- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Animal pigment patterns are excellent models to elucidate mechanisms of biological organization. Although theoretical simulations, such as Turing reaction–diffusion systems,
recapitulate many animal patterns, they are insufficient to account for those showing a high degree of spatial organization and reproducibility. Here, we study the coat of the African
striped mouse (_Rhabdomys pumilio_) to uncover how periodic stripes form. Combining transcriptomics, mathematical modelling and mouse transgenics, we show that the Wnt modulator _Sfrp2_
regulates the distribution of hair follicles and establishes an embryonic prepattern that foreshadows pigment stripes. Moreover, by developing in vivo gene editing in striped mice, we find
that _Sfrp2_ knockout is sufficient to alter the stripe pattern. Strikingly, mutants exhibited changes in pigmentation, revealing that _Sfrp2_ also regulates hair colour. Lastly, through
evolutionary analyses, we find that striped mice have evolved lineage-specific changes in regulatory elements surrounding _Sfrp2_, many of which may be implicated in modulating the
expression of this gene. Altogether, our results show that a single factor controls coat pattern formation by acting both as an orienting signalling mechanism and a modulator of
pigmentation. More broadly, our work provides insights into how spatial patterns are established in developing embryos and the mechanisms by which phenotypic novelty originates. Access
through your institution Buy or subscribe This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other
Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days cancel any time Learn more Subscribe to this journal Receive 12 digital issues and online
access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local
taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING
VIEWED BY OTHERS INFLUENCE OF SURVIVAL, PROMOTION, AND GROWTH ON PATTERN FORMATION IN ZEBRAFISH SKIN Article Open access 10 May 2021 PMEL IS INVOLVED IN SNAKE COLOUR PATTERN TRANSITION FROM
BLOTCHES TO STRIPES Article Open access 03 September 2024 REACTION-DIFFUSION IN A GROWING 3D DOMAIN OF SKIN SCALES GENERATES A DISCRETE CELLULAR AUTOMATON Article Open access 23 April 2021
DATA AVAILABILITY The bulk RNA-seq, scRNA-seq and ATAC-seq reads are submitted under an NCBI BioProject: PRJNA1004353.
https://figshare.com/projects/Data_repository_for_A_multifunctional_Wnt_regulator_underlies_the_evolution_of_rodent_stripe_patterns_/175200. Source data are provided with this paper. CODE
AVAILABILITY Code used for scRNA-seq analysis, bulk RNA-seq analysis and comparative genomics is deposited at
https://figshare.com/projects/Data_repository_for_A_multifunctional_Wnt_regulator_underlies_the_evolution_of_rodent_stripe_patterns_/175200. REFERENCES * Mills, M. G. & Patterson, L. B.
Not just black and white: pigment pattern development and evolution in vertebrates. _Semin. Cell Dev. Biol._ 20, 72–81 (2009). Article CAS PubMed Google Scholar * Caro, T. &
Mallarino, R. Coloration in mammals. _Trends Ecol. Evol._ 35, 357–366 (2020). Article PubMed Google Scholar * Cuthill, I. C. et al. The biology of color. _Science_ 357, eaan0221 (2017).
Article PubMed Google Scholar * Kratochwil, C. F. & Mallarino, R. Mechanisms underlying the formation and evolution of vertebrate color patterns. _Annu. Rev. Genet_.
https://doi.org/10.1146/annurev-genet-031423-120918 (2023). * Kondo, S. & Miura, T. Reaction–diffusion model as a framework for understanding biological pattern formation. _Science_ 329,
1616–1620 (2010). Article CAS PubMed Google Scholar * Kondo, S. An updated kernel-based Turing model for studying the mechanisms of biological pattern formation. _J. Theor. Biol._ 414,
120–127 (2017). Article PubMed Google Scholar * Turing, A. M. The chemical basis of morphogenesis. 1953. _Bull. Math. Biol._ 52, 153–197 (1990). Article CAS PubMed Google Scholar *
Vittadello, S. T., Leyshon, T., Schnoerr, D. & Stumpf, M. P. H. Turing pattern design principles and their robustness. _Philos. Trans. A_ 379, 20200272 (2021). Article Google Scholar *
Patterson, L. B. & Parichy, D. M. Zebrafish pigment pattern formation: insights into the development and evolution of adult form. _Annu. Rev. Genet._ 53, 505–530 (2019). Article CAS
PubMed Google Scholar * Kaelin, C. B., McGowan, K. A. & Barsh, G. S. Developmental genetics of color pattern establishment in cats. _Nat. Commun._ 12, 5127 (2021). Article CAS PubMed
PubMed Central Google Scholar * Mallarino, R. et al. Developmental mechanisms of stripe patterns in rodents. _Nature_ 539, 518–523 (2016). Article CAS PubMed PubMed Central Google
Scholar * Haupaix, N. & Manceau, M. The embryonic origin of periodic color patterns. _Dev. Biol._ 460, 70–76 (2020). Article CAS PubMed Google Scholar * Kaelin, C. B. et al.
Specifying and sustaining pigmentation patterns in domestic and wild cats. _Science_ 337, 1536–1541 (2012). Article CAS PubMed PubMed Central Google Scholar * Mallarino, R., Pillay, N.,
Hoekstra, H. E. & Schradin, C. African striped mice. _Curr. Biol._ 28, R299–R301 (2018). Article CAS PubMed Google Scholar * Hardy, M. H. The secret life of the hair follicle.
_Trends Genet._ 8, 55–61 (1992). Article CAS PubMed Google Scholar * Millar, S. E. Molecular mechanisms regulating hair follicle development. _J. Invest. Dermatol._ 118, 216–225 (2002).
Article CAS PubMed Google Scholar * Andl, T., Reddy, S. T., Gaddapara, T. & Millar, S. E. WNT signals are required for the initiation of hair follicle development. _Dev. Cell_ 2,
643–653 (2002). Article CAS PubMed Google Scholar * van Loon, K., Huijbers, E. J. M. & Griffioen, A. W. Secreted frizzled-related protein 2: a key player in noncanonical Wnt
signaling and tumor angiogenesis. _Cancer Metastasis Rev._ 40, 191–203 (2021). Article PubMed Google Scholar * Kim, M., Han, J. H., Kim, J.-H., Park, T. J. & Kang, H. Y. Secreted
frizzled-related protein 2 (sFRP2) functions as a melanogenic stimulator; the role of sFRP2 in UV-induced hyperpigmentary disorders. _J. Invest. Dermatol._ 136, 236–244 (2016). Article CAS
PubMed Google Scholar * Liang, C.-J. et al. SFRPs are biphasic modulators of Wnt-signaling-elicited cancer stem cell properties beyond extracellular control. _Cell Rep._ 28, 1511–1525
(2019). Article CAS PubMed Google Scholar * Lin, H. et al. sFRP2 activates Wnt/β-catenin signaling in cardiac fibroblasts: differential roles in cell growth, energy metabolism
extracellular matrix remodeling. _Am. J. Physiol. Cell Physiol._ 311, C710–C719 (2016). Article PubMed PubMed Central Google Scholar * Gupta, K. et al. Single-cell analysis reveals a
hair follicle dermal niche molecular differentiation trajectory that begins prior to morphogenesis. _Dev. Cell_ 48, 17–31 (2019). Article CAS PubMed Google Scholar * Sennett, R. et al.
An integrated transcriptome atlas of embryonic hair follicle progenitors, their niche, and the developing skin. _Dev. Cell_ 34, 577–591 (2015). Article CAS PubMed PubMed Central Google
Scholar * Rezza, A. et al. Signaling networks among stem cell precursors, transit-amplifying progenitors, and their niche in developing hair follicles. _Cell Rep._ 14, 3001–3018 (2016).
Article CAS PubMed PubMed Central Google Scholar * Sulic, A.-M. et al. Transcriptomic landscape of early hair follicle and epidermal development. _Cell Rep._ 42, 112643 (2023). Article
CAS PubMed Google Scholar * Saxena, N., Mok, K.-W. & Rendl, M. An updated classification of hair follicle morphogenesis. _Exp. Dermatol._ 28, 332–344 (2019). Article PubMed PubMed
Central Google Scholar * Tsai, S.-Y. et al. Wnt/β-catenin signaling in dermal condensates is required for hair follicle formation. _Dev. Biol._ 385, 179–188 (2014). Article CAS PubMed
Google Scholar * Gat, U., DasGupta, R., Degenstein, L. & Fuchs, E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. _Cell_ 95,
605–614 (1998). Article CAS PubMed Google Scholar * Yu, K. et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast
function and bone growth. _Development_ 130, 3063–3074 (2003). Article CAS PubMed Google Scholar * Šošić, D., Richardson, J. A., Yu, K., Ornitz, D. M. & Olson, E. N. Twist regulates
cytokine gene expression through a negative feedback loop that represses NF-κB activity. _Cell_ 112, 169–180 (2003). Article PubMed Google Scholar * Hiscock, T. W. & Megason, S. G.
Orientation of Turing-like patterns by morphogen gradients and tissue anisotropies. _Cell Syst._ 1, 408–416 (2015). Article CAS PubMed PubMed Central Google Scholar * Sick, S., Reinker,
S., Timmer, J. & Schlake, T. WNT and DKK determine hair follicle spacing through a reaction–diffusion mechanism. _Science_ 314, 1447–1450 (2006). Article CAS PubMed Google Scholar *
Van Gorder, R. A. Pattern formation from spatially heterogeneous reaction–diffusion systems. _Philos. Trans. A_ 379, 20210001 (2021). Article Google Scholar * Gierer, A. & Meinhardt,
H. A theory of biological pattern formation. _Kybernetik_ 12, 30–39 (1972). Article CAS PubMed Google Scholar * Yochelis, A., Tintut, Y., Demer, L. L. & Garfinkel, A. The formation
of labyrinths, spots and stripe patterns in a biochemical approach to cardiovascular calcification. _New J. Phys._ 10, 055002 (2008). Article Google Scholar * McKay, R. & Kolokolnikov,
T. Stability transitions and dynamics of mesa patterns near the shadow limit of reaction–diffusion systems in one space dimension. _Discret. Contin. Dyn. Syst. B_ 17, 191–220 (2012). Google
Scholar * Yoon, Y. et al. Streamlined ex vivo and in vivo genome editing in mouse embryos using recombinant adeno-associated viruses. _Nat. Commun._ 9, 412 (2018). * Iozumi, K., Hoganson,
G. E., Pennella, R., Everett, M. A. & Fuller, B. B. Role of tyrosinase as the determinant of pigmentation in cultured human melanocytes. _J. Invest. Dermatol._ 100, 806–811 (1993).
Article CAS PubMed Google Scholar * Edraki, A. et al. A compact, high-accuracy Cas9 with a dinucleotide PAM for in vivo genome editing. _Mol. Cell_ 73, 714–726 (2019). Article CAS
PubMed Google Scholar * Enshell-Seijffers, D., Lindon, C., Wu, E., Taketo, M. M. & Morgan, B. A. β-Catenin activity in the dermal papilla of the hair follicle regulates pigment-type
switching. _Proc. Natl Acad. Sci. USA_ 107, 21564–21569 (2010). Article CAS PubMed PubMed Central Google Scholar * Morgan, B. A. The dermal papilla: an instructive niche for epithelial
stem and progenitor cells in development and regeneration of the hair follicle. _Cold Spring Harb. Perspect. Med._ 4, a015180 (2014). Article PubMed PubMed Central Google Scholar *
Steingrímsson, E., Copeland, N. G. & Jenkins, N. A. Melanocytes and the microphthalmia transcription factor network. _Annu. Rev. Genet._ 38, 365–411 (2004). Article PubMed Google
Scholar * Jho, E.-H. et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. _Mol. Cell. Biol._ 22, 1172–1183 (2002).
Article CAS PubMed PubMed Central Google Scholar * Shtutman, M. et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. _Proc. Natl Acad. Sci. USA_ 96, 5522–5527
(1999). Article CAS PubMed PubMed Central Google Scholar * He, T. C. et al. Identification of c-MYC as a target of the APC pathway. _Science_ 281, 1509–1512 (1998). Article CAS PubMed
Google Scholar * Richardson, R. et al. The genomic basis of temporal niche evolution in a diurnal rodent. _Curr. Biol_. https://doi.org/10.1016/j.cub.2023.06.068 (2023). * Gao, F. et al.
EasyCodeML: a visual tool for analysis of selection using CodeML. _Ecol. Evol._ 9, 3891–3898 (2019). Article PubMed PubMed Central Google Scholar * Kaelin, C. B. & Barsh, G. S.
Genetics of pigmentation in dogs and cats. _Annu Rev. Anim. Biosci._ 1, 125–156 (2013). Article PubMed Google Scholar * Keller, S. H., Jena, S. G., Yamazaki, Y. & Lim, B. Regulation
of spatiotemporal limits of developmental gene expression via enhancer grammar. _Proc. Natl Acad. Sci. USA_ 117, 15096–15103 (2020). Article CAS PubMed PubMed Central Google Scholar *
Kaufman, M. H. _The Atlas of Mouse Development_ (Academic Press, 1992). * Wu, J. & Wang, X. Whole-mount in situ hybridization of mouse embryos using DIG-labeled RNA probes. _Methods Mol.
Biol._ 1922, 151–159 (2019). Article CAS PubMed Google Scholar * Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2.
_Genome Biol._ 15, 550 (2014). Article PubMed PubMed Central Google Scholar * Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets
and their properties. _Bioinformatics_ 33, 2938–2940 (2017). Article CAS PubMed PubMed Central Google Scholar * Burns, K. J., Vasil, G. M., Oishi, J. S., Lecoanet, D. & Brown, B.
P. Dedalus: a flexible framework for numerical simulations with spectral methods. _Phys. Rev. Res._ 2, 023068 (2020). Article CAS Google Scholar * Tuckerman, L. S. & Barkley, D. in
_Numerical Methods for Bifurcation Problems and Large-Scale Dynamical Systems_ (eds Doedel, E. & Tuckerman, L. S.) 453–466 (Springer, 2000). * Li, H. A statistical framework for SNP
calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. _Bioinformatics_ 27, 2987–2993 (2011). Article CAS PubMed PubMed
Central Google Scholar * Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. _Cell Syst._ 8, 281–291
(2019). * Fan, J. et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. _Nat. Methods_ 13, 241–244 (2016). Article CAS PubMed PubMed
Central Google Scholar * Stuart, T. et al. Comprehensive integration of single-cell data. _Cell_ 177, 1888–1902 (2019). Article CAS PubMed PubMed Central Google Scholar * Joost, S. et
al. The molecular anatomy of mouse skin during hair growth and rest. _Cell Stem Cell_ 26, 441–457 (2020). Article CAS PubMed Google Scholar * Beronja, S., Livshits, G., Williams, S.
& Fuchs, E. Rapid functional dissection of genetic networks via tissue-specific transduction and RNAi in mouse embryos. _Nat. Med._ 16, 821–827 (2010). Article CAS PubMed PubMed
Central Google Scholar * Aasen, T. & Izpisúa Belmonte, J. C. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem
cells. _Nat. Protoc._ 5, 371–382 (2010). Article CAS PubMed Google Scholar * Hahn, W. C. et al. Enumeration of the Simian virus 40 early region elements necessary for human cell
transformation. _Mol. Cell. Biol._ 22, 2111–2123 (2002). Article CAS PubMed PubMed Central Google Scholar * Concordet, J.-P. & Haeussler, M. CRISPOR: intuitive guide selection for
CRISPR/Cas9 genome editing experiments and screens. _Nucleic Acids Res._ 46, W242–W245 (2018). Article CAS PubMed PubMed Central Google Scholar * Stamatakis, A. RAxML version 8: a tool
for phylogenetic analysis and post-analysis of large phylogenies. _Bioinformatics_ 30, 1312–1313 (2014). Article CAS PubMed PubMed Central Google Scholar * Kowalczyk, A. et al.
RERconverge: an R package for associating evolutionary rates with convergent traits. _Bioinformatics_ 35, 4815–4817 (2019). Article CAS PubMed PubMed Central Google Scholar * Yang, Z.
PAML: a program package for phylogenetic analysis by maximum likelihood. _Comput. Appl. Biosci._ 13, 555–556 (1997). CAS PubMed Google Scholar * Álvarez-Carretero, S., Kapli, P. &
Yang, Z. Beginner’s guide on the use of PAML to detect positive selection. _Mol. Biol. Evol_. 40, msad041 (2023). * Corces, M. R. et al. An improved ATAC-seq protocol reduces background and
enables interrogation of frozen tissues. _Nat. Methods_ 14, 959–962 (2017). Article CAS PubMed PubMed Central Google Scholar * Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS).
_Genome Biol._ 9, R137 (2008). Article PubMed PubMed Central Google Scholar * Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features.
_Bioinformatics_ 26, 841–842 (2010). Article CAS PubMed PubMed Central Google Scholar * Shumate, A. & Salzberg, S. L. Liftoff: accurate mapping of gene annotations. _Bioinformatics_
37, 1639–1643 (2021). Article CAS PubMed PubMed Central Google Scholar * McLeay, R. C. & Bailey, T. L. Motif enrichment analysis: a unified framework and an evaluation on ChIP
data. _BMC Bioinform._ 11, 165 (2010). Article Google Scholar * Grant, C. E., Bailey, T. L. & Noble, W. S. FIMO: scanning for occurrences of a given motif. _Bioinformatics_ 27,
1017–1018 (2011). Article CAS PubMed PubMed Central Google Scholar * Gupta, S., Stamatoyannopoulos, J. A., Bailey, T. L. & Noble, W. S. Quantifying similarity between motifs.
_Genome Biol._ 8, R24 (2007). Article PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank members of the Mallarino laboratory; Princeton LAR (C. Dmytrow,
K. Gerhart, G. Barnett and J. McGuire) for help with striped mice husbandry; the LSI Genomics Core (W. Wang, J. M. Miller, J. Wiggins and J. Arley Volmar) for help with library preparation
and sequencing; the Nikon Center of Excellence Confocal Microscopy Core (S. Wang and G. Laevsky); and members of the Rivera-Perez laboratory (Y. Yoon and J. Gallant) for help with in vivo
genome editing experiments. We also thank E. F. Wieschaus, G. Deshpande and P. Holl for insights and discussion. This project was supported by an NIH grant to R.M. (R35GM133758). M.R.J. was
supported by an NIH fellowship (F32 GM139253). S.L. was supported by a Presidential Postdoctoral Research fellowship (Princeton University). B.J.B. was supported by an NIH training grant
(T32GM007388). C.Y.F. was supported by an NIH fellowship (F32 GM139240-01). C.F.G.-J. is partially supported by UC Irvine Chancellor’s ADVANCE Postdoctoral Fellowship Program. Q.N. was
partially supported by an NSF grant DMS1763272 and a Simons Foundation grant (594598). AUTHOR INFORMATION Author notes * These authors contributed equally: Matthew R. Johnson, Sha Li.
AUTHORS AND AFFILIATIONS * Department of Molecular Biology, Princeton University, Princeton, NJ, USA Matthew R. Johnson, Sha Li, Benjamin J. Brack, Sarah A. Mereby, Jorge A. Moreno, Charles
Y. Feigin, Jenna Gaska, Alexander Ploss, Stanislav Y. Shvartsman & Ricardo Mallarino * Carle Illinois College of Medicine, University of Illinois at Urbana-Champaign, Urbana, IL, USA
Christian F. Guerrero-Juarez * Department of Developmental and Cell Biology, University of California, Irvine, CA, USA Christian F. Guerrero-Juarez & Qing Nie * Department of
Mathematics, University of California, Irvine, CA, USA Christian F. Guerrero-Juarez & Qing Nie * Center for Complex Biological Systems, University of California, Irvine, CA, USA
Christian F. Guerrero-Juarez & Qing Nie * NSF-Simons Center for Multiscale Cell Fate Research, University of California, Irvine, CA, USA Christian F. Guerrero-Juarez & Qing Nie *
Center for Computational Biology, Flatiron Institute, New York, NY, USA Pearson Miller & Stanislav Y. Shvartsman * Frederick National Laboratory for Cancer Research, Frederick, MA, USA
Jaime A. Rivera-Perez * The Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ, USA Stanislav Y. Shvartsman Authors * Matthew R. Johnson View author
publications You can also search for this author inPubMed Google Scholar * Sha Li View author publications You can also search for this author inPubMed Google Scholar * Christian F.
Guerrero-Juarez View author publications You can also search for this author inPubMed Google Scholar * Pearson Miller View author publications You can also search for this author inPubMed
Google Scholar * Benjamin J. Brack View author publications You can also search for this author inPubMed Google Scholar * Sarah A. Mereby View author publications You can also search for
this author inPubMed Google Scholar * Jorge A. Moreno View author publications You can also search for this author inPubMed Google Scholar * Charles Y. Feigin View author publications You
can also search for this author inPubMed Google Scholar * Jenna Gaska View author publications You can also search for this author inPubMed Google Scholar * Jaime A. Rivera-Perez View author
publications You can also search for this author inPubMed Google Scholar * Qing Nie View author publications You can also search for this author inPubMed Google Scholar * Alexander Ploss
View author publications You can also search for this author inPubMed Google Scholar * Stanislav Y. Shvartsman View author publications You can also search for this author inPubMed Google
Scholar * Ricardo Mallarino View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.R.J. and R.M. conceived the project and designed experiments.
M.R.J. performed RNA-seq experiments and bulk RNA-seq analysis. S.L. performed the in vitro and in vivo genome editing in striped mice, with help from S.A.M. and J.A.R.-P. M.R.J. and S.L.
performed all downstream processing and analysis of genome edited animals. P.M. and S.Y.S. did the mathematical modelling. C.F.G.-J. led the scRNA-seq analysis, with support from M.R.J. and
Q.N. M.R.J., B.J.B. and R.M. performed in situ hybridizations. M.R.J., B.J.B., S.A.M. and R.M. performed the phenotypic characterization of striped mouse and laboratory mouse tissues,
including immunofluorescence and histology. M.R.J. and S.A.M. performed the melanocyte cell culture experiments. J.A.M. did the evolutionary analysis. C.Y.F. generated the rhabdomyzed _Mus_
genome and lift-over annotation. J.G. and A.P. generated the immortalized _Rhabdomys_ fibroblasts. M.R.J. and R.M. wrote the manuscript with input from all authors. CORRESPONDING AUTHOR
Correspondence to Ricardo Mallarino. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. PEER REVIEW PEER REVIEW INFORMATION _Nature Ecology & Evolution_
thanks Julien Debbache and Denis Headon for their contribution to the peer review of this work. Peer reviewer reports are available. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature
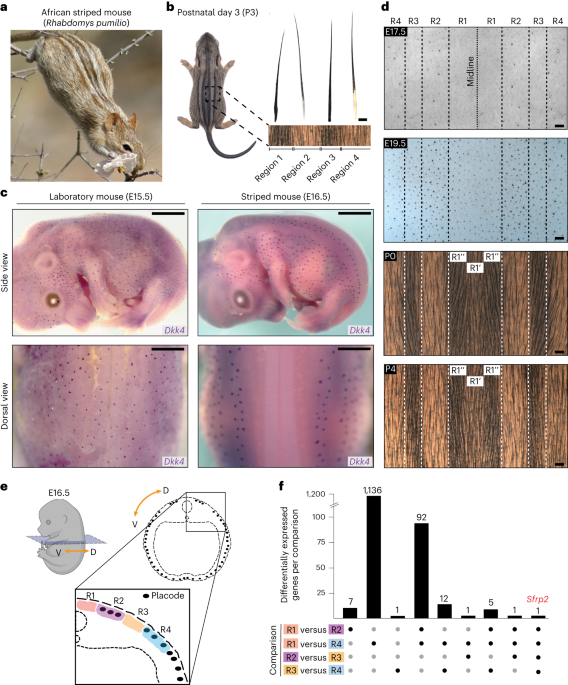
remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. EXTENDED DATA EXTENDED DATA FIG. 1 PATTERNS OF HAIR PLACODE FORMATION IN STRIPED MICE.
A, Side views of E13.5–E15.5 striped mouse embryos showing stages before the emergence of trunk hair placodes. Whole-mount _in situ_ hybridization for placode markers _Dkk4_ and _Ctnnb1_
shows the presence of whisker placodes (arrows), which develop before trunk placodes. No expression is detected in dorsal skin. B, Side views of E16.5 striped mouse embryos displaying
spatially restricted patterns of trunk hair placode formation, as visualized by whole-mount _in situ_ hybridization for placode markers _Wif1_, _Bmp4_, _Wnt10b_ _Dkk1_. C, Hematoxylin-Eosin
staining on cross-sections of striped mouse E18.5 embryos reveals both mature placodes (arrows) and nascent placodes (asterisks); the latter are evidenced by thickening of the epidermis. D,
Side views of E18.5 striped mouse embryos showing placode emergence in previously placode-barren regions, as visualized by whole-mount _in situ_ hybridization for placode markers _Dkk1_ and
_Ctnnb1_. E, Hematoxilin and Eosin (H&E) stains of longitudinal sections from different dorsal regions in striped mouse embryos. Placodes in Regions 1 (R1) and 3 (R3) emerge later than
those in Region 2 (R2). Scale bars: 5 mm in (A and B); 200 µm (zoomed out) and 50 µm (inset) in (C); 5 mm in (D); 100 µm in (E). For A-E, three individuals per stage per gene were analysed.
EXTENDED DATA FIG. 2 EXPRESSION OF SELECTED WNT MODULATORS IN E16.5 STRIPED MOUSE EMBRYOS. Fold expression changes of Wnt modulators in skin regions (R1, R2, R3, R4) dissected for bulk
RNA-seq analysis. Shown are selected modulators that are expressed in a dorsoventral gradient. Fold expression changes were calculated from average FPKM values (n = 3 biologically
independent samples. EXTENDED DATA FIG. 3 ANALYSIS OF HAIR PLACODE AND DERMAL CONDENSATE MARKERS. A-B, Plots showing the subset of cells that express established hair placode (a) and dermal
condensate (b) markers in the dorsal skin of E16.5 striped mice. C-D, Dot plots of hair placode25 (c) and dermal condensate26 (d) markers showing expression changes among the three different
dorsal regions sampled. The size of the dot encodes the percentage of cells within a dorsal region, while the colour encodes the average expression level across all cells within a dorsal
region (blue is high, red is low). Asterisks depict markers with high expression levels in Region 2 (R2), compared to Region 1 (R1) and Region 3 (R3). As described in the main text, R2 has
visible hair follicles at this developmental stage, whereas R1 and R3 do not. EXTENDED DATA FIG. 4 EXPRESSION OF _SFRP2_ IN DERMAL FIBROBLASTS. A, _Sfrp2_ expressing fibroblasts are
expressed primarily in the reticular (lower) dermis. Papillary (upper) and reticular (lower) dermis fibroblasts were defined based on previously established markers3; Papillary dermis:
_Ntn1, Pdpn, Ackr4, Lrig1, Apcdd1;_ Reticular dermis: _Tgm2, Cnn1, Cdh2, Mgp, Dlk1_. B, At E16.5, expression levels of _Sfrp2_ and the percentage of fibroblasts expressing _Sfrp2_ are
highest in Region 1 (R1) and lowest in Region 3 (R3), in agreement with the dorsoventral gradient revealed by the bulk RNA-seq data. In B, n = 3 biologically independent samples. Left panel:
bars represent average expression levels. Right panel: mean values (+/- SEM). EXTENDED DATA FIG. 5 HIGH EXPRESSION OF _SFRP2_ IN THE RETICULAR (LOWER) DERMIS COINCIDES WITH LOW EXPRESSION
OF LEF1. A, _In situ_ hybridization in striped mouse E16.5 embryos shows that _Sfrp2_ is primarily expressed in the reticular dermis. Right side image shows expression of _Sfrp2_ at
subcellular resolution. B-C, LEF1 immunostaining in staged matched striped (B) and laboratory (C) mouse embryos. Red boxes denote zoomed-in regions. Scale bars: 200 µm (zoomed out) and 100
µm (zoomed in) in A; 200 µm (zoomed out) and 50 µm (zoomed in) in B and C. NT = neural tube. For A-C, three different individuals were analysed. EXTENDED DATA FIG. 6 _DERMO1_ AND _SFRP2_
EXPRESSING FIBROBLASTS. A _Dermo-Cre_ mouse was used to drive Cre expression in dermal fibroblasts. As illustrated above, a subset of _Dermo1_ expressing fibroblasts express _Sfrp2_. Thus,
this mouse strain is adequate for driving expression of Cre in cells expressing _Sfrp2_. EXTENDED DATA FIG. 7 MATHEMATICAL SIMULATIONS. A, Schematic showing the role of _Sfrp2_ as an
inhibitor of Wnt signalling. B, Gradient steepness increases central stripe width independent of model. Each row depicts a schematic and equations governing a particular variant of our
modulator-activator-inhibitor system (left) and the resulting simulations of stripe spacing for different gradient steepness values using these models (right). In all cases, gradient
steepness affects stripe spacing. C, Predictions from an alternative model of positional information. Patterning based on positional information is inconsistent with our experimental
results. We illustrate this by considering two standard paradigms for stripe patterning by positional information. Under a classic ‘French Flag’ model (left, top), each stripe (marked in
grey) is assigned to a region of space in which a single morphogen gradient exists between two pathway-specific threshold concentrations (horizontal red lines). (top, left) Under such a
paradigm, a substantial reduction in morphogen expression, in this case by 80 percent, makes it impossible for the gradient to reach certain thresholds entirely, leading to stripe loss.
(bottom, left) Alternatively, stripes are frequently determined via an ‘opposing gradients’ motif via the interaction of multiple gradients. We depict one example, in which each stripe is
determined by two opposite facing gradients, such that a stripe forms in the region where each gradient exceeds a morphogen-specific threshold. (right, bottom) Major reduction of a single
morphogen eliminates one stripe while leaving the other unperturbed. EXTENDED DATA FIG. 8 GENERATION OF _IN VIVO_ GENOME EDITING IN STRIPED MOUSE. A, Schematic of the _Sfrp2_ locus (exons in
red) showing the transcriptional start site (TSS), protospacer adjacent motif (PAM) short guide RNA (sgRNA) target/sequence. Four types of deletions were achieved: 2 bp, 13 bp, 466 bp 527
bp (white boxes). All mutations are predicted to cause frameshift mutations. B, Representative western blot of individuals carrying different combinations of wild-type and a 13 bp deleted
allele (wild type: _Sfrp2__+/+_; heterozygous: _Sfrp2__+/-_; homozygous: (_Sfrp__-/-_). _Sfrp__-/-_ have no detectable SFRP2 Protein (green). Bands ~30 kDa correspond to SFRP2 protein.
b-TUBULIN (~50 kDa, red) was used as a loading control. In B, two different individuals from each genotype were analysed. EXTENDED DATA FIG. 9 PHENOTYPIC CHARACTERIZATION OF _SFRP2_ MUTANTS.
A and B, Whole-mount _in situ_ hybridization for _Dkk4_ in wild-type and _Sfrp2_ knockout E16.5 embryos (A) and corresponding width measurements of dorsal regions 1 and 3 (that is, R1 and
R3) (B). Note that _Dkk4_ expression diminishes in response to _Sfrp2_ knockout. C, Hair length measurements in postnatal day 3 wild-type and _Sfrp2_ knockout individuals. In B and C, n = 3
biologically independent samples for each _Sfrp2_ knockout and _Sfrp2_ wild-type individuals. Source data EXTENDED DATA FIG. 10 _SFRP2_ PROMOTES MELANOGENESIS BY ACTIVATING WNT SIGNALLING.
_In situ_ hybridization showing specific _Sfrp2_ expression in the dermal papilla of P4 striped mouse hair follicles. B, Melanocytes were stably transduced with either a control (LV-GFP) or
an experimental (LV-Sfrp2GFP) lentivirus and expression of Wnt targets and melanogenesis genes in stably transduced control and experimental cells, as determined was determined via qPCR (_P_
= 0.12026 (_Axin_); _P_ = 0.001816 (_C-myc_); _P_ = 0.006739 (_CyclinD_); _P_ = 0.001040 (_Mitf_); _P_ = 0.010712 (_Tyr_); ANOVA test; N = 4). C, Quantitative PCR (qPCR) showing _Sfrp2_
mRNA fold change levels along different dorsal skin regions in embryonic and postnatal stages (E16.5: _P_ = 0.0283 (R1vsR2); _P_ = 0.0062 (R1vsR3); _P_ = 0.3959 (R2vsR3); E19.5: _P_ = 0.8685
(R1vsR2); _P_ = 0.6319 (R1vsR3); _P_ = 0.9015 (R2vsR3); P0: _P_ = 0.9724 (R1vsR2); _P_ = 0.8207 (R1vsR3); _P_ = 0.6971 (R2vsR3); P4: _P_ = 0.0003 (R1vsR2); _P_ = 0.0022 (R1vsR3); _P_ =
0.0001 (R2vsR3); ANOVA test; N = 3 for E16.5, E19.5 P0, N = 4 for P4). Scale bars in A: 100 µm (left) and 25 µm (right). In A, three different individuals were analysed. In B and C, data are
presented as mean values +/− SEM. Source data SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION Supplementary Tables 1–5. REPORTING SUMMARY PEER REVIEW FILE SUPPLEMENTARY DATA 1
Differentially expressed genes between regions 1 and 4 of E16.5 striped mice skin. Differentially expressed genes were determined using DESeq2. _P_ value corrected for multiple testing
(_P_adj < 0.05). SUPPLEMENTARY DATA 2 Differentially expressed genes between Sfrp2high and Sfrp2l°w fibroblast populations. Differentially expressed genes were determined using DESeq2.
_P_ value corrected for multiple testing (_P_adj < 0.05). SOURCE DATA SOURCE DATA FIGS. 2E,M AND 4D,F AND EXTENDED DATA FIGS. 9A,B AND 10B,C. RIGHTS AND PERMISSIONS Springer Nature or its
licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the
accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Johnson, M.R., Li, S., Guerrero-Juarez, C.F. _et al._ A multifunctional Wnt regulator underlies the evolution of rodent stripe patterns. _Nat Ecol Evol_ 7, 2143–2159 (2023).
https://doi.org/10.1038/s41559-023-02213-7 Download citation * Received: 29 March 2023 * Accepted: 27 August 2023 * Published: 09 October 2023 * Issue Date: December 2023 * DOI:
https://doi.org/10.1038/s41559-023-02213-7 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative






