- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
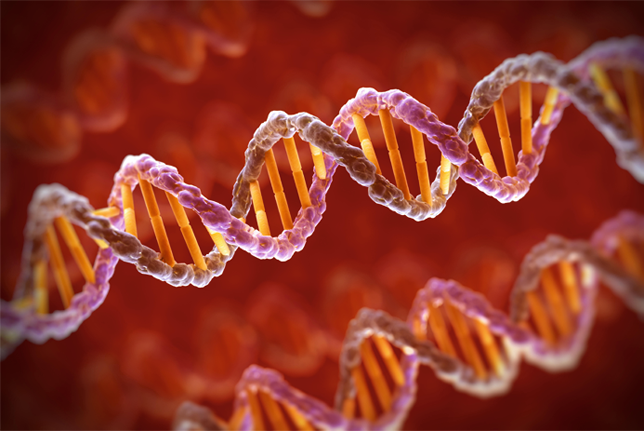
Early data from the 100,000 Genomes Project reveal to what extent patients with undiagnosed rare diseases can end their diagnostic odysseys by having their whole genomes sequenced. For the
pilot study, published in the New England Journal of Medicine, the UK National Health Service (NHS) collected data from 4,660 people from 2,183 families with a broad swath of rare diseases.
The pilot’s analysis of the pioneering project, run by Genomics England, revealed that a quarter of patients with rare diseases received a new diagnosis. Of these, 14% included genome
variants and coding variants in regions that would have been missed by exome sequencing and other genetic tests. For the study, the researchers collected clinical features data with Human
Phenotype Ontology terms, along with genome sequences obtained with HiSeq 2500 Illumina instruments. “The key to the successful diagnosis of the patients reported in this study is the design
of the analysis, where the investigators integrated the clinical features of the disease with the genome sequencing data,” says Bridget Bax, Reader in Rare Diseases, St George’s, University
of London.
Genomics England, wholly owned by the Department of Health and Social Care, was set up in 2013 to deliver the 100,000 Genomes Project. The pilot analysis shows that in 25% of those who
received a diagnosis, physicians could make immediate clinical decisions tailored to the patient’s condition. The researchers hope that their findings will help other health systems consider
the use of genome sequencing to care for patients with rare disease. The genomic and clinical data from the pilot study are freely accessible to members of a Genomics England Clinical
Interpretation Partnership.
Anyone you share the following link with will be able to read this content:




![[Withdrawn] Personal information charter - Department for Business, Energy & Industrial Strategy - GOV.UK](https://www.gov.uk/assets/static/govuk-opengraph-image-03837e1cec82f217cf32514635a13c879b8c400ae3b1c207c5744411658c7635.png)


