- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Transfer RNAs (tRNAs) have been historically viewed as non-dynamic adaptors that decode the genetic code into proteins. Recent work has uncovered dynamic regulatory roles for these
fascinating molecules. Advances in tRNA detection methods have revealed that specific tRNAs can become modulated upon DNA copy number and chromatin alterations and can also be perturbed by
oncogenic signalling and transcriptional regulators in cancer cells or the tumour microenvironment. Such alterations in the levels of specific tRNAs have been shown to causally impact cancer
progression, including metastasis. Moreover, sequencing methods have identified tRNA-derived small RNAs that influence various aspects of cancer progression, such as cell proliferation and
invasion, and could serve as diagnostic and prognostic biomarkers or putative therapeutic targets in various cancers. Finally, there is accumulating evidence, including from genetic models,
that specific tRNA synthetases — the enzymes responsible for charging tRNAs with amino acids — can either promote or suppress tumour formation. In this Review, we provide an overview of how
deregulation of tRNAs influences cancer formation and progression. Access through your institution Buy or subscribe This is a preview of subscription content, access via your institution
ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time
Learn more Subscribe to this journal Receive 12 print issues and online access $209.00 per year only $17.42 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access
to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our
FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS SNORNAS: FUNCTIONS AND MECHANISMS IN BIOLOGICAL PROCESSES, AND ROLES IN TUMOR PATHOPHYSIOLOGY Article Open access 12
May 2022 SMALL NON-CODING RNAS IN HUMAN CANCER: FUNCTION, CLINICAL UTILITY, AND CHARACTERIZATION Article 15 January 2021 ILLUMINATING THE NONCODING GENOME IN CANCER Article 14 September 2020
REFERENCES * Hoagland, M. B., Zamecnik, P. C. & Stephenson, M. L. Intermediate reactions in protein biosynthesis. _Biochim. Biophys. Acta_ 24, 215–216 (1957). Article CAS PubMed
Google Scholar * Ogata, K. & Nohara, H. The possible role of the ribonucleic acid (RNA) of the pH 5 enzyme in amino acid activation. _Biochim. Biophys. Acta_ 25, 659–660 (1957). Article
CAS PubMed Google Scholar * Holley, R. W. An alanine-dependent, ribonuclease-inhibited conversion of AMP to ATP, and its possible relationship to protein synthesis. _J. Am. Chem. Soc._
79, 658–662 (1957). THIS STUDY, TOGETHER WITH HOAGLAND ET AL. (1957) AND OGATA ET AL. (1957), IS ONE OF THE ORIGINAL STUDIES THAT DISCOVERED TRNAS. Article CAS Google Scholar * Söll, D.
& RajBhandary, U. L. _tRNA: Structure, Biosynthesis, and Function_ (Wiley, 1995). * Suzuki, T. The expanding world of tRNA modifications and their disease relevance. _Nat. Rev. Mol. Cell
Biol._ 22, 375–392 (2021). Article CAS PubMed Google Scholar * Suzuki, T., Nagao, A. & Suzuki, T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases.
_Annu. Rev. Genet._ 45, 299–329 (2011). THIS IS A COMPREHENSIVE REVIEW ON HUMAN MT-TRNAS. Article CAS PubMed Google Scholar * Orellana, E. A., Siegal, E. & Gregory, R. I. tRNA
dysregulation and disease. _Nat. Rev. Genet._ 23, 651–664 (2022). Article CAS PubMed Google Scholar * Holley, R. W. et al. Structure of a ribonucleic acid. _Science_ 147, 1462–1465
(1965). THIS PAPER DESCRIBES THE FIRST SEQUENCE OF A TRNA. Article CAS PubMed Google Scholar * Kim, S. H. et al. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA.
_Science_ 185, 435–440 (1974). Article CAS PubMed Google Scholar * Cramer, F., Erdmann, V. A., von der Haar, F. & Schlimme, E. Structure and reactivity of tRNA. _J. Cell Physiol._
74, 163–178 (1969). Article CAS Google Scholar * Ramsay, E. P. & Vannini, A. Structural rearrangements of the RNA polymerase III machinery during tRNA transcription initiation.
_Biochim. Biophys. Acta Gene Regul. Mech._ 1861, 285–294 (2018). Article CAS PubMed Google Scholar * Phizicky, E. M. & Hopper, A. K. tRNA biology charges to the front. _Genes Dev._
24, 1832–1860 (2010). Article PubMed PubMed Central Google Scholar * Rak, R. et al. Dynamic changes in tRNA modifications and abundance during T cell activation. _Proc. Natl Acad. Sci.
USA_ https://doi.org/10.1073/pnas.2106556118 (2021). * Torrent, M., Chalancon, G., de Groot, N. S., Wuster, A. & Madan Babu, M. Cells alter their tRNA abundance to selectively regulate
protein synthesis during stress conditions. _Sci. Signal._ 11, eaat6409 (2018). Article PubMed PubMed Central Google Scholar * Dittmar, K. A., Goodenbour, J. M. & Pan, T.
Tissue-specific differences in human transfer RNA expression. _PLoS Genet._ 2, e221 (2006). Article PubMed PubMed Central Google Scholar * Pinkard, O., McFarland, S., Sweet, T. &
Coller, J. Quantitative tRNA-sequencing uncovers metazoan tissue-specific tRNA regulation. _Nat. Commun._ 11, 4104 (2020). Article CAS PubMed PubMed Central Google Scholar * Kimura, S.,
Dedon, P. C. & Waldor, M. K. Comparative tRNA sequencing and RNA mass spectrometry for surveying tRNA modifications. _Nat. Chem. Biol._ 16, 964–972 (2020). Article CAS PubMed PubMed
Central Google Scholar * Gingold, H. et al. A dual program for translation regulation in cellular proliferation and differentiation. _Cell_ 158, 1281–1292 (2014). THIS PAPER CLASSIFIES
‘PROLIFERATION-SPECIFIC’ AND ‘DIFFERENTIATION-SPECIFIC’ TRNAS. Article CAS PubMed Google Scholar * Oler, A. J. et al. Human RNA polymerase III transcriptomes and relationships to Pol II
promoter chromatin and enhancer-binding factors. _Nat. Struct. Mol. Biol._ 17, 620–628 (2010). Article CAS PubMed PubMed Central Google Scholar * Barski, A. et al. Pol II and its
associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. _Nat. Struct. Mol. Biol._ 17, 629–634 (2010). Article CAS PubMed PubMed Central Google Scholar *
Goodarzi, H. et al. Modulated expression of specific tRNAs drives gene expression and cancer progression. _Cell_ 165, 1416–1427 (2016). Article CAS PubMed PubMed Central Google Scholar
* Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2021 update. _Nucleic Acids Res._ 50, D231–D235 (2022). Article CAS PubMed Google Scholar * Zhang, W., Foo, M.,
Eren, A. M. & Pan, T. tRNA modification dynamics from individual organisms to metaepitranscriptomics of microbiomes. _Mol. Cell_ 82, 891–906 (2022). THIS ARTICLE INCLUDES A SYSTEMATIC
ANALYSIS OF CURRENT TRNA PROFILING METHODS. Article CAS PubMed PubMed Central Google Scholar * Chujo, T. & Tomizawa, K. Human transfer RNA modopathies: diseases caused by
aberrations in transfer RNA modifications. _FEBS J._ 288, 7096–7122 (2021). Article CAS PubMed PubMed Central Google Scholar * Barbieri, I. & Kouzarides, T. Role of RNA
modifications in cancer. _Nat. Rev. Cancer_ 20, 303–322 (2020). Article CAS PubMed Google Scholar * Clark, W. C., Evans, M. E., Dominissini, D., Zheng, G. & Pan, T. tRNA base
methylation identification and quantification via high-throughput sequencing. _RNA_ 22, 1771–1784 (2016). Article CAS PubMed PubMed Central Google Scholar * Crick, F. H. Codon–anticodon
pairing: the wobble hypothesis. _J. Mol. Biol._ 19, 548–555 (1966). Article CAS PubMed Google Scholar * Chan, P. P. & Lowe, T. M. GtRNAdb 2.0: an expanded database of transfer RNA
genes identified in complete and draft genomes. _Nucleic Acids Res._ 44, D184–D189 (2016). Article CAS PubMed Google Scholar * Chan, P. P., Lin, B. Y., Mak, A. J. & Lowe, T. M.
tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. _Nucleic Acids Res._ 49, 9077–9096 (2021). THIS RESOURCE PAPER, TOGETHER WITH CHAN ET AL. (2016),
DESCRIBES THE GTRNADB DATABASE OF PREDICTED TRNA SEQUENCES. Article CAS PubMed PubMed Central Google Scholar * Pavon-Eternod, M. et al. tRNA over-expression in breast cancer and
functional consequences. _Nucleic Acids Res._ 37, 7268–7280 (2009). THIS PAPER IS ONE OF THE FIRST TO ATTEMPT GLOBAL PROFILING OF TRNA LEVELS IN CANCER USING MICROARRAYS. Article CAS
PubMed PubMed Central Google Scholar * Pavon-Eternod, M., Gomes, S., Rosner, M. R. & Pan, T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA
expression and increased proliferation in human epithelial cells. _RNA_ 19, 461–466 (2013). Article CAS PubMed PubMed Central Google Scholar * Earnest-Noble, L. B. et al. Two isoleucyl
tRNAs that decode synonymous codons divergently regulate breast cancer metastatic growth by controlling translation of proliferation-regulating genes. _Nat. Cancer_ 3, 1484–1497 (2022).
Article CAS PubMed Google Scholar * Taylor, M. W., Granger, G. A., Buck, C. A. & Holland, J. J. Similarities and differences among specific tRNA’s in mammalian tissues. _Proc. Natl
Acad. Sci. USA_ 57, 1712–1719 (1967). Article CAS PubMed PubMed Central Google Scholar * Gallo, R. C. Transfer RNA’s in human leukemia. _J. Cell Physiol._ 74, 149–153 (1969). Article
CAS Google Scholar * Baliga, B. S., Borek, E., Weinstein, I. B. & Srinivasan, P. R. Differences in the transfer RNA’s of normal liver and Novikoff hepatoma. _Proc. Natl Acad. Sci. USA_
62, 899–905 (1969). Article CAS PubMed PubMed Central Google Scholar * Sonenberg, N. & Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and
biological targets. _Cell_ 136, 731–745 (2009). Article CAS PubMed PubMed Central Google Scholar * Birch, J. et al. The initiator methionine tRNA drives cell migration and invasion
leading to increased metastatic potential in melanoma. _Biol. Open_ 5, 1371–1379 (2016). Article CAS PubMed PubMed Central Google Scholar * Ruggero, D. Translational control in cancer
etiology. _Cold Spring Harb. Perspect. Biol._ 5, a012336 (2013). Article PubMed PubMed Central Google Scholar * Silvera, D., Formenti, S. C. & Schneider, R. J. Translational control
in cancer. _Nat. Rev. Cancer_ 10, 254–266 (2010). Article CAS PubMed Google Scholar * Clarke, C. J. et al. The initiator methionine tRNA drives secretion of type II collagen from stromal
fibroblasts to promote tumor growth and angiogenesis. _Curr. Biol._ 26, 755–765 (2016). Article CAS PubMed PubMed Central Google Scholar * Yang, J. et al. SOX4-mediated repression of
specific tRNAs inhibits proliferation of human glioblastoma cells. _Proc. Natl Acad. Sci. USA_ 117, 5782–5790 (2020). Article CAS PubMed PubMed Central Google Scholar * Wang, B. et al.
miR-34a directly targets tRNAiMet precursors and affects cellular proliferation, cell cycle, and apoptosis. _Proc. Natl Acad. Sci. USA_ 115, 7392–7397 (2018). Article CAS PubMed PubMed
Central Google Scholar * Shigematsu, M. et al. YAMAT-seq: an efficient method for high-throughput sequencing of mature transfer RNAs. _Nucleic Acids Res._ 45, e70 (2017). CAS PubMed
PubMed Central Google Scholar * Behrens, A., Rodschinka, G. & Nedialkova, D. D. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by
mim-tRNAseq. _Mol. Cell_ 81, 1802–1815.e7 (2021). Article CAS PubMed PubMed Central Google Scholar * Gogakos, T. et al. Characterizing expression and processing of precursor and mature
human tRNAs by hydro-tRNAseq and PAR-CLIP. _Cell Rep._ 20, 1463–1475 (2017). Article CAS PubMed PubMed Central Google Scholar * Lucas, M. C. et al. Quantitative analysis of tRNA
abundance and modifications by nanopore RNA sequencing. _Nat. Biotechnol_. https://doi.org/10.1038/s41587-023-01743-6 (2023). Article PubMed Google Scholar * Iben, J. R. & Maraia, R.
J. tRNA gene copy number variation in humans. _Gene_ 536, 376–384 (2014). Article CAS PubMed Google Scholar * Darrow, E. M. & Chadwick, B. P. A novel tRNA variable number tandem
repeat at human chromosome 1q23.3 is implicated as a boundary element based on conservation of a CTCF motif in mouse. _Nucleic Acids Res._ 42, 6421–6435 (2014). Article CAS PubMed PubMed
Central Google Scholar * Parisien, M., Wang, X. & Pan, T. Diversity of human tRNA genes from the 1000-Genomes Project. _RNA Biol._ 10, 1853–1867 (2014). Article Google Scholar *
Berg, M. D. et al. Targeted sequencing reveals expanded genetic diversity of human transfer RNAs. _RNA Biol._ 16, 1574–1585 (2019). Article PubMed PubMed Central Google Scholar *
Thornlow, B. P. et al. Transfer RNA genes experience exceptionally elevated mutation rates. _Proc. Natl Acad. Sci. USA_ 115, 8996–9001 (2018). Article CAS PubMed PubMed Central Google
Scholar * Ishimura, R. et al. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. _Science_ 345, 455–459 (2014). THIS IS ONE OF THE FIRST REPORTS OF A
TRNA MUTATION CAUSING A TISSUE-SPECIFIC PHENOTYPE IN A MOUSE MODEL. Article CAS PubMed PubMed Central Google Scholar * Schoenmakers, E. et al. Mutation in human selenocysteine transfer
RNA selectively disrupts selenoprotein synthesis. _J. Clin. Invest._ 126, 992–996 (2016). Article PubMed PubMed Central Google Scholar * Santos, M. et al. Codon misreading tRNAs promote
tumor growth in mice. _RNA Biol._ 15, 773–786 (2018). PubMed PubMed Central Google Scholar * Doran, J. L., Bingle, W. H. & Roy, K. L. Two human genes encoding tRNA. _Gene_ 65, 329–336
(1988). Article CAS PubMed Google Scholar * Sprague, K. U., Larson, D. & Morton, D. 5′ Flanking sequence signals are required for activity of silkworm alanine tRNA genes in
homologous in vitro transcription systems. _Cell_ 22, 171–178 (1980). Article CAS PubMed Google Scholar * Park, J. L. et al. Epigenetic regulation of noncoding RNA transcription by
mammalian RNA polymerase III. _Epigenomics_ 9, 171–187 (2017). Article CAS PubMed Google Scholar * White, R. J. Transcription by RNA polymerase III: more complex than we thought. _Nat.
Rev. Genet._ 12, 459–463 (2011). Article CAS PubMed Google Scholar * Larminie, C. G. et al. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. _EMBO J._
16, 2061–2071 (1997). Article CAS PubMed PubMed Central Google Scholar * Cairns, C. A. & White, R. J. p53 is a general repressor of RNA polymerase III transcription. _EMBO J._ 17,
3112–3123 (1998). Article CAS PubMed PubMed Central Google Scholar * Crighton, D. et al. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter
occupancy by TFIIIB. _EMBO J._ 22, 2810–2820 (2003). Article CAS PubMed PubMed Central Google Scholar * Gomez-Roman, N., Grandori, C., Eisenman, R. N. & White, R. J. Direct
activation of RNA polymerase III transcription by c-Myc. _Nature_ 421, 290–294 (2003). Article CAS PubMed Google Scholar * Kenneth, N. S. et al. TRRAP and GCN5 are used by c-Myc to
activate RNA polymerase III transcription. _Proc. Natl Acad. Sci. USA_ 104, 14917–14922 (2007). Article CAS PubMed PubMed Central Google Scholar * Willis, I. M. & Moir, R. D.
Signaling to and from the RNA polymerase III transcription and processing machinery. _Annu. Rev. Biochem._ 87, 75–100 (2018). Article CAS PubMed PubMed Central Google Scholar * Raha, D.
et al. Close association of RNA polymerase II and many transcription factors with Pol III genes. _Proc. Natl Acad. Sci. USA_ 107, 3639–3644 (2010). Article CAS PubMed PubMed Central
Google Scholar * Zhang, J. et al. SOX4 inhibits GBM cell growth and induces G0/G1 cell cycle arrest through Akt–p53 axis. _BMC Neurol._ 14, 207 (2014). Article PubMed PubMed Central
Google Scholar * Smith, D. K., Yang, J., Liu, M. L. & Zhang, C. L. Small molecules modulate chromatin accessibility to promote NEUROG2-mediated fibroblast-to-neuron reprogramming. _Stem
Cell Rep._ 7, 955–969 (2016). Article CAS Google Scholar * Lee, H., Goodarzi, H., Tavazoie, S. F. & Alarcon, C. R. TMEM2 is a SOX4-regulated gene that mediates metastatic migration
and invasion in breast cancer. _Cancer Res._ 76, 4994–5005 (2016). Article CAS PubMed PubMed Central Google Scholar * Vervoort, S. J., van Boxtel, R. & Coffer, P. J. The role of
SRY-related HMG box transcription factor 4 (SOX4) in tumorigenesis and metastasis: friend or foe? _Oncogene_ 32, 3397–3409 (2013). Article CAS PubMed Google Scholar * Besser, D. et al.
DNA methylation inhibits transcription by RNA polymerase III of a tRNA gene, but not of a 5S rRNA gene. _FEBS Lett._ 269, 358–362 (1990). Article CAS PubMed Google Scholar * Bartke, T.
et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. _Cell_ 143, 470–484 (2010). Article CAS PubMed PubMed Central Google Scholar * Rossello-Tortella, M.,
Bueno-Costa, A., Martinez-Verbo, L., Villanueva, L. & Esteller, M. DNA methylation-associated dysregulation of transfer RNA expression in human cancer. _Mol. Cancer_ 21, 48 (2022).
Article CAS PubMed PubMed Central Google Scholar * Tavtigian, S. V. et al. A candidate prostate cancer susceptibility gene at chromosome 17p. _Nat. Genet._ 27, 172–180 (2001). Article
CAS PubMed Google Scholar * Xu, B., Tong, N., Li, J. M., Zhang, Z. D. & Wu, H. F. ELAC2 polymorphisms and prostate cancer risk: a meta-analysis based on 18 case-control studies.
_Prostate Cancer Prostatic Dis._ 13, 270–277 (2010). Article CAS PubMed PubMed Central Google Scholar * Lubas, M., Chlebowski, A., Dziembowski, A. & Jensen, T. H. Biochemistry and
function of RNA exosomes. _Enzymes_ 31, 1–30 (2012). Article CAS PubMed Google Scholar * Kadaba, S. et al. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in _S.
cerevisiae_. _Genes Dev._ 18, 1227–1240 (2004). Article CAS PubMed PubMed Central Google Scholar * Whipple, J. M., Lane, E. A., Chernyakov, I., D’Silva, S. & Phizicky, E. M. The
yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. _Genes Dev._ 25, 1173–1184 (2011). Article CAS PubMed PubMed Central
Google Scholar * Alexandrov, A. et al. Rapid tRNA decay can result from lack of nonessential modifications. _Mol. Cell_ 21, 87–96 (2006). Article CAS PubMed Google Scholar * De Zoysa,
T. & Phizicky, E. M. Hypomodified tRNA in evolutionarily distant yeasts can trigger rapid tRNA decay to activate the general amino acid control response, but with different
consequences. _PLoS Genet._ 16, e1008893 (2020). Article PubMed PubMed Central Google Scholar * Wilusz, J. E., Whipple, J. M., Phizicky, E. M. & Sharp, P. A. tRNAs marked with CCACCA
are targeted for degradation. _Science_ 334, 817–821 (2011). Article CAS PubMed PubMed Central Google Scholar * Orellana, E. A. et al. METTL1-mediated m7G modification of Arg-TCT tRNA
drives oncogenic transformation. _Mol. Cell_ 81, 3323–3338.e14 (2021). Article CAS PubMed PubMed Central Google Scholar * Han, H. et al. N7-methylguanosine tRNA modification promotes
esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. _Nat. Commun._ 13, 1478 (2022). Article CAS PubMed PubMed Central Google Scholar * Chen, J. et al.
Aberrant translation regulated by METTL1/WDR4-mediated tRNA N7-methylguanosine modification drives head and neck squamous cell carcinoma progression. _Cancer Commun._ 42, 223–244 (2022).
Article Google Scholar * Dai, Z. et al. N7-methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. _Mol. Cell_ 81,
3339–3355.e8 (2021). Article CAS PubMed Google Scholar * Chen, Z. et al. METTL1 promotes hepatocarcinogenesis via m7G tRNA modification-dependent translation control. _Clin. Transl.
Med._ 11, e661 (2021). Article CAS PubMed PubMed Central Google Scholar * Pandolfini, L. et al. METTL1 promotes let-7 microRNA processing via m7G methylation. _Mol. Cell_ 74,
1278–1290.e9 (2019). Article CAS PubMed PubMed Central Google Scholar * Cheng, W., Gao, A., Lin, H. & Zhang, W. Novel roles of METTL1/WDR4 in tumor via m7G methylation. _Mol. Ther.
Oncolytics_ 26, 27–34 (2022). Article CAS PubMed PubMed Central Google Scholar * Girstmair, H. et al. Depletion of cognate charged transfer RNA causes translational frameshifting within
the expanded CAG stretch in huntingtin. _Cell Rep._ 3, 148–159 (2013). Article CAS PubMed Google Scholar * Guzy, R. D. et al. Mitochondrial complex III is required for hypoxia-induced
ROS production and cellular oxygen sensing. _Cell Metab._ 1, 401–408 (2005). Article CAS PubMed Google Scholar * Bell, E. L. et al. The Qo site of the mitochondrial complex III is
required for the transduction of hypoxic signaling via reactive oxygen species production. _J. Cell Biol._ 177, 1029–1036 (2007). Article CAS PubMed PubMed Central Google Scholar *
Nawrot, B., Sochacka, E. & Duchler, M. tRNA structural and functional changes induced by oxidative stress. _Cell Mol. Life Sci._ 68, 4023–4032 (2011). Article CAS PubMed PubMed
Central Google Scholar * Endres, L. et al. Alkbh8 regulates selenocysteine-protein expression to protect against reactive oxygen species damage. _PLoS ONE_ 10, e0131335 (2015). Article
PubMed PubMed Central Google Scholar * Chan, C. T. et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. _Nat.
Commun._ 3, 937 (2012). Article PubMed Google Scholar * Netzer, N. et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. _Nature_ 462, 522–526
(2009). Article CAS PubMed PubMed Central Google Scholar * Schwenzer, H. et al. Oxidative stress triggers selective tRNA retrograde transport in human cells during the integrated stress
response. _Cell Rep._ 26, 3416–3428.e5 (2019). Article CAS PubMed PubMed Central Google Scholar * Thompson, D. M., Lu, C., Green, P. J. & Parker, R. tRNA cleavage is a conserved
response to oxidative stress in eukaryotes. _RNA_ 14, 2095–2103 (2008). Article CAS PubMed PubMed Central Google Scholar * Goodarzi, H. et al. Endogenous tRNA-derived fragments suppress
breast cancer progression via YBX1 displacement. _Cell_ 161, 790–802 (2015). Article CAS PubMed PubMed Central Google Scholar * Huh, D. et al. A stress-induced tyrosine-tRNA depletion
response mediates codon-based translational repression and growth suppression. _EMBO J._ 40, e106696 (2021). Article CAS PubMed Google Scholar * Yamasaki, S., Ivanov, P., Hu, G. F. &
Anderson, P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. _J. Cell Biol._ 185, 35–42 (2009). Article CAS PubMed PubMed Central Google Scholar * Masson,
G. R. Towards a model of GCN2 activation. _Biochem. Soc. Trans._ 47, 1481–1488 (2019). Article CAS PubMed PubMed Central Google Scholar * Pataskar, A. et al. Tryptophan depletion
results in tryptophan-to-phenylalanine substitutants. _Nature_ 603, 721–727 (2022). Article CAS PubMed PubMed Central Google Scholar * Hsu, D. J. et al. Arginine limitation drives a
directed codon-dependent DNA sequence evolution response in colorectal cancer cells. _Sci. Adv._ 9, eade9120 (2023). Article PubMed PubMed Central Google Scholar * Passarelli, M. C. et
al. Leucyl-tRNA synthetase is a tumour suppressor in breast cancer and regulates codon-dependent translation dynamics. _Nat. Cell Biol._ 24, 307–315 (2022). Article CAS PubMed PubMed
Central Google Scholar * Turvey, A. K., Horvath, G. A. & Cavalcanti, A. R. O. Aminoacyl-tRNA synthetases in human health and disease. _Front. Physiol._ 13, 1029218 (2022). Article
PubMed PubMed Central Google Scholar * Hyeon, D. Y. et al. Evolution of the multi-tRNA synthetase complex and its role in cancer. _J. Biol. Chem._ 294, 5340–5351 (2019). Article CAS
PubMed PubMed Central Google Scholar * Wang, J. & Yang, X. L. Novel functions of cytoplasmic aminoacyl-tRNA synthetases shaping the hallmarks of cancer. _Enzymes_ 48, 397–423 (2020).
Article PubMed Google Scholar * Sung, Y., Yoon, I., Han, J. M. & Kim, S. Functional and pathologic association of aminoacyl-tRNA synthetases with cancer. _Exp. Mol. Med._ 54, 553–566
(2022). Article CAS PubMed PubMed Central Google Scholar * Guo, M. & Schimmel, P. Essential nontranslational functions of tRNA synthetases. _Nat. Chem. Biol._ 9, 145–153 (2013).
Article PubMed PubMed Central Google Scholar * Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. _Cell_ 144, 646–674 (2011). Article CAS PubMed Google
Scholar * Wang, J. et al. Multi-omics database analysis of aminoacyl-tRNA synthetases in cancer. _Genes_ 11, 1384 (2020). Article CAS PubMed PubMed Central Google Scholar * Thandapani,
P. et al. Valine tRNA levels and availability regulate complex I assembly in leukaemia. _Nature_ 601, 428–433 (2022). Article CAS PubMed Google Scholar * Su, Z., Wilson, B., Kumar, P.
& Dutta, A. Noncanonical roles of tRNAs: tRNA fragments and beyond. _Annu. Rev. Genet._ 54, 47–69 (2020). Article CAS PubMed PubMed Central Google Scholar * Borek, E. et al. High
turnover rate of transfer RNA in tumor tissue. _Cancer Res._ 37, 3362–3366 (1977). CAS PubMed Google Scholar * Speer, J., Gehrke, C. W., Kuo, K. C., Waalkes, T. P. & Borek, E. tRNA
breakdown products as markers for cancer. _Cancer_ 44, 2120–2123 (1979). Article CAS PubMed Google Scholar * Kumar, P., Anaya, J., Mudunuri, S. B. & Dutta, A. Meta-analysis of tRNA
derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. _BMC Biol._ 12, 78 (2014). Article PubMed PubMed
Central Google Scholar * Pliatsika, V. et al. MINTbase v2.0: a comprehensive database for tRNA-derived fragments that includes nuclear and mitochondrial fragments from all the cancer
genome atlas projects. _Nucleic Acids Res._ 46, D152–D159 (2018). Article CAS PubMed Google Scholar * Yao, D. et al. OncotRF: an online resource for exploration of tRNA-derived fragments
in human cancers. _RNA Biol._ 17, 1081–1091 (2020). Article CAS PubMed PubMed Central Google Scholar * Lee, S. R. & Collins, K. Starvation-induced cleavage of the tRNA anticodon
loop in _Tetrahymena thermophila_. _J. Biol. Chem._ 280, 42744–42749 (2005). Article CAS PubMed Google Scholar * Tao, E. W., Cheng, W. Y., Li, W. L., Yu, J. & Gao, Q. Y. tiRNAs: a
novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. _J. Cell Physiol._ 235, 683–690 (2020). Article CAS PubMed Google Scholar
* Fu, H. et al. Stress induces tRNA cleavage by angiogenin in mammalian cells. _FEBS Lett._ 583, 437–442 (2009). Article CAS PubMed Google Scholar * Donovan, J., Rath, S.,
Kolet-Mandrikov, D. & Korennykh, A. Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. _RNA_ 23, 1660–1671 (2017).
Article CAS PubMed PubMed Central Google Scholar * Nechooshtan, G., Yunusov, D., Chang, K. & Gingeras, T. R. Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in
the extracellular environment. _Nucleic Acids Res._ 48, 8035–8049 (2020). Article CAS PubMed PubMed Central Google Scholar * Lee, Y. S., Shibata, Y., Malhotra, A. & Dutta, A. A
novel class of small RNAs: tRNA-derived RNA fragments (tRFs). _Genes Dev._ 23, 2639–2649 (2009). Article CAS PubMed PubMed Central Google Scholar * Oberbauer, V. & Schaefer, M. R.
tRNA-derived small RNAs: biogenesis, modification, function and potential impact on human disease development. _Genes_ 9, 607 (2018). Article PubMed PubMed Central Google Scholar *
Guzzi, N. & Bellodi, C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. _RNA Biol._ 17, 1214–1222 (2020). Article CAS PubMed PubMed Central
Google Scholar * Fagan, S. G., Helm, M. & Prehn, J. H. M. tRNA-derived fragments: a new class of non-coding RNA with key roles in nervous system function and dysfunction. _Prog.
Neurobiol._ 205, 102118 (2021). Article CAS PubMed Google Scholar * Thompson, D. M. & Parker, R. Stressing out over tRNA cleavage. _Cell_ 138, 215–219 (2009). Article CAS PubMed
Google Scholar * Maute, R. L. et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. _Proc. Natl Acad. Sci. USA_ 110,
1404–1409 (2013). Article CAS PubMed PubMed Central Google Scholar * Balatti, V. et al. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia.
_Proc. Natl Acad. Sci. USA_ 112, 2169–2174 (2015). Article CAS PubMed PubMed Central Google Scholar * Pekarsky, Y. et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in
human cancer. _Proc. Natl Acad. Sci. USA_ 113, 5071–5076 (2016). Article CAS PubMed PubMed Central Google Scholar * Wang, H. et al. The tRNA-derived fragment tRF-24-V29K9UV3IU functions
as a miRNA-like RNA to prevent gastric cancer progression by inhibiting GPR78 expression. _J. Oncol._ 2022, 8777697 (2022). PubMed PubMed Central Google Scholar * Dong, D. D., Zhou, H.
& Li, G. GPR78 promotes lung cancer cell migration and metastasis by activation of Gαq-Rho GTPase pathway. _BMB Rep._ 49, 623–628 (2016). Article CAS PubMed PubMed Central Google
Scholar * Shi, Z. & Barna, M. Translating the genome in time and space: specialized ribosomes, RNA regulons, and RNA-binding proteins. _Annu. Rev. Cell Dev. Biol._ 31, 31–54 (2015).
Article CAS PubMed Google Scholar * Falconi, M. et al. A novel 3′-tRNAGlu-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin. _FASEB J._ 33, 13228–13240
(2019). Article CAS PubMed Google Scholar * Guzzi, N. et al. Pseudouridine-modified tRNA fragments repress aberrant protein synthesis and predict leukaemic progression in myelodysplastic
syndrome. _Nat. Cell Biol._ 24, 299–306 (2022). Article CAS PubMed PubMed Central Google Scholar * Guzzi, N. et al. Pseudouridylation of tRNA-derived fragments steers translational
control in stem cells. _Cell_ 173, 1204–1216.e26 (2018). Article CAS PubMed Google Scholar * Pan, L. et al. Inflammatory cytokine-regulated tRNA-derived fragment tRF-21 suppresses
pancreatic ductal adenocarcinoma progression. _J. Clin. Invest._ 131, e148130 (2021). Article CAS PubMed PubMed Central Google Scholar * Tong, L. et al. The tRNA-derived fragment-3017A
promotes metastasis by inhibiting NELL2 in human gastric cancer. _Front. Oncol._ 10, 570916 (2020). Article PubMed Google Scholar * Zhang, F. et al. A 3′-tRNA-derived fragment enhances
cell proliferation, migration and invasion in gastric cancer by targeting FBXO47. _Arch. Biochem. Biophys._ 690, 108467 (2020). Article CAS PubMed Google Scholar * Han, L. et al. A
5′-tRNA halve, tiRNA-Gly promotes cell proliferation and migration via binding to RBM17 and inducing alternative splicing in papillary thyroid cancer. _J. Exp. Clin. Cancer Res._ 40, 222
(2021). Article CAS PubMed PubMed Central Google Scholar * Liu, X. et al. A pro-metastatic tRNA fragment drives nucleolin oligomerization and stabilization of its bound metabolic mRNAs.
_Mol. Cell_ 82, 2604–2617.e8 (2022). Article CAS PubMed PubMed Central Google Scholar * Lampson, B. L. et al. Rare codons regulate KRas oncogenesis. _Curr. Biol._ 23, 70–75 (2013).
Article CAS PubMed Google Scholar * Pershing, N. L. et al. Rare codons capacitate Kras-driven de novo tumorigenesis. _J. Clin. Invest._ 125, 222–233 (2015). Article PubMed Google
Scholar * Li, S. & Counter, C. M. Signaling levels mold the RAS mutation tropism of urethane. _eLife_ 10, e67172 (2021). Article CAS PubMed PubMed Central Google Scholar * Ali, M.
et al. Codon bias imposes a targetable limitation on KRAS-driven therapeutic resistance. _Nat. Commun._ 8, 15617 (2017). Article PubMed PubMed Central Google Scholar * Keller, T. L. et
al. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. _Nat. Chem. Biol._ 8, 311–317 (2012). Article CAS PubMed PubMed Central Google Scholar * Kim, Y. et
al. Aminoacyl-tRNA synthetase inhibition activates a pathway that branches from the canonical amino acid response in mammalian cells. _Proc. Natl Acad. Sci. USA_ 117, 8900–8911 (2020).
Article CAS PubMed PubMed Central Google Scholar * Kim, S. H., Bae, S. & Song, M. Recent development of aminoacyl-tRNA synthetase inhibitors for human diseases: a future
perspective. _Biomolecules_ 10, 1625 (2020). Article CAS PubMed PubMed Central Google Scholar * Zuko, A. et al. tRNA overexpression rescues peripheral neuropathy caused by mutations in
tRNA synthetase. _Science_ 373, 1161–1166 (2021). Article CAS PubMed PubMed Central Google Scholar * Wang, J. et al. AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice.
_Nature_ 604, 343–348 (2022). Article CAS PubMed PubMed Central Google Scholar * Albers, S. et al. Engineered tRNAs suppress nonsense mutations in cells and in vivo. _Nature_ 618,
842–848 (2023). Article CAS PubMed PubMed Central Google Scholar * Chang, J. C., Temple, G. F., Trecartin, R. F. & Kan, Y. W. Suppression of the nonsense mutation in homozygous β0
thalassaemia. _Nature_ 281, 602–603 (1979). Article CAS PubMed Google Scholar * Winkle, M., El-Daly, S. M., Fabbri, M. & Calin, G. A. Noncoding RNA therapeutics — challenges and
potential solutions. _Nat. Rev. Drug Discov._ 20, 629–651 (2021). Article CAS PubMed PubMed Central Google Scholar * Papadimitriou, M. A. et al. tRNA-derived fragments (tRFs) in bladder
cancer: increased 5′-tRF-LysCTT results in disease early progression and patients’ poor treatment outcome. _Cancers_ 12, 3661 (2020). Article PubMed PubMed Central Google Scholar *
Wang, J. et al. Circulating tRNA-derived small RNAs (tsRNAs) signature for the diagnosis and prognosis of breast cancer. _NPJ Breast Cancer_ 7, 4 (2021). Article CAS PubMed PubMed Central
Google Scholar * Wang, X. et al. Identification of tRNA-derived fragments expression profile in breast cancer tissues. _Curr. Genomics_ 20, 199–213 (2019). Article CAS PubMed PubMed
Central Google Scholar * Sun, C. et al. tRNA-derived fragments as novel predictive biomarkers for trastuzumab-resistant breast cancer. _Cell. Physiol. Biochem._ 49, 419–431 (2018). Article
CAS PubMed Google Scholar * Veneziano, D. et al. Dysregulation of different classes of tRNA fragments in chronic lymphocytic leukemia. _Proc. Natl Acad. Sci. USA_ 116, 24252–24258
(2019). Article CAS PubMed PubMed Central Google Scholar * Nientiedt, M. et al. Identification of aberrant tRNA-halves expression patterns in clear cell renal cell carcinoma. _Sci.
Rep._ 6, 37158 (2016). Article CAS PubMed PubMed Central Google Scholar * Wu, Y. et al. 5′-tRF-GlyGCC: a tRNA-derived small RNA as a novel biomarker for colorectal cancer diagnosis.
_Genome Med._ 13, 20 (2021). Article CAS PubMed PubMed Central Google Scholar * Zhu, Y. et al. Comprehensive analysis of a tRNA-derived small RNA in colorectal cancer. _Front. Oncol._
11, 701440 (2021). Article CAS PubMed PubMed Central Google Scholar * Shao, Y. et al. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. _Chem. Biol.
Drug Des._ 90, 730–738 (2017). Article CAS PubMed PubMed Central Google Scholar * Li, J., Cao, C., Fang, L. & Yu, W. Serum transfer RNA-derived fragment tRF-31-79MP9P9NH57SD acts as
a novel diagnostic biomarker for non-small cell lung cancer. _J. Clin. Lab. Anal_. https://doi.org/10.1002/jcla.24492 (2022). Article PubMed PubMed Central Google Scholar * Huang, Y. et
al. Elucidating the role of serum tRF-31-U5YKFN8DYDZDD as a novel diagnostic biomarker in gastric cancer (GC). _Front. Oncol._ 11, 723753 (2021). Article CAS PubMed PubMed Central
Google Scholar * Panoutsopoulou, K. et al. tRNAGlyGCC-derived internal fragment (i-tRF-GlyGCC) in ovarian cancer treatment outcome and progression. _Cancers_ 14, 24 (2021). Article PubMed
PubMed Central Google Scholar * Olvedy, M. et al. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. _Oncotarget_ 7, 24766–24777 (2016). Article PubMed PubMed
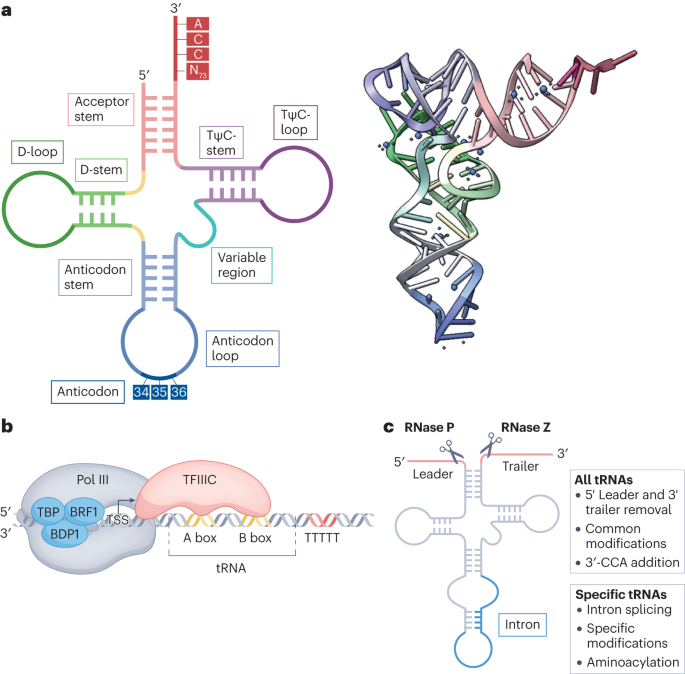
Central Google Scholar * Rich, A. & RajBhandary, U. L. Transfer RNA: molecular structure, sequence, and properties. _Annu. Rev. Biochem._ 45, 805–860 (1976). Article CAS PubMed
Google Scholar * Crothers, D. M., Seno, T. & Söll, D. G. Is there a discriminator site in transfer RNA? _Proc. Natl Acad. Sci. USA_ 69, 3063–3067 (1972). Article CAS PubMed PubMed
Central Google Scholar * Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A. & Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. _Nucleic Acids Res._ 26, 148–153
(1998). Article CAS PubMed PubMed Central Google Scholar * Shi, H. & Moore, P. B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure
revisited. _RNA_ 6, 1091–1105 (2000). Article CAS PubMed PubMed Central Google Scholar * Galli, G., Hofstetter, H. & Birnstiel, M. L. Two conserved sequence blocks within eukaryotic
tRNA genes are major promoter elements. _Nature_ 294, 626–631 (1981). Article CAS PubMed Google Scholar * Hofstetter, H., Kressmann, A. & Birnstiel, M. L. A split promoter for a
eucaryotic tRNA gene. _Cell_ 24, 573–585 (1981). Article CAS PubMed Google Scholar * Takaku, H., Minagawa, A., Takagi, M. & Nashimoto, M. A candidate prostate cancer susceptibility
gene encodes tRNA 3′ processing endoribonuclease. _Nucleic Acids Res._ 31, 2272–2278 (2003). Article CAS PubMed PubMed Central Google Scholar * Rideout, E. J., Marshall, L. &
Grewal, S. S. _Drosophila_ RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. _Proc. Natl Acad.
Sci. USA_ 109, 1139–1144 (2012). Article CAS PubMed PubMed Central Google Scholar * Chu, A. et al. Large-scale profiling of microRNAs for the cancer genome atlas. _Nucleic Acids Res._
44, e3 (2016). Article PubMed Google Scholar * Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. _Nat.
Methods_ 12, 879–884 (2015). Article CAS PubMed PubMed Central Google Scholar * Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. _Nat. Methods_ 12, 835–837
(2015). Article CAS PubMed PubMed Central Google Scholar * Smith, A. M., Abu-Shumays, R., Akeson, M. & Bernick, D. L. Capture, unfolding, and detection of individual tRNA molecules
using a nanopore device. _Front. Bioeng. Biotechnol._ 3, 91 (2015). Article PubMed PubMed Central Google Scholar * Thomas, N. K. et al. Direct nanopore sequencing of individual full
length tRNA strands. _ACS Nano_ 15, 16642–16653 (2021). Article CAS PubMed PubMed Central Google Scholar * Hughes, L. A. et al. Copy number variation in tRNA isodecoder genes impairs
mammalian development and balanced translation. _Nat. Commun._ 14, 2210 (2023). THIS IS ONE OF THE FIRST STUDIES TO SYSTEMATICALLY GENERATE SINGLE AND COMBINATORIAL DELETIONS OF RELATED TRNA
ISODECODER GENES AND ASSESS THEIR CONSEQUENCES AT AN ORGANISMAL LEVEL IN MICE. Article CAS PubMed PubMed Central Google Scholar * Koukuntla, R., Ramsey, W. J., Young, W. B. & Link,
C. J. U6 promoter-enhanced GlnUAG suppressor tRNA has higher suppression efficacy and can be stably expressed in 293 cells. _J. Gene Med._ 15, 93–101 (2013). Article CAS PubMed Google
Scholar Download references ACKNOWLEDGEMENTS The authors thank the members of the Tavazoie laboratory for critically reading the manuscript and for their helpful suggestions and comments.
The authors apologize to all the scientists whose work could not be cited owing to space limitations. S.F.T. and A.M.P. were supported by grants from the National Cancer Institute of the
National Institutes of Health under award numbers R01CA257153, R35CA274446 and U54CA261701, as well as the Black Family Metastasis Center and the Breast Cancer Research Foundation. Molecular
graphics were performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco, with support from
National Institutes of Health R01-GM129325 and the Office of Cyber Infrastructure and Computational Biology, National Institute of Allergy and Infectious Diseases. AUTHOR INFORMATION AUTHORS
AND AFFILIATIONS * Laboratory of Systems Cancer Biology, The Rockefeller University, New York, NY, USA Alexandra M. Pinzaru & Sohail F. Tavazoie Authors * Alexandra M. Pinzaru View
author publications You can also search for this author inPubMed Google Scholar * Sohail F. Tavazoie View author publications You can also search for this author inPubMed Google Scholar
CONTRIBUTIONS The authors contributed equally to all aspects of the article. CORRESPONDING AUTHORS Correspondence to Alexandra M. Pinzaru or Sohail F. Tavazoie. ETHICS DECLARATIONS COMPETING
INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Reviews Cancer_ thanks Pierre Close and the other, anonymous, reviewer(s) for their
contribution to the peer review of this work. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. RELATED LINKS RCSB PROTEIN DATA BANK: https://www.rcsb.org/ UCSF CHIMERAX SOFTWARE: https://www.rbvi.ucsf.edu/chimerax GLOSSARY * Argonaute 2 (AGO2) cleavage assay An in vitro
assay that uses recombinant AGO2 to show specific cleavage of target oligonucleotides. * Epistatic interactions Interactions between two or more genes that influence a particular phenotype.
* Lineage-negative haematopoietic stem and progenitor cells Immature blood cells that lack surface protein markers found on mature blood cells. * Rare codons Codons found with the lowest
frequency in genes compared with synonymous codons in the same group. * Ribosome An essential ribonucleoprotein complex that performs protein synthesis in cells. * Ribosome profiling A
technique that provides a snapshot of all ribosome-bound messenger RNAs in cells with codon-level resolution. * RNA polymerase III (Pol III). An RNA polymerase complex that translates short,
non-coding RNAs, including transfer RNAs, 5S ribosomal RNA, U6 small nuclear RNA, vault RNA and other small RNAs. * tRNA misacylation The charging of a transfer RNA with the wrong amino
acid, not the amino acid cognate to the anticodon of the transfer RNA. * Wobble pairing Non-Watson–Crick base pairing of the first anticodon nucleoside: inosine can pair with adenosine,
uridine or cytidine; guanosine can pair with cytidine or uridine; uridine can pair with adenosine or guanosine. RIGHTS AND PERMISSIONS Springer Nature or its licensor (e.g. a society or
other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of
this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Pinzaru, A.M., Tavazoie, S.F.
Transfer RNAs as dynamic and critical regulators of cancer progression. _Nat Rev Cancer_ 23, 746–761 (2023). https://doi.org/10.1038/s41568-023-00611-4 Download citation * Accepted: 28 July
2023 * Published: 09 October 2023 * Issue Date: November 2023 * DOI: https://doi.org/10.1038/s41568-023-00611-4 SHARE THIS ARTICLE Anyone you share the following link with will be able to
read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative