- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Intratumoral heterogeneity is a critical factor when diagnosing and treating patients with cancer. Marked differences in the genetic and epigenetic backgrounds of cancer cells have
been revealed by advances in genome sequencing, yet little is known about the phenotypic landscape and the spatial distribution of intratumoral heterogeneity within solid tumours. Here, we
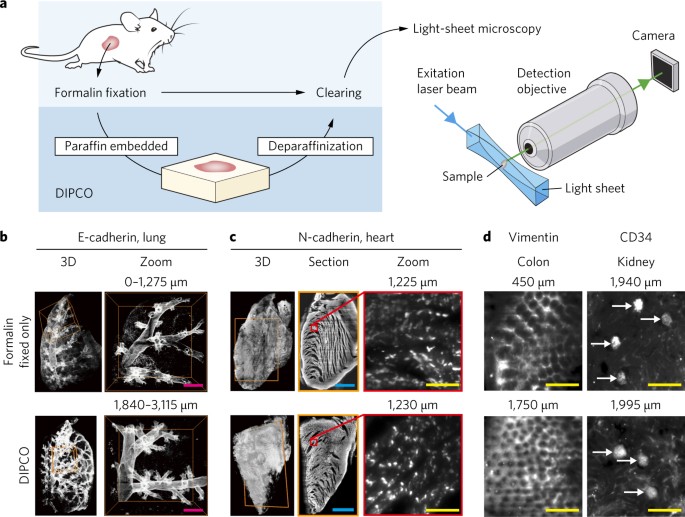
show that three-dimensional light-sheet microscopy of cleared solid tumours can identify unique patterns of phenotypic heterogeneity, in the epithelial-to-mesenchymal transition and in
angiogenesis, at single-cell resolution in whole formalin-fixed paraffin-embedded (FFPE) biopsy samples. We also show that cleared FFPE samples can be re-embedded in paraffin after
examination for future use, and that our tumour-phenotyping pipeline can determine tumour stage and stratify patient prognosis from clinical samples with higher accuracy than current
diagnostic methods, thus facilitating the design of more efficient cancer therapies. Access through your institution Buy or subscribe This is a preview of subscription content, access via
your institution ACCESS OPTIONS Access through your institution Access Nature and 54 other Nature Portfolio journals Get Nature+, our best-value online-access subscription $29.99 / 30 days
cancel any time Learn more Subscribe to this journal Receive 12 digital issues and online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on
SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about
institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS HIGH RESOLUTION MAPPING OF THE TUMOR MICROENVIRONMENT USING INTEGRATED
SINGLE-CELL, SPATIAL AND IN SITU ANALYSIS Article Open access 19 December 2023 A MULTI-MODAL SINGLE-CELL AND SPATIAL EXPRESSION MAP OF METASTATIC BREAST CANCER BIOPSIES ACROSS
CLINICOPATHOLOGICAL FEATURES Article Open access 30 October 2024 SPATIALLY RESTRICTED DRIVERS AND TRANSITIONAL CELL POPULATIONS COOPERATE WITH THE MICROENVIRONMENT IN UNTREATED AND
CHEMO-RESISTANT PANCREATIC CANCER Article Open access 22 August 2022 CHANGE HISTORY * _ 02 JANUARY 2018 In this Article originally published, owing to a technical error, author affiliations
were incorrectly assigned in the HTML version; the PDF was correct. These errors have now been corrected. _ REFERENCES * Gerlinger, M. et al. Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. _N. Engl. J. Med._ 366, 883–892 (2012). Article CAS PubMed PubMed Central Google Scholar * Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour
heterogeneity: a looking glass for cancer? _Nat. Rev. Cancer_ 12, 323–334 (2012). Article CAS PubMed Google Scholar * Junttila, M. R. & de Sauvage, F. J. Influence of tumour
micro-environment heterogeneity on therapeutic response. _Nature_ 501, 346–354 (2013). Article CAS PubMed Google Scholar * Zhang, J. et al. Intratumor heterogeneity in localized lung
adenocarcinomas delineated by multiregion sequencing. _Science_ 346, 256–259 (2014). Article CAS PubMed PubMed Central Google Scholar * Vogelstein, B. et al. Cancer genome landscapes.
_Science_ 339, 1546–1558 (2013). Article CAS PubMed PubMed Central Google Scholar * McGranahan, N. & Swanton, C. Biological and therapeutic impact of intratumor heterogeneity in
cancer evolution. _Cancer Cell_ 27, 15–26 (2015). Article CAS PubMed Google Scholar * Waclaw, B. et al. A spatial model predicts that dispersal and cell turnover limit intratumour
heterogeneity. _Nature_ 525, 261–264 (2015). Article CAS PubMed PubMed Central Google Scholar * Polyak, K. & Weinberg, R. A. Transitions between epithelial and mesenchymal states:
acquisition of malignant and stem cell traits. _Nat. Rev. Cancer_ 9, 265–273 (2009). Article CAS PubMed Google Scholar * Plaks, V., Kong, N. & Werb, Z. The cancer stem cell niche:
how essential is the niche in regulating stemness of tumor cells? _Cell Stem Cell_ 16, 225–238 (2015). Article CAS PubMed PubMed Central Google Scholar * Wang, K. et al. Whole-genome
sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. _Nat. Genet._ 46, 573–582 (2014). Article CAS PubMed Google Scholar * Andor, N. et al.
Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. _Nat. Med._ 22, 105–113 (2016). Article CAS PubMed Google Scholar * Saliba, A. E., Westermann, A. J.,
Gorski, S. A. & Vogel, J. Single-cell RNA-seq: advances and future challenges. _Nucleic Acids Res._ 42, 8845–8860 (2014). Article CAS PubMed PubMed Central Google Scholar * Patel,
A. P. et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. _Science_ 344, 1396–1401 (2014). Article CAS PubMed PubMed Central Google Scholar *
Lawson, D. A. et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. _Nature_ 526, 131–135 (2015). Article CAS PubMed PubMed Central Google
Scholar * Tabassum, D. P. & Polyak, K. Tumorigenesis: it takes a village. _Nat. Rev. Cancer_ 15, 473–483 (2015). Article CAS PubMed Google Scholar * Erturk, A. et al.
Three-dimensional imaging of solvent-cleared organs using 3DISCO. _Nat. Protoc._ 7, 1983–1995 (2012). Article CAS PubMed Google Scholar * Ke, M. T., Fujimoto, S. & Imai, T. SeeDB: a
simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. _Nat. Neurosci._ 16, 1154–1161 (2013). Article CAS PubMed Google Scholar * Chung, K. &
Deisseroth, K. CLARITY for mapping the nervous system. _Nat. Methods_ 10, 508–513 (2013). Article CAS PubMed Google Scholar * Susaki, E. A. et al. Whole-brain imaging with single-cell
resolution using chemical cocktails and computational analysis. _Cell_ 157, 726–739 (2014). Article CAS PubMed Google Scholar * Yang, B. et al. Single-cell phenotyping within transparent
intact tissue through whole-body clearing. _Cell_ 158, 945–958 (2014). Article CAS PubMed PubMed Central Google Scholar * Hama, H. et al. ScaleS: an optical clearing palette for
biological imaging. _Nat. Neurosci._ 18, 1518–1529 (2015). Article CAS PubMed Google Scholar * Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and
high-resolution imaging of intact tissues. _Nat. Protoc._ 9, 1682–1697 (2014). Article CAS PubMed PubMed Central Google Scholar * Tainaka, K. et al. Whole-body imaging with single-cell
resolution by tissue decolorization. _Cell_ 159, 911–924 (2014). Article CAS PubMed Google Scholar * Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples
for volume imaging. _Cell_ 159, 896–910 (2014). Article CAS PubMed Google Scholar * Belle, M. et al. A simple method for 3D analysis of immunolabeled axonal tracts in a transparent
nervous system. _Cell Rep._ 9, 1191–1201 (2014). Article CAS PubMed Google Scholar * Susaki, E. A. et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging.
_Nat. Protoc._ 10, 1709–1727 (2015). Article CAS PubMed Google Scholar * Eliceiri, K. W. et al. Biological imaging software tools. _Nat. Methods_ 9, 697–710 (2012). Article CAS PubMed
PubMed Central Google Scholar * Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. _Nat. Methods_ 9, 671–675 (2012). Article CAS
PubMed PubMed Central Google Scholar * Peng, H., Bria, A., Zhou, Z., Iannello, G. & Long, F. Extensible visualization and analysis for multidimensional images using Vaa3D. _Nat.
Protoc._ 9, 193–208 (2014). Article CAS PubMed Google Scholar * Salois, G. & Smith, J. S. Housing complexity alters GFAP-immunoreactive astrocyte morphology in the rat dentate gyrus.
_Neural Plast._ 2016, 3928726 (2016). Article PubMed PubMed Central Google Scholar * Peinado, H., Olmeda, D. & Cano, A. Snail, Zeb and bHLH factors in tumour progression: an
alliance against the epithelial phenotype? _Nat. Rev. Cancer_ 7, 415–428 (2007). Article CAS PubMed Google Scholar * Maier, J., Traenkle, B. & Rothbauer, U. Visualizing
epithelial–mesenchymal transition using the chromobody technology. _Cancer Res._ 76, 5592–5596 (2016). Article CAS PubMed Google Scholar * Savagner, P. The epithelial–mesenchymal
transition (EMT) phenomenon. _Ann. Oncol._ 21, vii89–vii92 (2010). Article PubMed Google Scholar * Connor, J. et al. Regression of bladder tumors in mice treated with interleukin 2
gene-modified tumor cells. _J. Exp. Med._ 177, 1127–1134 (1993). Article CAS PubMed Google Scholar * Matsumoto, K. et al. Intravesical interleukin-15 gene therapy in an orthotopic
bladder cancer model. _Human Gene Ther._ 22, 1423–1432 (2011). Article CAS Google Scholar * Kobayashi, T., Owczarek, T. B., McKiernan, J. M. & Abate-Shen, C. Modelling bladder cancer
in mice: opportunities and challenges. _Nat. Rev. Cancer_ 15, 42–54 (2015). Article CAS PubMed PubMed Central Google Scholar * Dobosz, M., Ntziachristos, V., Scheuer, W. & Strobel,
S. Multispectral fluorescence ultramicroscopy: three-dimensional visualization and automatic quantification of tumor morphology, drug penetration, and antiangiogenic treatment response.
_Neoplasia_ 16, 1–13 (2014). Article PubMed PubMed Central Google Scholar * Dodt, H. U. et al. Ultramicroscopy: development and outlook. _Neurophotonics_ 2, 041407 (2015). Article
PubMed PubMed Central Google Scholar * Frampton, G. M. et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. _Nat.
Biotechnol._ 31, 1023–1031 (2013). Article CAS PubMed PubMed Central Google Scholar * Zheng, Z. et al. Anchored multiplex PCR for targeted next-generation sequencing. _Nat. Med._ 20,
1479–1484 (2014). Article CAS PubMed Google Scholar * Brat, D. J. et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. _N. Engl. J. Med._ 372, 2481–2498
(2015). Article CAS PubMed Google Scholar * Maley, C. C. et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. _Nat. Genet._ 38, 468–473 (2006). Article CAS
PubMed Google Scholar * Landau, D. A. et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. _Cell_ 152, 714–726 (2013). Article CAS PubMed PubMed Central
Google Scholar * Bochtler, T. et al. Clonal heterogeneity as detected by metaphase karyotyping is an indicator of poor prognosis in acute myeloid leukemia. _J. Clin. Oncol._ 31, 3898–3905
(2013). Article PubMed Google Scholar * Mroz, E. A., Tward, A. D., Hammon, R. J., Ren, Y. & Rocco, J. W. Intra-tumor genetic heterogeneity and mortality in head and neck cancer:
analysis of data from the Cancer Genome Atlas. _PLoS Med._ 12, e1001786 (2015). Article PubMed PubMed Central Google Scholar * Vergote, I. et al. Re: new guidelines to evaluate the
response to treatment in solid tumors [ovarian cancer]. Gynecologic Cancer Intergroup. _J. Natl Cancer Inst._ 92, 1534–1535 (2000). Article CAS PubMed Google Scholar * Tomer, R. et al.
SPED light sheet microscopy: fast mapping of biological system structure and function. _Cell_ 163, 1796–1806 (2015). Article CAS PubMed PubMed Central Google Scholar * von Dadelszen, P.
et al. Prediction of adverse maternal outcomes in pre-eclampsia: development and validation of the fullPIERS model. _Lancet_ 377, 219–227 (2011). Article Google Scholar * Silvestri, G. A.
et al. A bronchial genomic classifier for the diagnostic evaluation of lung cancer. _N. Engl. J. Med._ 373, 243–251 (2015). Article CAS PubMed PubMed Central Google Scholar *
Stutchfield, P., Whitaker, R. & Russell, I. Antenatal betamethasone and incidence of neonatal respiratory distress after elective caesarean section: pragmatic randomised trial. _BMJ_
331, 662 (2005). Article PubMed PubMed Central Google Scholar * Tierney, W. M., McDonald, C. J., Hui, S. L. & Martin, D. K. Computer predictions of abnormal test results. _Effects
Outpatient Test. JAMA_ 259, 1194–1198 (1988). CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS The authors would like to thank J. Szumiło, Department of Clinical
Pathomorphology, Medical University of Lublin, Lublin, Poland for kindly providing human tissue samples. This study was supported by the Swedish Research Council (grants 2009-3364, 2010-4392
and 2013-3189 to P.U.), the Swedish Cancer Society (grant CAN2013/802 and CAN2016/801 to P.U.), the Swedish Brain Foundation (grant FO2017/0107 to P.U.), the Linnaeus Center in
Developmental Biology for Regenerative Medicine (DBRM) (P.U.), a Knut and Alice Wallenberg Foundation Grant to the Center for Live Imaging of Cells at the Karolinska (CLICK) Institutet
(P.U.), the Royal Swedish Academy of Sciences (P.U.), the David and Astrid Hagelén Foundation (N.T.), the Takeda Science Foundation (N.T.), the Scandinavia-Japan Sasakawa Foundation (N.T.
and S.K.), and the Wenner-Gren Foundation (S.K.). The light-sheet microscopy infrastructure used in this work received grants from the Strategic Research Area in Neuroscience – StratNeuro
and the Strategic Research Area in Stem Cells and Regenerative Medicine – StratRegen supported by the Swedish government. AUTHOR INFORMATION Author notes * Nobuyuki Tanaka and Shigeaki
Kanatani contributed equally to this work. AUTHORS AND AFFILIATIONS * Department of Medical Biochemistry and Biophysics, Karolinska Institutet, SE-17177, Stockholm, Sweden Nobuyuki Tanaka,
Shigeaki Kanatani, Dagmara Kaczynska, Lauri Louhivuori, Ayako Miyakawa & Per Uhlén * Department of Urology, Keio University School of Medicine, Tokyo, 160-8582, Japan Nobuyuki Tanaka,
Kazuhiro Matsumoto & Mototsugu Oya * Department of Biological Sciences, Columbia University, New York, NY, 10027, USA Raju Tomer * Turku Centre for Biotechnology, University of Turku and
Faculty of Science and Engineering, Åbo Akademi University, FI-20521, Turku, Finland Cecilia Sahlgren * Institute for Complex Molecular Systems, Eindhoven University of Technology,
Eindhoven, MB, 5600, The Netherlands Cecilia Sahlgren * Institute of Biomedicine/Pathology, University of Turku, FI-20014, Turku, Finland Pauliina Kronqvist * Department of
Oncology-Pathology, Karolinska Institutet, SE-17176, Stockholm, Sweden Lorand Kis, Claes Lindh, Sara Corvigno, Johan Hartman, Artur Mezheyeuski, Carina Strell, Joseph W. Carlson, Hanna
Dahlstrand & Arne Östman * Department of Pathology and Cytology, Karolinska University Hospital, SE-17176, Stockholm, Sweden Lorand Kis, Claes Lindh, Johan Hartman & Joseph W.
Carlson * Department of Urology and Oncological Urology, Medical University in Lublin, ul. Jaczewskiego 8, 20-954, Lublin, Poland Przemysław Mitura * Department of Biochemistry and Molecular
Biology, Medical University in Lublin, ul. Chodzki 1, 20-093, Lublin, Poland Andrzej Stepulak * Department of Immunology, Genetics and Pathology, Uppsala University, SE-75185, Uppsala,
Sweden Patrick Micke & Hanna Dahlstrand * Department of Laboratory Medicine, Division of Pathology, Karolinska Institutet, Karolinska University Hospital, SE-14186, Stockholm, Sweden
Carlos Fernández Moro * Department of Clinical Pathology/Cytology, Karolinska University Hospital, SE-14186, Stockholm, Sweden Carlos Fernández Moro * Department of Oncology-Pathology,
Karolinska University Hospital, SE-17176, Stockholm, Sweden Hanna Dahlstrand * Department of Molecular Medicine and Surgery, Karolinska Institutet, SE-17177, Stockholm, Sweden Peter Wiklund
& Ayako Miyakawa * Department of Urology, Karolinska University Hospital, SE-17176, Stockholm, Sweden Peter Wiklund & Ayako Miyakawa * Howard Hughes Medical Institute, W080 Clark
Center, Stanford University, 318 Campus Drive West, Stanford, CA, 94305, USA Karl Deisseroth * Department of Bioengineering, W080 Clark Center, Stanford University, 318 Campus Drive West,
Stanford, CA, 94305, USA Karl Deisseroth * Department of Psychiatry and Behavioral Sciences, W080 Clark Center, Stanford University, 318 Campus Drive West, Stanford, CA, 94305, USA Karl
Deisseroth Authors * Nobuyuki Tanaka View author publications You can also search for this author inPubMed Google Scholar * Shigeaki Kanatani View author publications You can also search for
this author inPubMed Google Scholar * Raju Tomer View author publications You can also search for this author inPubMed Google Scholar * Cecilia Sahlgren View author publications You can
also search for this author inPubMed Google Scholar * Pauliina Kronqvist View author publications You can also search for this author inPubMed Google Scholar * Dagmara Kaczynska View author
publications You can also search for this author inPubMed Google Scholar * Lauri Louhivuori View author publications You can also search for this author inPubMed Google Scholar * Lorand Kis
View author publications You can also search for this author inPubMed Google Scholar * Claes Lindh View author publications You can also search for this author inPubMed Google Scholar *
Przemysław Mitura View author publications You can also search for this author inPubMed Google Scholar * Andrzej Stepulak View author publications You can also search for this author
inPubMed Google Scholar * Sara Corvigno View author publications You can also search for this author inPubMed Google Scholar * Johan Hartman View author publications You can also search for
this author inPubMed Google Scholar * Patrick Micke View author publications You can also search for this author inPubMed Google Scholar * Artur Mezheyeuski View author publications You can
also search for this author inPubMed Google Scholar * Carina Strell View author publications You can also search for this author inPubMed Google Scholar * Joseph W. Carlson View author
publications You can also search for this author inPubMed Google Scholar * Carlos Fernández Moro View author publications You can also search for this author inPubMed Google Scholar * Hanna
Dahlstrand View author publications You can also search for this author inPubMed Google Scholar * Arne Östman View author publications You can also search for this author inPubMed Google
Scholar * Kazuhiro Matsumoto View author publications You can also search for this author inPubMed Google Scholar * Peter Wiklund View author publications You can also search for this author
inPubMed Google Scholar * Mototsugu Oya View author publications You can also search for this author inPubMed Google Scholar * Ayako Miyakawa View author publications You can also search
for this author inPubMed Google Scholar * Karl Deisseroth View author publications You can also search for this author inPubMed Google Scholar * Per Uhlén View author publications You can
also search for this author inPubMed Google Scholar CONTRIBUTIONS N.T., S.K., A.Mi. and P.U. designed the study. N.T., S.K., D.K., L.L. and K.M. performed the experiments. N.T., S.K. and
R.T. performed 3D image processing. R.T. and K.D. developed the custom-built light-sheet microscope system. C.Sa., P.K., L.K., C.L., P.M., A.S., S.C., J.H., P.M., A.Me., C.St., J.W.C.,
C.F.M., H.D. and A.Mi. provided human tumour samples. P.W., M.O., A.Ö. and K.D. provided conceptual advice. N.T. and P.U. wrote the manuscript. CORRESPONDING AUTHOR Correspondence to Per
Uhlén. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing financial interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE: Springer Nature remains neutral with regard to
jurisdictional claims in published maps and institutional affiliations. A correction to this article is available online at https://doi.org/10.1038/s41551-017-0162-1. SUPPLEMENTARY
INFORMATION SUPPLEMENTARY INFORMATION Supplementary figures, tables and video legends. LIFE SCIENCES REPORTING SUMMARY SUPPLEMENTARY VIDEO 1 Three-dimensional volume reconstruction of
hTumour 1immunostained for E-cadherin. SUPPLEMENTARY VIDEO 2 Three-dimensional volume reconstruction of hTumour 3 immunostained for N-cadherin. SUPPLEMENTARY VIDEO 3 Three-dimensional volume
reconstruction of hTumour 6 immunostained for CD34. SUPPLEMENTARY VIDEO 4 Three-dimensional volume reconstruction of the CD34 signal. SUPPLEMENTARY VIDEO 5 Single-cell 3D volume
reconstruction of hTumour 7 immunostained for Vimentin. MATLAB SCRIPT 1 Generation of centroids list (point cloud) from Hmaxima images. MATLAB SCRIPT 2 Calculation of mean intensity value of
each dots area. MATLAB SCRIPT 3 Generation of binary images from XYZ coordinates. SUPPLEMENTARY TABLE 1 Clinicopathological characteristics of 50 human urothelial FFPE samples.
SUPPLEMENTARY TABLE 2 Clinicopathological characteristics of 16 human ovarian cancer FFPE samples. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Tanaka, N., Kanatani, S., Tomer, R. _et al._ Whole-tissue biopsy phenotyping of three-dimensional tumours reveals patterns of cancer heterogeneity. _Nat Biomed Eng_ 1, 796–806 (2017).
https://doi.org/10.1038/s41551-017-0139-0 Download citation * Received: 11 January 2017 * Accepted: 31 August 2017 * Published: 02 October 2017 * Issue Date: October 2017 * DOI:
https://doi.org/10.1038/s41551-017-0139-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative