- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT We have identified six patients harbouring distinct germline _BAP1_ mutations. In this study, we functionally characterise known _BAP1_ pathogenic and likely benign germline
variants out of these six patients to aid in the evaluation and classification of unknown _BAP1_ germline variants. We found that pathogenic germline variants tend to encode truncated
proteins, show diminished expression of epithelial-mesenchymal transition (EMT) markers, are localised in the cytosol and have reduced deubiquitinase capabilities. We show that these
functional assays are useful for _BAP1_ variant curation and may be added in the American College of Medical Genetics and Genomics (ACMG) criteria for BAP1 variant classification. This will
allow clinicians to distinguish between _BAP1_ pathogenic and likely benign variants reliably and may aid to quickly benchmark newly identified _BAP1_ germline variants. Classification of
novel _BAP1_ germline variants allows clinicians to inform predisposed patients and relevant family members regarding potential cancer risks, with appropriate clinical interventions
implemented if required. SIMILAR CONTENT BEING VIEWED BY OTHERS FUNCTIONAL EVALUATION AND CLINICAL CLASSIFICATION OF _BRCA2_ VARIANTS Article Open access 08 January 2025 INTEGRATION OF
FUNCTIONAL ASSAY DATA RESULTS PROVIDES STRONG EVIDENCE FOR CLASSIFICATION OF HUNDREDS OF _BRCA1_ VARIANTS OF UNCERTAIN SIGNIFICANCE Article Open access 22 October 2020 AN INTEGRATIVE MODEL
FOR THE COMPREHENSIVE CLASSIFICATION OF _BRCA1_ AND _BRCA2_ VARIANTS OF UNCERTAIN CLINICAL SIGNIFICANCE Article Open access 03 June 2022 INTRODUCTION BAP1 (BRCA1-associated protein) is a
nuclear localised deubiquitinating enzyme that is made up of a ubiquitin carboxyl hydrolase (UCH) domain, a host cell factor 1 (HCF1) binding domain, a C-terminal domain with a coiled-coil
motif and a nuclear localisation signal1,2,3. BAP1 is known to be a tumour suppressor that is involved in many cellular processes including apoptosis, metabolism, DNA damage repair,
regulation of gene transcription and removal of ubiquitin from histones (H2AK119ub)4,5,6,7,8,9,10. The polycomb group repressive deubiqutinase complex (PR-DUB) containing BAP1 and ASXL1/2
acts in opposition to polycomb repressive complexes (PRC)2. Together PRCs and PR-DUBs ubiquitinate and deubiquitinate histones, respectively, to fine-tune gene transcription in the cells.
Loss-of-function mutations in _BAP1_ result in loss of deubiquitination activities, disrupting various cellular processes including cell cycle, therefore driving tumourigenesis1 in many
cancers such as renal cell carcinoma, mesothelioma, uveal melanoma, small cell and non-small-cell lung cancers and cholangiocarcinoma11,12,13,14. _BAP1_ germline variants predispose
individuals to high risks of development of aggressive cancers, such as, uveal melanoma, mesothelioma, cutaneous melanoma and renal cell carcinoma at a younger age with poor prognosis15.
Current therapeutic approaches for BAP1-deficient tumours are not particularly promising16,17,18,19,20. An increasing number of _BAP1_ variants have been reported. For example, in 2018, 181
families have been discovered to carry a _BAP1_ variant, of which 140 of the variants are unique21. Classification of missense _BAP1_ variants is more difficult as compared to frameshift,
nonsense and canonical splice variants even with in silico predictions21. As of April 2020, 466 _BAP1_ _germline_ variants of uncertain significance (VUS) have been reported on ClinVar, with
447 of them having single nucleotide variations and 404 of them resulting in missense mutations22. More of such _BAP1_ variants are expected to be discovered as clinical laboratories have
increasingly adopted next-generation sequencing to diagnose hereditary disorders due to its cost-effectiveness, high throughput and accuracy23. Given the increased risks and aggressiveness
of cancer development and progression in patients with _BAP1_ germline variants, understanding how variants in _BAP1_ affect function will aid interpretation of uncertain variants.
Classification of VUS allows us to inform predisposed patients and affected family members regarding potential cancer risks, with appropriate clinical interventions implemented if necessary.
Here, we study and characterise various _BAP1_ germline variants identified in our patients. These patients have family histories of kidney cancers and other cancers such as breast cancer.
Epidemiological, genetic and functional evidence are required to classify variants, and functional assays are ideal to elucidate pathogenicity status especially in rare variants24. To our
knowledge, there are currently no recommended assays to reliably evaluate the functional impact of the germline mutations on BAP1 cellular function in the assessment of variant
pathogenicity. Therefore, by using assays such as deubiquitination assays and immunofluorescence staining, we functionally compare _BAP1_ germline VUS from our patient cohort against known
pathogenic and likely benign variants, which may aid in classification of these variants. RESULTS CLINICOPATHOLOGICAL CHARACTERISTICS OF PATIENTS WITH GERMLINE _BAP1_ VARIANTS Through
clinical genetic testing, we identified six patients harbouring germline _BAP1_ mutations (Table 1, Supplementary Data 1). Three patients are carriers of pathogenic variants, two with
variants of uncertain significance (VUS) and one with a likely benign variant. One of our patients had an additional VUS identified in _MSH6_ (Supplementary Data 1). However, we prioritised
the investigation of _BAP1_ VUS in this study and the possible role of this _MSH6_ VUS in tumorigenesis was not evaluated. Among the pathogenic variant carriers, cancer spectrum was variable
but consistent with _BAP1_ tumor predisposition syndrome (Table 1): one presented mesothelioma, two with breast cancers, of whom one also presented multiple cancers including clear cell
renal cell carcinoma (ccRCC). The median age at diagnosis is 39 (range: 36–61 years). Both VUS carriers were diagnosed with ccRCC in their late thirties, whereas the likely benign carrier
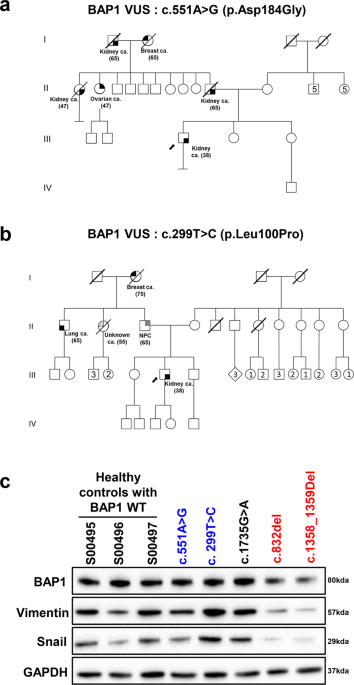
presented thymoma and adrenal cortical cancer. All patients demonstrated strong family history of cancer (Table 1). Pedigrees for the cases identified with _BAP1_ germline VUS show strong
family history of cancers from the paternal side and early onset cancers, including renal and breast cancers (Fig. 1a, b). PATIENT-DERIVED LYMPHOBLASTOID CELL LINES WITH _BAP1_ PATHOGENIC
VARIANTS SHOW REDUCED EXPRESSION OF FULL-LENGTH BAP1 AND EMT MARKERS BAP1 is known to regulate migration, epithelial to mesenchymal transition (EMT) and therefore metastasis in various
cancers including cervical cancer, breast, osteosarcoma and kidney25,26,27,28,29,30. Using patient-derived lymphoblastoid cell lines, we found that carriers of pathogenic _BAP1_ germline
variants (c.852_del and c.1358_1359del) had reduced expression of full-length BAP1, Vimentin and Snail, as compared to controls with wild-type BAP1 (BAP1 WT) (Fig. 1c), whereas _BAP1_
germline VUS (c.299 T > C and c.551 A > G) or likely benign carriers were not significantly different from wild-type _BAP1_ WT. Vimentin is a type III intermediate filament that drives
EMT when overexpressed. Similarly, Snail also promotes EMT by repressing the expression of the adhesion protein E-cadherin. Loss of BAP1 has been reported to downregulate the expression of
Snail, promoting ccRCC cells towards mesenchymal-epithelial transition25. Our results suggest compromised BAP1 function among the pathogenic variants carriers, which results in altered
migration capabilities of the cancer cells, akin to the loss of BAP1. EFFECTS OF _BAP1_ VARIANTS ON CELLULAR LOCALISATION Known mutation positions of _BAP1_ pathogenic variants, _BAP1_
benign variants and _BAP1_ VUS are mapped as shown in Fig. 2a. _BAP1_ pathogenic variants are mostly located before the nuclear localisation signal (NLS) and are predominately frameshift and
nonsense mutations. Missense mutations observed in _BAP1_ pathogenic variants are all found in the UCH domain of BAP1 (Fig. 2b). _BAP1_ VUS appear to be distributed across the gene (Fig.
2a). While the majority of the missense mutations are not in the UCH domain of BAP1; 20–30% of such mutations are in the UCH domain of which function has not yet been established (Fig. 2b).
To investigate if the genetic alterations lead to changes in the cellular localisation of BAP1, we expressed the different _BAP1_ germline variants in HK-2 _BAP1_ KO cell line. In addition
to the variants identified in our patients, we also characterised the _BAP1_ c.1166 G > A variant (p.R389H) that is not located in any known important domains of BAP1 (Fig. 2a) and _BAP1_
c.271 T > G, c.272 G > C variant (p.C91A), a known mutation that abrogates the UCH function31. The nuclear localization of BAP1 is essential to its function, with the loss of nuclear
localization observed in many cancers including uveal melanoma, mesothelioma and ccRCC11,13,32,33,34,35. _BAP1_ variants that lead to protein truncation or frameshift before the nuclear
localisation signal will change the localisation of BAP1, promoting oncogenic pathways36,37. Indeed, we observed that all three of our _BAP1_ truncating variant carriers (c.588 G > A,
c.852_del and c.1358_1359del) displayed cytoplasmic localisation of BAP1, which was not seen for the likely benign variant (c.1735G > A) or WT _BAP1_ (Fig. 3), suggesting that the
cytoplasmic localisation of BAP1 is consistent with the pathogenic classification for these variants. Of note, nuclear localisation of both _BAP1_ VUS carriers (c.299 T > C and c.551 A
> G) were unaffected (Fig. 3). EFFECT OF GENE MUTATIONS ON DEUBIQUITINASE FUNCTION We performed homology modelling of the UCH domain of BAP1 using UCH-L5 (3ris.pdb) as a template (Fig.
4a). _BAP1_ c.551 A > G (p.D184G) is in the catalytic triad of UCH (made up of cysteine, histidine and aspartic acid that abrogates its deubiquitinase function38. Deubiquitination by UCH
domain takes place in two steps39 (Fig. 4b). The thiol of cysteine residue will first be deprotonated by histidine, followed by a nucleophilic attack by the deprotonated thiol on the
carbonyl carbon of the substrate. The aspartic acid residue is likely important to keep the active site residues in a favourable geometry and to stabilise the ion pair formation of the
catalytic residues40. All remaining _BAP1_ variants are not located on the catalytic triad of the UCH domain, hence their effects on the deubiquitinase function of BAP1 protein are unknown
(Fig. 4a). Our homology model of UCH domain of BAP1 using UCH-L5 (3ris.pdb) as a template shows that L100 of BAP1 is located close to the catalytic triad (Fig. 4a). To investigate the
effects of the _BAP1_ VUS p.L100P on the deubiquitinase function of BAP1 protein due to the conformational change of protein 3D structure, we would like to perform molecular dynamics (MD)
simulations for BAP1 homology model. However, because of no structure of template covered, a long unstructured loop (10 amino acid) was modelled for Q156-M165. This region is close to H169
of catalytic triad and may affect the dynamic behaviour of catalytic triad. Instead, we performed MD simulations of its template/homologous protein, UCH-L5, for that the corresponding
residue of L100 and adjacent residues of BAP1 is highly conserved among different species of BAP1 and human UCH-L5 (Supplementary Fig. 1). The corresponding position of L100 of BAP1 on
UCH-L5 is L97 (Supplementary Fig. 1). We performed MD simulations for both wild-type and L97P of UCH-L5. The convergent and comparable Cα RMSDs over 500 ns simulations (Supplementary Fig. 2)
implied that the structures of mutant UCH-L5 was as intact and stable as wild-type during simulations. However, during the last 200 ns stable simulations, the substantial conformational
changes lasting ~30 ns were sampled on L97P mutant UCH-L5. The distance between the two members of catalytic triad (Nε2 of H164 and Oδ of D179) from L97P UCH-L5 was extended to >5 Å
compared to the constant and stable ~3 Å in wild-type protein (Fig. 4c). The falling apart between catalytic triad may affect the enzymatic activity and implies that the highly conserved
leucine (L100 of BAP1 and L97 of UCH-L5) may play an important structural role in supporting the spatial positions of catalytic triad of UCH domain. In addition, we performed in vitro
deubiquitination assay on H2AK119ub, a well-known substrate of BAP1, to assess the effect that the various mutations have on the deubiquitination function of BAP1 (Fig. 4d). _BAP1_ p.C91A
(c.271 T > G, c.272 G > C), a known amino acid mutation that abrogates the UCH function31 serves as our positive control. _BAP1_ variants within the UCH domain, including the _BAP1_
VUS p.L100P, displayed defective deubiquitination capabilities consistent with our positive control (Fig. 4d), suggesting defective UCH function. Interestingly, we found that the pathogenic
_BAP1_ germline variants also have impaired UCH function, even though not positioned within the UCH domain (Fig. 4d). By calculating the ratio of H2AK119ub band intensity compared to H2A
band intensity, normalised to FLAG expressing sample, the pathogenic germline variants showed the greatest disruption to the deubiquitination activity of BAP1, while the likely benign
variant showed the least disruption (Fig. 4d). Our results suggest that pathogenicity of the _BAP1_ germline variant may be related to the integrity of the catalytic activities. Our _BAP1_
germline VUS also showed impaired deubiquitinase function, albeit moderate compared to pathogenic _BAP1_ germline variants (Fig. 4d). FUNCTIONAL CHARACTERISATION OF _BAP1_ MISSENSE GERMLINE
VARIANTS WITH CONFLICTING INTERPRETATIONS Overall, we have functionally characterised _BAP1_ germline VUS and compared them with known _BAP1_ pathogenic and likely benign variants. In 2015,
the American College of Medical Genetics and Genomics (ACMG) has published a standard framework containing variant curation criteria to guide assessment of variant pathogenicity24. The
variants are assessed based on the strength of evidence of pathogenicity in these categories: population data, computational and predictive data, functional data, segregation data, allelic
data, de novo data, other database and other data24 Subsequently, the Clinical Genome Resource (ClinGen) Sequence Variant Interpretation Working Group (SVI) proposed to refine criteria for
variant interpretation from functional assays, recommending different levels of strength of pathogenicity to be assigned based on factors such as the type and number of controls used41.
Based on results of the functional assays and number of controls used in the study, we have summarised our findings in a flowchart to aid in the determination of the functionality of a novel
_BAP1_ germline variant (Fig. 5a). These functional assays may be useful as supporting evidence for pathogenicity in the computational and predictive data and functional data categories of
the ACMG criteria and supporting level evidence for pathogenicity according to the criteria proposed by ClinGen. Any functional abnormality detected using the functional assays described in
this study will be considered evidence for pathogenicity as per ACMG guidelines; if in agreement with other evidence, variants with abnormality detected in one assay are possibly pathogenic
and in two or more assays are likely pathogenic (Fig. 5b). Our results show that pathogenic variants tend to lead to the truncation of BAP1 and associated with reduced expression of
full-length BAP1 and epithelial-mesenchymal transition (EMT) markers in patient-derived lymphoblastoid cell lines. Pathogenic _BAP1_ variants also adversely affect deubiquitination
capabilities and cellular localisation of BAP1. Combining the profiles of these assays will allow us to reliably distinguish between _BAP1_ pathogenic and likely benign variants and
therefore aid to benchmark newly identified _BAP1_ germline variants. 17 _BAP1_ missense germline variants with conflicting interpretations are listed on ClinVar as of July 2020. Five of
these variants were picked for functional characterisation (Fig. 6a), to yield additional insights for these variants based on the proposed functional assays. Two of these variants are
located in the UCH domain, two are located in the BRCA1 binding domain and one is located in region without any known domains (Fig. 6a). The nuclear localisation of these _BAP1_ variants
were not affected (Fig. 6b). In vitro deubiquitination assay showed that _BAP1_ c.121 G > A and BAP1 c.341 G > A had impaired deubiquitinase functions, even though moderate compared to
positive control (Fig. 6c). Our results provide functional evidence that _BAP1_ c.121 G > A and _BAP1_ c.341 G > A are possibly pathogenic variants and demonstrate that the proposed
functional assays can be utilised to rapidly evaluate and facilitate classification of _BAP1_ variants of uncertain significance to resolve conflicting interpretations for these variants.
DISCUSSION _BAP1_ tumour predisposition syndrome is a hereditary tumour syndrome that is associated with germline pathogenic mutations in _BAP1_21 and increased susceptibility for uveal
melanoma, mesothelioma, cutaneous melanoma and renal cell carcinoma42. An increasing number of other cancers including cholangiocarcinoma, breast cancers and neuroendocrine tumours have also
been shown to be associated with the syndrome15,43,44,45. The mode of inheritance in _BAP1_ tumour predisposition syndrome is autosomal dominant and a majority of the germline pathogenic
variants in _BAP1_ are truncating46. Cancer patients with _BAP1_ tumour predisposition syndrome are generally associated with poorer prognosis compared to patients without
predisposition15,43,45. Germline carriers of _BAP1_ pathogenic variants demonstrate earlier onset (e.g. median age of onset in uveal melanoma patients with germline _BAP1_ mutations is 51
years compared to 62 years in the general population47) and more aggressive cancers15. Several therapeutic approaches have been explored for BAP1-deficient tumours, including
chemotherapeutic drugs (e.g. gemcitabine), radiotherapy and small molecule inhibitors (e.g. EZH2 inhibitors) with limited success16,17,18,19,20. Due to the increasingly widespread use of
next-generation sequencing in clinical laboratories23, more novel _BAP1_ variants are expected to be discovered21,22. Functional studies have emerged as one of the indispensable methods to
help classify _BAP1_ VUS in addition to family cancer history and genetic evidence. Variant classification is important as it may have impact on subsequent cancer screening, follow-up and
therapeutic strategies. Unfortunately, no standard protocol of functional assays is used to aid the classification of _BAP1_ VUS. In this study, we functionally assay each _BAP1_ germline
variant found in our patient cohort, including benign, pathogenic and VUS, using in vitro deubiquitination assay, cellular localisation determination using immunofluorescence staining and
expression of EMT markers. These assays are only valid when the _BAP1_ germline variant of interest can be expressed in cell models. By analysing known BAP1 pathogenic variants listed in
ClinVar, we found that they are mostly located before the nuclear localisation signal (NLS) and are predominately frameshift and nonsense mutations (Fig. 2), which is consistent with that
from our patient cohort. These types of mutations introduce a premature stop codon and are predicted to be target of degradation by nonsense-mediated mRNA decay (NMD) to protect the cells
from the presence of abnormal peptides48,49. While we cannot exclude the possibility that our curated pathogenic frameshift and nonsense variants will undergo NMD, we have shown that
truncated proteins can be expressed from these variants (Fig. 4d). With the exception of analysing the expression of EMT markers in the LCLs, our functional assays are based on introducing
the _BAP1_ germline variant into cell models and therefore cannot be used to investigate effects of loss of _BAP1_ expression. We found that pathogenic _BAP1_ germline variants show
defective deubiquitination function, are aberrantly localised in the cytosol or display low expression of EMT markers such as Vimentin and Snail, allowing them to be clearly distinguished
from likely benign variants. Based on our deubiquitination assay, _BAP1_ VUS c.299 T > C and c.551 A > T, both of which located in the UCH domain of BAP1, were observed to have
decreased deubiquitinase activities. These patients also have strong family history of _BAP1_-related cancers. Therefore, _BAP1_ VUS c.299 T > C (Supplementary Fig. 3a) and c.551 A > T
(Supplementary Fig. 3b) are functionally abnormal. We have proposed the use of the functional assays used in the study to augment the ACMG criteria and ClinGen recommendations for assessing
_BAP1_ variant pathogenicity (Fig. 5). Given these lines of evidence and based on the level of evidence in the other categories in the ACMG framework, _BAP1_ VUS c.299 T > C and c.551 A
> T may therefore be considered to be reclassified as likely pathogenic (Table 2). Although our validation set of _BAP1_ germline variants is limited in size, the profiles demonstrated by
our functional assays could clearly distinguish the functional impact of pathogenic from benign variants and may be used to provide a line of functional evidence in determining the
pathogenicity of newly identified _BAP1_ variants. In conclusion, our study may allow clinicians to quickly benchmark newly identified _BAP1_ variants against known pathogenic and likely
benign variants. Even though the penetrance of having a _BAP1_ germline variant has been shown incomplete and the type of _BAP1-_related cancers has been observed to vary among different
family members42, our findings contribute to providing functional support for resolution of _BAP1_ variants with uncertain significance and facilitating clinical interpretation by clinicians
so that appropriate prognosis, therapeutic intervention strategies and care may be delivered to patients with _BAP1_ tumour predisposition syndrome. METHODS VARIANT CURATION Patients
selected for this study were referred to our Cancer Genetics Service at National Cancer Centre Singapore for clinical genetic testing on multi-gene panels (including _BAP1)_ by commercial
laboratories accredited by CLIA/CAP. Classification of identified _BAP1_ variants were performed by the respective accredited laboratories in accordance with the American College of Medical
Genetics/Association for Molecular Pathology (ACMG/AMP) guidelines24. Patients included for analysis were identified to harbour germline _BAP1_ variants, regardless of classification.
Patient clinical history and family history of cancer were assessed by a clinical geneticist or genetic counsellor at our service. This study was approved by SingHealth Centralized
Institutional Review Board (CIRB 2011/826/B) with signed informed consent from all participants. COMMERCIAL CELL LINES AND CELL CULTURE All commercial cell lines were purchased from ATCC.
HEK293T is cultured in DMEM + 10% FBS. HK-2 _BAP1_ KO cell is kind gift from Prof Teh Bin Tean’s lab and cultured in RPMI 1640 + 10% FBS. Patient-derived cell lines are cultured in RPMI 1640
+ 10% FBS. All cell lines were maintained at 37 °C in a humidified chamber in the presence of 5% CO2 and routinely confirmed to be free of mycoplasma. Cells used for experiments were
between 3 and 8 passages from thawing. At least three biological replicates were performed for each experiment. IMMUNOBLOTTING Cell pellets were collected and washed twice in cold PBS,
before they are lysed in RIPA buffer supplemented with protease and phosphatase inhibitor cocktail (Roche). The lysed cells were then and sonicated for 10 cycles at 30 s on and 30 s off,
high settings using a Biorupter (Diagenode) to ensure complete lysis. The lysates were clarified by centrifugation at 16,000 × _g_ r.c.f for 20 min and the protein extracts were quantified
using BCA assay (ThermoFisher Scientific Corporation, Waltham, MA, USA). 5–10 μg of protein was loaded onto 4–12% gradient gel, electrophoresed by SDS-PAGE and transferred to 0.2 μm
nitrocellulose membrane (Biorad). The membranes were then blocked in 5% non-fat milk diluted in 1X PBST and incubated overnight with primary antibody diluted in blocking buffer at 4 °C. The
following primary antibodies were used: Monoclonal Anti-Flag® M2 antibody (Sigma–Aldrich: F1804, 1:2000), anti- ubiquityl-Histone H2A (Cell Signaling, Beverly, MA; #2577, 1:1000),
anti-Histone H2A antibody (Abcam, Cambridge, MA, USA: ab18255, 1:1000), anti-BAP1 (Santa Cruz Biotechnology: sc-28383, 1;200), anti-Vimentin (Cell Signaling, Beverly, MA: #5741, 1:1000),
anti-Snail (Cell Signaling, Beverly, MA: #3879, 1:1000) and GAPDH (Santa Cruz Biotechnology: sc-166545, 1;1000). Membranes were washed three times for 10 min each with 1× PBST then incubated
with secondary HRP-conjugated antibody anti-rabbit or anti-mouse immunoglobulin (Promega Corporation: W4011 or W4021, 1:10,000) for 1 h. After washing three times for 10 min each with 1X
PBST after incubation, the immunoreactivity was detected with SuperSignal West Femto Maximum Sensitivity Substrate (ThermoFisher Scientific Corporation, Waltham, MA, USA). All blots were
derived from the same experiments and processed in parallel. Un-cropped images of blots are shown in Supplementary Fig. 4. IMMUNOFLUORESCENCE STAINING Transfected HK-2 BAP1 KO cells were
plated on µ-Dish 35 mm imaging dishes (ibidi). Fixation was performed for 10 min using 4% paraformaldehyde (Sigma–Aldrich) at room temperature, followed by permeabilization using 0.3%
Triton-X 100 (Sigma–Aldrich) on ice for 10 min. The cells were then blocked in 5% BSA in PBS for 1 h and then incubated overnight with Monoclonal Anti-Flag® M2 antibody (Sigma–Aldrich:
F1804, 1:500). Slides were washed three times with 1X PBS for 10 min and then incubated AlexaFluor A488 conjugated secondary antibodies (ThermoFisher Scientific Corporation, Waltham, MA,
USA: A11008, 1:1000). Hoechst 33342 was used to stain the nucleus. After incubation for 1 h at room temperature, the slides were washed 3 times with PBS and mounted with Prolong Gold
antifade reagent (ThermoFisher Scientific Corporation, Waltham, MA, USA). Images were acquired by confocal microscope (Zeiss LSM700). HOMOLOGY MODELING Protein sequences of human BAP1
(Q92560) and human UCH-L5 (Q9Y5K5) were obtained from Uniprot50. The sequence identity of UCH domain between human BAP1 and UCH-L5 is 44%. The homology model of UCH domain of human BAP1 was
built by MODELLER51 according to the template 3D structure of human UCH-L5 (3ris.pdb52). MD SIMULATION To investigate the effect of VUS on deubiquitination function due to conformational
change of UCH domain, we implemented the computational approach, molecular dynamics (MD) simulation. The initial 3D structure of human UCH-L5 (3ris.pdb52 chainA, amino acids 7–227) was
obtained from Protein Data Bank53. The structure of six missing residues (amino acid 155–160) were modeled by MODELLER51. Explicit hydrogen atoms and protonation states of titratable
residues of UCH-L5 were determined by the web server PDB2PQR54 (http://nbcr-222.ucsd.edu/pdb2pqr_2.1.1/). The systems of MD simulations were prepared using AmberTools package55. We adopted
ff14SB force fields for the UCH-L5. The UCH-L5 was solvated in virtual cubic boxes filled with TIP3P water molecules. To maintain charge neutralization of the systems, proper numbers of
counter ions (Na+ or Cl−) were added into the simulation boxes. All energy minimizations and dynamics simulations in this study were performed using AMBER16 package55. To release steric
clashes between atoms, we energy minimized the systems by the following protocol. The first 5000 cycles of energy minimization were performed by steepest descent method while the backbone
heavy atoms of UCH-L5 were restrained by harmonic potentials (force constant was set as 10.0 kcal/mol-Å2). The second 5000 cycles of energy minimization were followed without any restraints.
Periodic boundary condition was employed during dynamics simulations while long range electrostatic interactions were treated by the particle mesh Ewald method and short range nonbonded
cutoff was set as 12.0 Å. SHAKE algorithm was used to constrain bond length so that the simulations of production run allow a larger time step of 2 fs. To heat up the system, we ran 100 ps
simulations using time step of 1 fs. During the heating process, backbone heavy atoms of UCH-L5 were restrained by a harmonic potential with force constant of 10.0 kcal/mol-Å2. The dynamics
simulations in this study were in the NPT ensemble. The pressure was coupled to 1.0 bar and temperature was coupled to 298.15 K by Berendsen’s coupling methods with both coupling constants
of 1 ps. Backbone restraints were relaxed to 5.0 kcal/mol-Å2 for additional 100 ps simulation before production run. During the simulations of production run, we performed 200 ns simulations
for both systems with larger time step of 2 fs. The temperature coupling method of production simulations was switched to Langevin thermostat with the collision frequency as 5 ps−1 whereas
Berendsen’s method was retained for pressure coupling. The simulations of production run were performed for 500 ns. All data analyses except heavy atom RMSD were based on the trajectories
belonging to the period from 300 ns to 500 ns. IN VITRO DEUBIQUITINATION ASSAY The indicated BAP1 variants were fused in frame with N-terminal FLAG in pFLAG-CMV1 expression vector
(Sigma–Aldrich). HEK293T cells were transfected with 1 µg each of the indicated plasmids using Lipofectamine 3000 (ThermoFisher Scientific Corporation, Waltham, MA, USA). After 3 days, the
cell pellets of the transfected cells were harvested, washed in PBS and lysed (50 mM Tris HCl, pH 7.4, 1% Triton X-100, with 150 mM NaCl). The FLAG tagged BAP1 variants were allowed to bind
to Dynabeads™ Protein G (ThermoFisher Scientific Corporation, Waltham, MA, USA) that had been conjugated with Monoclonal Anti-Flag® M2 antibody overnight. The FLAG tagged BAP1 variants bound
to the beads were washed five times (0.5 M Tris HCl, pH 7.4, with 1.5 M NaCl). In vitro deubiquitination assay was performed as described56 with modifications. The washed beads with bound
FLAG tagged BAP1 variants were resuspended in deubiquitination buffer (50 mM Tris-HCl, pH 7.3, 1 mM MgCl2, 50 mM NaCl, and 1 mM DTT, 1 mM EDTA and 1% Triton X-100). 200 ng of H2AK119UB
(EpiCyher) was added to the mixture and deubiquitination was carried out at 37 °C for 3 h with gentle shaking. The reaction was quenched by adding 2× Laemmli buffer and analysed by
immunoblotting. REPORTING SUMMARY Further information on research design is available in the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY All data generated or
analysed during this study are included in this published article (and its supplementary information files). CODE AVAILABILITY Algorithms used for molecular dynamics stimulation detailed
are in the Methods section. REFERENCES * Daou, S. et al. The BAP1/ASXL2 histone H2A deubiquitinase complex regulates cell proliferation and is disrupted in cancer. _J. Biol. Chem._ 290,
28643–28663 (2015). Article CAS PubMed PubMed Central Google Scholar * Murali, R., Wiesner, T. & Scolyer, R. A. Tumours associated with BAP1 mutations. _Pathology_ 45, 116–126
(2013). Article CAS PubMed Google Scholar * Okino, Y., Machida, Y., Frankland-Searby, S. & Machida, Y. J. BRCA1-associated protein 1 (BAP1) deubiquitinase antagonizes the
ubiquitin-mediated activation of FoxK2 target genes. _J. Biol. Chem._ 290, 1580–1591 (2015). Article PubMed CAS Google Scholar * Bononi, A. et al. BAP1 regulates IP3R3-mediated Ca(2+)
flux to mitochondria suppressing cell transformation. _Nature_ 546, 549–553 (2017). Article CAS PubMed PubMed Central Google Scholar * Eletr, Z. M., Yin, L. & Wilkinson, K. D. BAP1
is phosphorylated at serine 592 in S-phase following DNA damage. _FEBS Lett._ 587, 3906–3911 (2013). Article CAS PubMed PubMed Central Google Scholar * Ji, Z. et al. The forkhead
transcription factor FOXK2 acts as a chromatin targeting factor for the BAP1-containing histone deubiquitinase complex. _Nucleic Acids Res._ 42, 6232–6242 (2014). Article CAS PubMed
PubMed Central Google Scholar * White, A. E. & Harper, J. W. Cancer. Emerging anatomy of the BAP1 tumor suppressor system. _Science_ 337, 1463–1464 (2012). Article CAS PubMed Google
Scholar * Yu, H. et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. _Proc. Natl Acad. Sci. USA_ 111, 285–290 (2014). Article CAS PubMed Google
Scholar * Zarrizi, R., Menard, J. A., Belting, M. & Massoumi, R. Deubiquitination of gamma-tubulin by BAP1 prevents chromosome instability in breast cancer cells. _Cancer Res._ 74,
6499–6508 (2014). Article CAS PubMed Google Scholar * Zhang, Y. et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. _Nat. Cell Biol._ 20, 1181–1192 (2018).
Article CAS PubMed PubMed Central Google Scholar * Carbone, M. et al. BAP1 and cancer. _Nat. Rev. Cancer_ 13, 153–159 (2013). Article CAS PubMed PubMed Central Google Scholar *
Jusakul, A. et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. _Cancer Discov._ 7, 1116–1135 (2017). Article CAS PubMed PubMed Central
Google Scholar * Ventii, K. H. et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. _Cancer Res._ 68, 6953–6962
(2008). Article CAS PubMed PubMed Central Google Scholar * Yu, H. et al. The ubiquitin carboxyl hydrolase BAP1 forms a ternary complex with YY1 and HCF-1 and is a critical regulator of
gene expression. _Mol. Cell. Biol._ 30, 5071–5085 (2010). Article CAS PubMed PubMed Central Google Scholar * Rai, K., Pilarski, R., Cebulla, C. M. & Abdel-Rahman, M. H.
Comprehensive review of BAP1 tumor predisposition syndrome with report of two new cases. _Clin. Genet._ 89, 285–294 (2016). Article CAS PubMed Google Scholar * Chau, C. et al. Families
with BAP1-tumor predisposition syndrome in The Netherlands: path to identification and a proposal for genetic screening guidelines. _Cancers._ https://doi.org/10.3390/cancers11081114 (2019).
* Guazzelli, A. et al. BAP1 status determines the sensitivity of malignant mesothelioma cells to gemcitabine treatment. _Int. J. Mol. Sci._ https://doi.org/10.3390/ijms20020429 (2019). *
Ladanyi, M., Sanchez Vega, F. & Zauderer, M. Loss of BAP1 as a candidate predictive biomarker for immunotherapy of mesothelioma. _Genome Med._ 11, 18 (2019). Article PubMed PubMed
Central Google Scholar * Shrestha, R. et al. BAP1 haploinsufficiency predicts a distinct immunogenic class of malignant peritoneal mesothelioma. _Genome Med._ 11, 8 (2019). Article PubMed
PubMed Central Google Scholar * Zauderer, M. G. et al. Phase 2, multicenter study of the EZH2 inhibitor tazemetostat as monotherapy in adults with relapsed or refractory (R/R) malignant
mesothelioma (MM) with BAP1 inactivation. _J. Clin. Oncol._ 36, 8515–8515 (2018). Article Google Scholar * Walpole, S. et al. Comprehensive study of the clinical phenotype of germline BAP1
variant-carrying families worldwide. _J. Natl Cancer Inst._ 110, 1328–1341 (2018). Article PubMed PubMed Central CAS Google Scholar * Landrum, M. J. et al. ClinVar: improving access to
variant interpretations and supporting evidence. _Nucleic Acids Res._ 46, D1062–d1067 (2018). Article CAS PubMed Google Scholar * Hartman, P. et al. Next generation sequencing for
clinical diagnostics: five year experience of an academic laboratory. _Mol. Genet. Metab. Rep._ 19, 100464 (2019). Article CAS PubMed PubMed Central Google Scholar * Richards, S. et al.
Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for
Molecular Pathology. _Genet. Med._ 17, 405–424 (2015). Article PubMed PubMed Central Google Scholar * Chen, P. et al. Loss of BAP1 results in growth inhibition and enhances
mesenchymal-epithelial transition in kidney tumor cells. _Mol. Cell. Proteom._ 18, 1320–1329 (2019). Article CAS Google Scholar * Gao, S., Sun, H., Cheng, C. & Wang, G.
BRCA1-associated protein-1 suppresses osteosarcoma cell proliferation and migration through regulation PI3K/Akt pathway. _DNA Cell Biol._ 36, 386–393 (2017). Article CAS PubMed Google
Scholar * Onken, M. D., Li, J. & Cooper, J. A. Uveal melanoma cells utilize a novel route for transendothelial migration. _PloS ONE_ 9, e115472 (2014). Article PubMed PubMed Central
CAS Google Scholar * Qin, J. et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. _Nat. Commun._ 6, 8471 (2015). Article CAS PubMed Google
Scholar * Wang, N., Li, Y. & Zhou, J. miR-31 functions as an oncomir which promotes epithelial-mesenchymal transition via regulating BAP1 in cervical cancer. _BioMed. Res. Int._ 2017,
6361420 (2017). PubMed PubMed Central Google Scholar * Yue, H. et al. Calpastatin participates in the regulation of cell migration in BAP1-deficient uveal melanoma cells. _Int. J.
Ophthalmol._ 12, 1680–1687 (2019). Article PubMed PubMed Central Google Scholar * Misaghi, S. et al. Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell
cycle regulator host cell factor 1. _Mol. Cell. Biol._ 29, 2181–2192 (2009). Article CAS PubMed PubMed Central Google Scholar * Abdel-Rahman, M. H. et al. Germline BAP1 mutation
predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. _J. Med. Genet._ 48, 856–859 (2011). Article CAS PubMed Google Scholar * Harbour, J. W. et al. Frequent
mutation of BAP1 in metastasizing uveal melanomas. _Science_ 330, 1410–1413 (2010). Article CAS PubMed PubMed Central Google Scholar * Song, H. et al. Loss of nuclear BAP1 expression
is associated with poor prognosis in oral mucosal melanoma. _Oncotarget_ 8, 29080–29090 (2017). Article PubMed PubMed Central Google Scholar * Wi, Y. C. et al. Loss of nuclear BAP1
expression is associated with high WHO/ISUP grade in clear cell renal cell carcinoma. _J. Pathol. Transl. Med._ 52, 378–385 (2018). Article PubMed PubMed Central Google Scholar *
Bhattacharya, S., Hanpude, P. & Maiti, T. K. Cancer associated missense mutations in BAP1 catalytic domain induce amyloidogenic aggregation: a new insight in enzymatic inactivation.
_Sci. Rep._ 5, 18462 (2015). Article CAS PubMed PubMed Central Google Scholar * Mori, T. et al. Somatic alteration and depleted nuclear expression of BAP1 in human esophageal squamous
cell carcinoma. _Cancer Sci._ 106, 1118–1129 (2015). Article CAS PubMed PubMed Central Google Scholar * Jensen, D. E. et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1
RING finger and enhances BRCA1-mediated cell growth suppression. _Oncogene_ 16, 1097–1112 (1998). Article CAS PubMed Google Scholar * Elsässer, B. et al. Distinct roles of catalytic
cysteine and histidine in the protease and ligase mechanisms of human legumain as revealed by DFT-based QM/MM simulations. _ACS Catal._ 7, 5585–5593 (2017). Article PubMed PubMed Central
CAS Google Scholar * Vernet, T. et al. Structural and functional roles of asparagine 175 in the cysteine protease papain. _J. Biol. Chem._ 270, 16645–16652 (1995). Article CAS PubMed
Google Scholar * Brnich, S. E. et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. _Genome
Med._ 12, 3 (2019). Article PubMed PubMed Central Google Scholar * Pilarski R. et al. BAP1 Tumor Predisposition Syndrome. 2016 Oct 13 [Updated 2020 Sep 17]. In: Adam MP, Ardinger HH,
Pagon RA, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2020. Available from: https://www.ncbi.nlm.nih.gov/books/NBK390611/. * Gupta, M. P.
et al. Clinical characteristics of uveal melanoma in patients with germline BAP1 mutations. _JAMA Ophthalmol._ 133, 881–887 (2015). Article PubMed Google Scholar * Helgadottir, H. &
Hoiom, V. The genetics of uveal melanoma: current insights. _Appl. Clin. Genet._ 9, 147–155 (2016). Article CAS PubMed PubMed Central Google Scholar * Wang, A., Papneja, A., Hyrcza, M.,
Al-Habeeb, A. & Ghazarian, D. Gene of the month: BAP1. _J. Clin. Pathol._ 69, 750–753 (2016). Article CAS PubMed Google Scholar * Masoomian, B., Shields, J. A. & Shields, C. L.
Overview of BAP1 cancer predisposition syndrome and the relationship to uveal melanoma. _J. Curr. Ophthalmol._ 30, 102–109 (2018). Article PubMed PubMed Central Google Scholar * Singh,
A. D., Turell, M. E. & Topham, A. K. Uveal melanoma: trends in incidence, treatment, and survival. _Ophthalmology_ 118, 1881–1885 (2011). Article PubMed Google Scholar * Frischmeyer,
P. A. & Dietz, H. C. Nonsense-mediated mRNA decay in health and disease. _Hum. Mol. Genet._ 8, 1893–1900 (1999). Article CAS PubMed Google Scholar * Keeling, K. M. & Bedwell, D.
M. Suppression of nonsense mutations as a therapeutic approach to treat genetic diseases. _Wiley Interdiscip. Rev. RNA_ 2, 837–852 (2011). Article CAS PubMed PubMed Central Google
Scholar * Consortium, U. UniProt: a worldwide hub of protein knowledge. _Nucleic Acids Res._ 47, D506–D515 (2019). Article CAS Google Scholar * Webb, B. & Sali, A. Comparative
protein structure modeling using MODELLER. _Curr. Protoc. Bioinforma._ 54, 5.6. 1–5.6. 37 (2016). Article Google Scholar * Maiti, T. K. et al. Crystal structure of the catalytic domain of
UCHL5, a proteasome‐associated human deubiquitinating enzyme, reveals an unproductive form of the enzyme. _FEBS J._ 278, 4917–4926 (2011). Article CAS PubMed PubMed Central Google
Scholar * Burley, S. K. et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy.
_Nucleic Acids Res._ 47, D464–D474 (2019). Article CAS PubMed Google Scholar * Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. PDB2PQR: an automated pipeline for the
setup of Poisson–Boltzmann electrostatics calculations. _Nucleic Acids Res._ 32, W665–W667 (2004). Article CAS PubMed PubMed Central Google Scholar * Case, D. A. et al. _AMBER 2016_
(University of California, San Francisco 810, 2016). * Masclef, L. et al. In vitro ubiquitination and deubiquitination assays of nucleosomal histones. _J. Vis. Exp._
https://doi.org/10.3791/59385 (2019). Article PubMed Google Scholar * Zhou, X. et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. _Nat. Genet._ 48, 4–6 (2016).
Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We would like to thank the patients and research participants for their contribution to the study.
This work was supported by the Khoo Postdoctoral Fellowship Award (Duke-NUS-KPFA/2019/0034 to J.H.H), the National Medical Research Council Open Fund—Young Individual Research Grant
(MOH-000232 to J.H.H), the National Medical Research Council Open Fund—Individual Research Grant (MOH-000144 to B.T.T), the National Medical Research Council Singapore Translational Research
Investigator Award (MOH-000248 to B.T.T.), the NRF-NSFC Joint Research Grant (Data Science) (NRF2016NRF-NSFC001–057 to B.T.T), the LKC Startup Grant (LKC MOE to J.N.), the National Research
Foundation Singapore under its Clinical Scientist Award (NMRC/CSA-INV/0017/2017) and administered by the Singapore Ministry of Health’s National Medical Research Council (to J.N.). This
work is also partially funded by NCC Research Fund, NCC Cancer Fund, Terry Fox and Lee Foundation supporting funds to J.N. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Cancer and Stem Cell
Biology Program, Duke-NUS Medical School, Singapore, 169857, Singapore Jing Han Hong, Hong Lee Heng & Bin Tean Teh * Institute of Molecular and Cellular Biology, Agency for Science,
Technology and Research, Singapore, 138673, Singapore Jing Han Hong, Bin Tean Teh & Joanne Ngeow * Cancer Genetics Service, Division of Medical Oncology, National Cancer Center,
Singapore, 169610, Singapore Siao Ting Chong, Nur Diana Binte Ishak, Sock Hoai Chan & Joanne Ngeow * Cancer Science Institute of Singapore, National University of Singapore, Singapore,
117599, Singapore Po-Hsien Lee & Bin Tean Teh * Genome Institute of Singapore, Agency for Science, Technology and Research, Singapore, 138672, Singapore Po-Hsien Lee & Bin Tean Teh *
Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center of Cancer Medicine, 510060, Guangzhou, Guangdong, China Jing Tan *
Laboratory of Cancer Epigenome, Division of Medical Sciences, National Cancer Centre Singapore, Singapore, 169610, Singapore Jing Tan, Hong Lee Heng & Bin Tean Teh * SingHealth/Duke-NUS
Institute of Precision Medicine, National Heart Centre, Singapore, Singapore Bin Tean Teh * Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore, 308232, Singapore
Joanne Ngeow * Oncology Academic Clinical Program, Duke-NUS Medical School, Singapore, 169857, Singapore Joanne Ngeow Authors * Jing Han Hong View author publications You can also search for
this author inPubMed Google Scholar * Siao Ting Chong View author publications You can also search for this author inPubMed Google Scholar * Po-Hsien Lee View author publications You can
also search for this author inPubMed Google Scholar * Jing Tan View author publications You can also search for this author inPubMed Google Scholar * Hong Lee Heng View author publications
You can also search for this author inPubMed Google Scholar * Nur Diana Binte Ishak View author publications You can also search for this author inPubMed Google Scholar * Sock Hoai Chan View
author publications You can also search for this author inPubMed Google Scholar * Bin Tean Teh View author publications You can also search for this author inPubMed Google Scholar * Joanne
Ngeow View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Conceptualization, J.N., B.T.T., J.H.H.; Methodology, J.H.H., S.T.C., H.P.L, T.J.,
H.L.H.; Investigation, J.H.H., S.T.C., H.P.L., T.J., H.L.H., N.D.B.I.; Writing—original draft, J.H.H., J.N; Writing—-review and editing, J.H.H., J.N., S.T.C., P.H.L., S.H.C., T.J., B.T.T.;
Funding acquisition, J.N., B.T.T., J.H.H.; Resources, J.N., B.T.T., J.H.H.; Supervision, J.N. and B.T.T. CORRESPONDING AUTHORS Correspondence to Bin Tean Teh or Joanne Ngeow. ETHICS
DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in
published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION SUPPLEMENTARY DATA 1 REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article is
licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give
appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in
this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative
Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a
copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Hong, J.H., Chong, S.T., Lee, PH. _et al._ Functional
characterisation guides classification of novel _BAP1_ germline variants. _npj Genom. Med._ 5, 50 (2020). https://doi.org/10.1038/s41525-020-00157-6 Download citation * Received: 05 June
2020 * Accepted: 20 October 2020 * Published: 19 November 2020 * DOI: https://doi.org/10.1038/s41525-020-00157-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to
read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative