARY INFORMATION SUPPLEMENTARY METHODS, RESULTS AND DISCUSSION. REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution
4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and
the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s
Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not
permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Sarrazin-Gendron, R., Ghasemloo Gheidari, P., Butyaev, A. _et al._ Improving
microbial phylogeny with citizen science within a mass-market video game. _Nat Biotechnol_ 43, 76–84 (2025). https://doi.org/10.1038/s41587-024-02175-6 Download citation * Received: 21
August 2023 * Accepted: 05 February 2024 * Published: 15 April 2024 * Issue Date: January 2025 * DOI: https://doi.org/10.1038/s41587-024-02175-6 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative




:max_bytes(150000):strip_icc():focal(216x0:218x2)/jessica-simpson-435-1-1d4aefc159624d78a704ad541e01b69b.jpg)