- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Nirmatrelvir, an oral antiviral agent that targets a SARS-CoV-2 main protease (3CLpro), is clinically useful against infection with SARS-CoV-2 including its omicron variants. Since
most omicron subvariants have reduced sensitivity to many monoclonal antibody therapies, potential SARS-CoV-2 resistance to nirmatrelvir is a major public health concern. Several amino acid
substitutions have been identified as being responsible for reduced susceptibility to nirmatrelvir. Among them, we selected L50F/E166V and L50F/E166A/L167F in the 3CLpro because these
combinations of substitutions are unlikely to affect virus fitness. We prepared and characterized delta variants possessing Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F. Both mutant viruses
showed decreased susceptibility to nirmatrelvir and their growth in VeroE6/TMPRSS2 cells was delayed. Both mutant viruses showed attenuated phenotypes in a male hamster infection model,
maintained airborne transmissibility, and were outcompeted by wild-type virus in co-infection experiments in the absence of nirmatrelvir, but less so in the presence of the drug. These
results suggest that viruses possessing Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F do not become dominant in nature. However, it is important to closely monitor the emergence of
nirmatrelvir-resistant SARS-CoV-2 variants because resistant viruses with additional compensatory mutations could emerge, outcompete the wild-type virus, and become dominant. SIMILAR CONTENT
BEING VIEWED BY OTHERS IN VITRO AND IN VIVO CHARACTERIZATION OF SARS-COV-2 RESISTANCE TO ENSITRELVIR Article Open access 15 July 2023 MULTIPLE PATHWAYS FOR SARS-COV-2 RESISTANCE TO
NIRMATRELVIR Article Open access 09 November 2022 MOLECULAR MECHANISMS OF SARS-COV-2 RESISTANCE TO NIRMATRELVIR Article 11 September 2023 INTRODUCTION Severe acute respiratory syndrome
coronavirus 2 (SARS-CoV-2), the cause of COVID-19, is still prevalent three years since its emergence and continues to spread around the world. It accumulates amino acid substitutions
frequently and many variants of concern (VOCs) have appeared and caused several waves of infection. Among these variants, omicron (lineage B.1.1.529) was identified at the end of 2021 and
became globally dominant1. Omicron variants possess more than 30 amino acid substitutions in the spike protein, and its subvariants BA.4/5, BA.4.6, and BA.2.75 are less susceptible to
therapeutic monoclonal antibodies including REGEN-COV [casirivimab (REGN10933) plus imdevimab (REGN10987)], bamlanivimab (LY-CoV555) plus etesevimab (LY-CoV016), evusheld [tixagevimab
(COV2-2196) plus cilgavimab (COV2-2130)], and xevudy [sotrovimab (S309)]2,3,4,5,6. BA.4/5, BA.4.6, and BA.2.75 are susceptible to bebtelovimab (LY-CoV1404) but the latest subvariants BQ.1.1
and XBB escape from it7. The antiviral compounds nirmatrelvir, remdesivir, molnupiravir, and ensitrelvir, which target virus proteins other than the spike protein, are effective against such
variants and subvariants. Several antivirals have been widely used for to treat COVID-19 including GS-441524 (remdesivir), EIDD-1931 (molnupiravir), PF-07321332 (nirmatrelvir), and S-217622
(ensitrelvir). Remdesivir and molnupiravir inhibit virus RNA-dependent RNA polymerase, whereas nirmatrelvir and ensitrelvir interfere with 3CL protease [3CLpro; also known as main protease
(Mpro) and nonstructural protein 5 (nsp5)]. As these drugs are increasingly used in clinical settings, there is concern about the emergence of viruses with reduced sensitivity or resistance.
Amino acid substitutions associated with resistance were reported for remdesivir8,9 and nirmatrelvir10,11. Nirmatrelvir (PF-07321332) is co-formulated with the pharmacokinetic enhancer
ritonavir (the co-formulated product is called paxlovid)12. When treatment is initiated within three days of onset, approximately 90% protection against severe COVID-19 and hospitalization
has been reported13. Amino acid substitutions that are responsible for resistance to nirmatrelvir have been reported in 3CLpro (nsp5). In silico analysis showed that the N142L, E166M, Q189E,
Q189I, and Q192T substitutions reduced the potency of nirmatrelvir in vitro14. A screen using a VSV-based system revealed that the Y54C, G138S, L167F, and Q192R substitutions confer
resistance to nirmatrelvir15. Naturally occurring amino acid substitutions such as S144A, A173V, and E166D/G also contribute to resistance to nirmatrelvir10,11,16. Deep mutational analysis
has shown that the E166V, P252L, and T304I substitutions reduce the IC50 value to nirmatrelvir17,18. In another study, resistant viruses possessing the E166V and T304I substitutions in
combination with other amino acid substitutions in nsp5 were selected through virus passaging in the presence of nirmatrelvir in cell culture19. The E166V and T304I substitutions conferred
strong resistance but resulted in a loss of viral fitness; viral fitness was restored by compensatory substitutions such as L50F and T21I18. The L50F and T21I substitutions had little effect
on the sensitivity to nirmatrelvir18. Another resistant virus selection experiment in vitro identified the combination of the L50F, E166A, and L167F substitutions as being associated with
resistance to nirmatrelvir20. Each E166A and L167F substitution conferred reduced sensitivity to nirmatrelvir20. Recent studies have reported that the virus fitness of viruses possessing the
L50F and E166V substitutions and the L50F, E166A, and L167F substitutions is comparable to that of wild-type virus in cell culture19,20. Another preprint reported that a recombinant virus
possessing the L50F, E166A, and L167F substitutions showed similar pathogenicity in hamsters and transmitted to co-housed hamsters21. Therefore, among the many resistant mutations reported,
we chose to study Nsp5-L50F/E166V19 and Nsp5-L50F/E166A/L167F20 because they appeared unlikely to affect virus fitness in vitro. By using reverse-genetics based on the delta variant, we
prepared mutant viruses possessing these substitution combinations and characterized their replication in vitro, their pathogenicity in hamsters, and their airborne transmissibility in
hamsters. We also assessed their replication in co-infection experiments with wild-type virus in the presence or absence of nirmatrelvir in the hamster model. RESULTS SENSITIVITY OF MUTANT
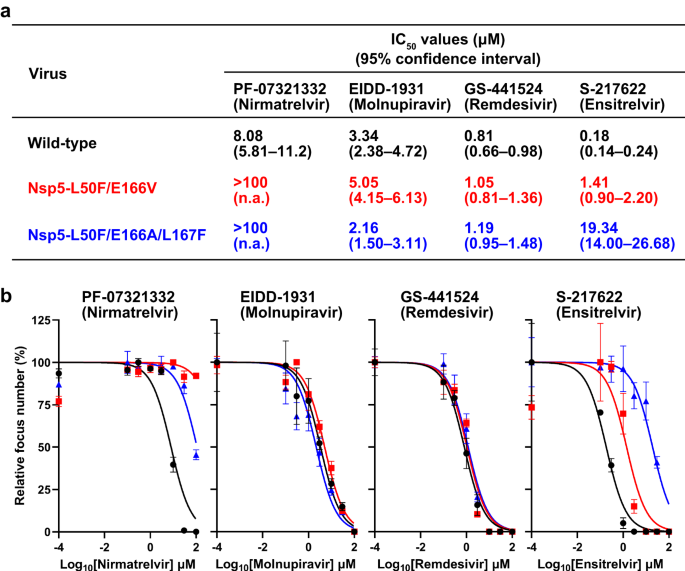
SARS-COV-2 VIRUSES TO ANTIVIRAL COMPOUNDS To characterize mutant viruses possessing amino acids mutations that are responsible for reduced sensitivity to nirmatrelvir, we prepared a
wild-type delta (B.1.617.2) variant and its mutant viruses possessing the L50F and E166V substitutions in Nsp5 (Nsp5-L50F/E166V) or the L50F, E166A, and L167F substitutions
(Nsp5-L50F/E166A/L167F) by using a BAC-based reverse genetics system. No nucleotide mixtures of more than 10% were identified in the three stock viruses. These three viruses were tested for
sensitivity to the antivirals PF-07321332 (nirmatrelvir), EIDD-1931 (molnupiravir), GS-441524 (remdesivir), and S-217622 (ensitrelvir) in a focus reduction assay (Fig. 1a and 1b). The IC50
value of PF-07321332 (nirmatrelvir) against wild-type virus was 8.08 μM (95% CI, 5.81–11.2 μM), whereas those against Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F were greater than 100 μM.
Although EIDD-1931 (molnupiravir) and GS-441524 (remdesivir) showed similar IC50 values against all three viruses tested, the sensitivity of Nsp5-L50F/E166V to S-217622 (ensitrelvir) was
moderately reduced and that of Nsp5-L50F/E166A/L167F was appreciably reduced; the IC50 values were 1.41 μM (95% CI, 0.90–2.20 μM) and 19.34 μM (95% CI, 14.00–26.68 μM). These results
indicate that both mutant viruses have reduced sensitivity to PF-07321332 (nirmatrelvir) and show reduced sensitivity to S-217622 (ensitrelvir) as well. PROPAGATION OF MUTANT VIRUSES IN THE
PRESENCE OR ABSENCE OF NIRMATRELVIR IN VITRO We evaluated the replicative ability of Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F in vitro. VeroE6/TMPRSS2 cells were infected with each virus at
an MOI of 0.001 and virus titers were determined at the indicated timepoints (Fig. 2). In the absence of nirmatrelvir, the growth of Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F was
significantly delayed compared with that of wild-type virus, but the maximum virus titers were similar (Fig. 2a). In the presence of nirmatrelvir, wild-type virus grew poorly, reaching only
3.8 × 104 PFU at 72 h post-infection, whereas Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F replicated similarly to in the absence of nirmatrelvir (Fig. 2b). These results indicate that these
amino acid substitutions that are responsible for resistance to nirmatrelvir decrease the efficiency of virus replication. PATHOGENICITY OF MUTANT VIRUSES IN HAMSTERS Next, we compared the
pathogenicity of Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F with that of wild-type virus. Hamsters were infected with these viruses and body weight, respiratory functions, and virus titers in
the nasal turbinate and lungs were measured at each timepoint. Mock-infected hamsters gradually gained body weight, whereas hamsters infected with wild-type virus lost body weight until 7
days post-infection (Fig. 3a). The body weight of the hamsters infected with Nsp5-L50F/E166V did not change until 6 days post-infection and then gradually increased. Infection with
Nsp5-L50F/E166A/L167F did not affect body weight, resulting in a similar body weight change as that seen with mock-infected hamsters. The respiratory function of the hamsters was assessed by
measuring Penh and Rpef, which are surrogate markers for bronchoconstriction and airway obstruction, respectively, by using a whole-body plethysmography system. Wild-type virus
significantly impaired Penh and Rpef at 5 dpi, whereas both mutant viruses moderately impaired in Penh and Rpef (Fig. 3b). We next compared virus titers in the nasal turbinates and lungs of
infected hamsters at 3 and 6 days post-infection. At 3 days post-infection, wild-type virus and both mutant viruses replicated in the nasal turbinates to a similar level, whereas the virus
titers in the lungs of hamsters infected with Nsp5-L50F/E166V or Nsp5-L50F/E166A/L167F were significantly lower than those of wild-type virus (Fig. 3c). At 6 days post-infection, virus
titers in the nasal turbinates of the mutant virus-infected groups were lower than those in the wild-type virus-infected group, although no significant difference in lung virus titer was
found among the three groups. Taken together, these data suggest that Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F are slightly attenuated compared with wild-type virus. TRANSMISSION OF
NSP5-L50F/E166V AND NSP5-L50F/E166A/L167F BETWEEN HAMSTERS We examined whether Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F maintain airborne transmissibility in hamsters. Naïve hamsters were
exposed to infected hamsters at one day after infection and virus titers were measured in the nasal turbinates and lungs of the infected and exposed hamsters at 4 days after infection or 3
days after exposure. The infected and exposed hamsters were separated by a double-layer partition to avoid direct contact. Among the infected hamsters, the virus titers for the wild-type,
Nsp5-L50F/E166V, and Nsp5-L50F/E166A/L167F were higher in the lungs than in the nasal turbinates (Fig. 4, infected). For the exposed hamsters, wild-type virus was detected in 100% (5/5) and
80% (4/5) of the nasal turbinates and lungs, respectively, indicating that wild-type virus was transmitted to all pairs tested (Fig. 4, exposed). Nsp5-L50F/E166V was detected in 60% (3/5)
and 20% (1/5) of the nasal turbinates and lungs, respectively, whereas Nsp5-L50F/E166A/L167F was detected in 80% (4/5) of both the nasal turbinates and lungs. The transmission efficiency was
not statistically different among the wild-type (100%), Nsp5-L50F/E166V (60%), and Nsp5-L50F/E166A/L167F (80%). These results suggest that Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F maintain
airborne transmissibility in the hamster model, although their efficiency may be slightly reduced. COMPETITIVE GROWTH OF MUTANT AND WILD-TYPE VIRUSES IN HAMSTERS We next examined the
competitive growth capability of each virus by co-infecting the wild-type virus with a mutant virus at a ratio of 3:7 based on virus infectious titers. The ratios of each mutant virus to
wild-type virus in the inoculum based on deep sequence analysis were 48:52 (wild-type: Nsp5-L50F/E166V) and 42:58 (wild-type: Nsp5-L50F/E166A/L167F) (top panels of Fig. 5). At 4 days
post-infection, the frequency of each virus in the nasal turbinates and lungs of the co-infected hamsters was determined. Wild-type virus clearly dominated in the lungs and nasal turbinates
of all five hamsters co-infected with wild-type virus and Nsp5-L50F/E166V. Similarly, the proportion of Nsp5-L50F/E166A/L167F relative to the wild-type virus was markedly decreased in the
nasal turbinates and lungs of all five animals, although Nsp5-L50F/E166A/L167F was present at 10 and 18% in the nasal turbinates of two hamsters. Next, we performed a coinfection experiment
in hamsters treated with nirmatrelvir. The ratios of each mutant virus to wild-type virus in the inoculum were 45:55 (wild-type: Nsp5-L50F/E166V) and 43:57 (wild-type: Nsp5-L50F/E166A/L167F)
(bottom panels of Fig. 5). Although the proportion of Nsp5-L50F/E166V or Nsp5-L50F/E166A/L167F was significantly increased in the nasal turbinates (_p_ = 0.00794 or _p_ = 0.00794) and lungs
((_p_ = 0.00794 or _p_ = 0.0476) of the treated hamsters compared to the untreated animals, the wild-type virus was predominant in most of the hamster nasal turbinates. However, in the
lungs of three hamsters, Nsp5-L50F/E166V became dominant, and in the lungs of one hamster, Nsp5-L50F/E166A/L167F became dominant. These results demonstrate that although Nsp5-L50F/E166V and
Nsp5-L50F/E166A/L167F are less fit than the wild-type virus, Nsp5-L50F/E166V could be problematic by dominating the wild-type virus in lungs in the presence of nirmatrelvir. DISCUSSION The
emergence of viruses resistant to anti-SARS-CoV-2 drugs is a public health concern. Indeed, monoclonal antibody therapies face increasing resistance from viruses with amino acid
substitutions in their spike protein. In contrast, antiviral compounds targeting virus proteins other than the spike protein are becoming more prevalent and important. However, amino acid
substitutions that reduce virus sensitivity to such compounds have been found in viruses isolated from COVID-19 patients8,9,10,11 and in laboratory experiments14,15,16,17,18,22,23. Here, we
attempted to characterize mutant viruses carrying amino acid substitutions linked to resistance to the oral antiviral nirmatrelvir, which targets 3CLpro (nsp5), since it is the antiviral
drug used most often to treat COVID-1918. Of the nirmatrelvir-resistant substitutions reported, we chose Nsp5-L50F/E166V19 and Nsp5-L50F/E166A/L167F20 because they appear to maintain virus
fitness in vitro. As expected, both mutant viruses reached a similar virus titer to that of the wild-type virus and were not inhibited by nirmatrelvir, although virus growth in cultured
cells was delayed. In the hamster infection model, the two resistant viruses were slightly attenuated, maintained airborne transmissibility with slightly reduced efficiency, and were unable
to compete with the growth of the wild-type virus in the absence of nirmatrelvir; however, although Nsp5-L50F/E166V was dominated by the wild-type virus in the nasal turbinate in the
presence of the drug, it dominated in three of five animals in the lungs. A previous study showed that Nsp5-L50F/E166A/L167F replicates in and damages lungs at similar levels to wild-type
virus and directly transmits from infected hamsters to naïve hamsters21. Thus, Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F show similar or slightly reduced pathogenicity in hamsters. However,
even though these nirmatrelvir-resistant mutants emerge, wild-type viruses outcompete them in the absence of nirmatrelvir, whereas in the presence of the drug, Nsp5-L50F/E166V may outcompete
the wild-type virus in the lungs to some extent. Viruses, especially RNA viruses, generate variants. Although SARS-CoV-2 has a proofreading mechanism, many people have been infected and
each individual producing a large amount of virus could lead to the emergence of viruses resistant to monoclonal antibodies or antivirals when used as monotherapy. In influenza patients,
viruses resistant to neuraminidase (NA) or PA inhibitors have been detected after treatment with the antivirals24. Some of these drug-resistant viruses are less fit than the wild-type
virus25, but others are not26,27,28. Regardless of whether viral fitness is reduced, such resistant viruses rarely spread worldwide. If a resistant virus does spread globally, either the
selective pressure is extremely high due to the widespread use of the antiviral (which is unlikely), or the resistant virus acquires greater fitness through the acquisition of additional
amino acid substitutions25,29. Combination therapy is thought to be a better strategy to treat viral infections and reduce the likelihood of emergence of resistant viruses30, but it is
possible for viruses resistant to both antivirals to emerge31,32. In conclusion, Nsp5-L50F/E166V and Nsp5-L50F/E166A/L167F are unlikely to dominate in nature. However, secondary adaptive
mutations may make these mutant viruses dominant to wild-type virus. Moreover, mutant viruses possessing other resistance-conferring substitutions could emerge and dominate in the human
population. Therefore, we need to closely monitor the amino acid mutations that confer resistance to nirmatrelvir and secondary adaptive mutations by undertaking active virus surveillance to
detect such viruses as soon as possible after emergence. METHODS ETHICS All animal experiments were conducted in accordance with the University of Tokyo’s Regulations for Animal Care and
Use, which were approved by the Animal Experiment Committee of the Institute of Medical Science, the University of Tokyo. The committee acknowledged and accepted both the legal and ethical
responsibility for the animals, as specified in the Fundamental Guidelines for Proper Conduct of Animal Experiment and Related Activities in Academic Research Institutions under the
jurisdiction of the Ministry of Education, Culture, Sports, Science, and Technology of Japan. All experiments with SARS-CoV-2 were performed in enhanced biosafety level 3 (BSL3) containment
laboratories at the University of Tokyo, which are approved for such use by the Ministry of Agriculture, Forestry, and Fisheries, Japan. CELLS VeroE6/TMPRSS2 (JCRB 1819) cells were
propagated in the presence of 1 mg/ml geneticin (G418; Invivogen) and 5 μg/ml plasmocin prophylactic (Invivogen) in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS.
VeroE6-TMPRSS2-T2A-ACE2 cells were cultured in DMEM containing 10% Fetal Bovine Serum (FBS), 100 U/mL penicillin–streptomycin, and 10 µg/mL puromycin. HEK293T cells were cultured in
Dulbecco’s modified Eagle medium supplemented with 10% FBS. All cells were maintained at 37 °C with 5% CO2. The cells were regularly tested for mycoplasma contamination by using PCR and
confirmed to be mycoplasma free. ANTIVIRALS Active components of remdesivir and molnupiravir (i.e., GS-441524 and EIDD-1931), and nirmatrelvir (PF-07321332) were purchased from
MedChemExpress. S-217622 (ensitrelvir) was kindly provided by Shionogi Co., Ltd. Compounds were dissolved in dimethyl sulfoxide (in vitro) or 0.5% methylcellulose (in vivo) prior to use.
VIRUSES The full-genome nucleotide sequence of SARS-CoV-2 (hCoV-19/USA/WI-UW-5250/2021, B.1.617.2) was assembled into the pBeloBAC11 vector to generate infectious cDNA clones under the
control of a cytomegalovirus (CMV) promoter as described previously33,34. The mutations responsible for the L50F/E166V and L50F/E166A/L167F substitutions in 3CLpro were introduced during the
PCR step. To rescue these viruses, pBeloBAC11 encoding the wild-type virus [hCoV-19/USA/WI-UW-5250/2021 (B.1.617.2, delta variant), GenBank Accession No. OR116091], Nsp5-L50F/E166V
(Accession No. OR116092), or Nsp5-L50F/E166A/L167F (Accession No. OR116093) was transfected into HEK293T cells. At 3 days post-transfection, the supernatant containing the viruses was
collected and inoculated onto VeroE6/TMPRSS2 at 37 °C to prepare virus stocks. The stock viruses were deep sequenced to confirm the absence of unwanted mutations, and no position contained
unwanted nucleotides that exceeded 10% of the population in all stock viruses. DEEP SEQUENCE ANALYSIS The whole genome of SARS-CoV-2 was amplified by using a modified ARTIC network protocol
in which some primers were replaced or added35. In brief, viral RNA was extracted by using a QIAamp Viral RNA Mini Kit (QIAGEN). cDNA was synthesized by using a Lunar-Script RT SuperMix Kit
(New England BioLabs) and subjected to a multiplexed PCR in two pools using ARTIC-N1 primers v5 and Q5 Hot Start DNA polymerase (New England BioLabs). The DNA libraries for Illumina NGS were
prepared from pooled amplicons by using a QIAseq FX DNA Library Kit (QIAGEN) and then analyzed by using the iSeq100 System in 150-bp paired-end mode using an iSeq 100 i1 Reagent v2
(300-cycle) kit (Illumina). The reads were assembled by CLC Genomics Workbench (version 22, Qiagen). The average coverage depths of the wild-type virus, L50F/E166V, and L50F/E166A/L167F were
8,078×, 10,742×, and 8,731×, respectively. FOCUS REDUCTION ASSAY Antiviral susceptibilities were determined by using a focus reduction assay as previously reported2,3,4,36. Briefly,
VeroE6-TMPRSS2-T2A-ACE2 cells in 96-well plates were infected with SARS-CoV-2 at 100–400 focus forming unit/well. After a 1-h incubation at 37 °C, the inoculum was replaced with 1% Methyl
Cellulose 400 (FUJIFILM Wako Pure Chemical Corporation) in culture medium containing serial dilutions of the antiviral compounds. The cells were incubated for 18 h at 37 °C and then fixed
with formalin. The cells were stained with a mouse monoclonal antibody against SARS-CoV-2 nucleoprotein, clone N45 (TAUNS Laboratories, Inc., Japan), followed by a horseradish
peroxidase-labeled goat anti-mouse immunoglobulin (Jackson ImmunoResearch Laboratories Inc.). Foci were visualized by using TrueBlue Substrate (SeraCare Life Sciences). The focus numbers
were quantified by using an ImmunoSpot S6 Analyzer, ImmunoCapture software, and BioSpot software (Cellular Technology). The 50% inhibitory concentration (IC50) values and 95% confidence
intervals were calculated by using GraphPad Prism 9.3.0 (GraphPad Software). GROWTH KINETICS IN VITRO VeroE6/TMPRSS2 cells were infected with the indicated virus at a multiplicity of
infection (MOI) of 0.001. After a 1-h incubation at 37 °C, the inoculum was replaced with medium ± 20 μM nirmatrelvir. Cell culture supernatants were collected at 8, 16, 24, 32, 40, 48, 56,
and 72 h post-infection. Virus titers were determined by the use of a plaque assay in VeroE6/TMPRSS2 cells. EXPERIMENTAL INFECTION OF SYRIAN HAMSTERS Five- to six-week-old male Syrian
hamsters (Japan SLC) were used in this study. Since SARS-CoV-2 occasionally fails to replicate in female hamsters, we used only male hamsters in this study. Under isoflurane anesthesia, five
hamsters per group were intranasally inoculated with 105 plaque forming unit (PFU) of the indicated virus. Body weights were measured daily before inoculation and 10 days after inoculation.
Respiratory parameters [Penh (a nonspecific assessment of breathing patterns) and Rpef (a measure of airway obstruction)] were also measured by using a whole-body plethysmography system
(PrimeBioscience) as previously described37,38. For the virus titration, five hamsters per group were intranasally infected with 105 PFU of the indicated virus. At 3 and 6 days
post-infection, the animals were euthanized and nasal turbinates and lungs were collected. The virus titers in these organs were determined by use of plaque assays on VeroE6/TMPRSS2 cells.
The transmission study was performed as previously described39,40. Briefly, five hamsters per group were intranasally infected with 105 PFU of each indicated virus and placed in cages. One
day after infection, naïve hamsters were placed in adjacent cages, 5-cm apart, separated by a double-layer partition with unidirectional air flow. Infected and exposed hamsters were
euthanized at 4 days post-infection or 3 days after exposure. Nasal turbinates and lungs were collected for virus titration on VeroE6/TMPRSS2 cells. For the co-infection study, wild-type
virus and Nsp5-L50F/E166V or Nsp5-L50F/E166A/L167F were mixed at a ratio of 3:7 based on their titers, and the virus mixture (total 2 × 105 PFU) was inoculated into ten hamsters per group.
One day after inoculation, five hamsters per group were treated orally with nirmatrelvir at 250 mg/kg twice daily as previously described41,42. The remaining five hamsters per group were
left untreated. The animals were euthanized at 4 dpi and their nasal turbinates and lungs were collected for deep sequence analysis. The ratio of wild-type to Nsp5-L50F/E166V or
Nsp5-L50F/E166A/L167F was calculated from the differences at positions 10202 and 10551 or 10202, 10551, and 10555, respectively. Samples with read depths of more than 1000 were analyzed.
STATISTICAL ANALYSIS GraphPad Prism software version 9.3.0 was used to calculate _P_ values. We compared virus titers in hamsters with the control by using a one-way ANOVA followed by
Tukey’s multiple comparisons. Body weight and virus growth kinetics were compared by using a two-way ANOVA followed by Tukey’s multiple comparisons. Transmission efficiency was compared by
using a two-sided Fisher’s exact test. The frequency of wild-type virus in the nasal turbinate and lungs of the treated hamsters was compared with that of the untreated hamsters by using the
Mann–Whitney test followed by the two-stage step-up procedure of the Benjamini, Krieger, and Yekutieli test. Differences between groups were considered significant for _P_ values < 0.05.
REAGENT AVAILABILITY All reagents described in this paper are available through Material Transfer Agreements. REPORTING SUMMARY Further information on research design is available in the
Nature Portfolio Reporting Summary linked to this article. DATA AVAILABILITY All data supporting the findings of this study are available within the paper and are provided in the Source data
file. There are no restrictions to obtaining access to the primary data. The virus genome sequences of the wild-type virus, Nsp5-L50F/E166V, and Nsp5-L50F/E166A/L167F are available in
GenBank (Accession Nos. OR116091, OR116092, and OR116093, respectively). CODE AVAILABILITY No code was used during the data acquisition or analysis. REFERENCES * Callaway, E. Heavily mutated
Omicron variant puts scientists on alert. _Nature_ 600, 21 (2021). Article ADS CAS PubMed Google Scholar * Takashita, E. et al. Efficacy of antiviral agents against the Omicron
subvariant BA.2.75. _N. Engl. J. Med._ 387, 1236–1238 (2022). Article PubMed Google Scholar * Takashita, E. et al. In vitro efficacy of antiviral agents against Omicron subvariant BA.4.6.
_N. Engl. J. Med._ 387, 2094–2097 (2022). Article PubMed Google Scholar * Takashita, E. et al. Efficacy of antibodies and antiviral drugs against Omicron BA.2.12.1, BA.4, and BA.5
subvariants. _N. Engl. J. Med._ 387, 468–470 (2022). Article PubMed Google Scholar * Takashita, E. et al. Efficacy of antiviral agents against the SARS-CoV-2 Omicron subvariant BA.2. _N.
Engl. J. Med._ 386, 1475–1477 (2022). Article PubMed Google Scholar * Takashita, E. et al. Efficacy of antibodies and antiviral drugs against Covid-19 Omicron variant. _N. Engl. J. Med._
386, 995–998 (2022). Article PubMed Google Scholar * Imai, M. et al. Efficacy of antiviral agents against Omicron subvariants BQ.1.1 and XBB. _N. Engl. J. Med._ 388, 89–91 (2023). Article
PubMed Google Scholar * Gandhi, S. et al. De novo emergence of a remdesivir resistance mutation during treatment of persistent SARS-CoV-2 infection in an immunocompromised patient: a
case report. _Nat. Commun._ 13, 1547 (2022). Article ADS CAS PubMed PubMed Central Google Scholar * Focosi, D., Maggi, F., McConnell, S. & Casadevall, A. Very low levels of
remdesivir resistance in SARS-COV-2 genomes after 18 months of massive usage during the COVID19 pandemic: A GISAID exploratory analysis. _Antiviral Res_ 198, 105247 (2022). Article CAS
PubMed PubMed Central Google Scholar * Hu, Y. et al. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. _bioRxiv_,
https://doi.org/10.1101/2022.06.28.497978 (2022). * Lee, J. T. et al. Genetic surveillance of SARS-CoV-2 M(pro) reveals high sequence and structural conservation prior to the introduction of
protease inhibitor paxlovid. _mBio_ 13, e0086922 (2022). Article PubMed Google Scholar * Owen, D. R. et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of
COVID-19. _Science_ 374, 1586–1593 (2021). Article ADS CAS PubMed Google Scholar * Mahase, E. Covid-19: Pfizer’s paxlovid is 89% effective in patients at risk of serious illness,
company reports. _BMJ_ 375, n2713 (2021). Article PubMed Google Scholar * Sasi, V. M. et al. Predicting antiviral resistance mutations in SARS-CoV-2 main protease with computational and
experimental screening. _Biochemistry_ 61, 2495–2505 (2022). Article CAS PubMed Google Scholar * Heilmann, E. et al. SARS-CoV-2 3CL(pro) mutations selected in a VSV-based system confer
resistance to nirmatrelvir, ensitrelvir, and GC376. _Sci. Transl. Med._ 15, eabq7360 (2023). Article CAS PubMed Google Scholar * Moghadasi, S. A. _et al_. Transmissible SARS-CoV-2
variants with resistance to clinical protease inhibitors. _bioRxiv_, https://doi.org/10.1101/2022.08.07.503099 (2022). * Iketani, S. et al. Functional map of SARS-CoV-2 3CL protease reveals
tolerant and immutable sites. _Cell Host Microbe_ 30, 1354–1362.e1356 (2022). Article CAS PubMed PubMed Central Google Scholar * Iketani, S. et al. Multiple pathways for SARS-CoV-2
resistance to nirmatrelvir. _Nature_ 613, 558–564 (2023). Article ADS CAS PubMed Google Scholar * Zhou, Y. et al. Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an
infectious cell culture system. _Sci. Adv._ 8, eadd7197 (2022). Article CAS PubMed PubMed Central Google Scholar * Jochmans, D. et al. The Substitutions L50F, E166A, and L167F in
SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir. _mBio_ 14, e0281522 (2023). Article PubMed Google Scholar * Abdelnabi, R. et al.
Nirmatrelvir-resistant SARS-CoV-2 is efficiently transmitted in Syrian hamsters. _bioRxiv_, https://doi.org/10.1101/2022.09.28.509903 (2022). * Checkmahomed, L. et al. In vitro selection of
remdesivir-resistant SARS-CoV-2 demonstrates high barrier to resistance. _Antimicrob. Agents Chemother._ 66, e0019822 (2022). Article PubMed Google Scholar * Stevens, L. J. et al.
Mutations in the SARS-CoV-2 RNA-dependent RNA polymerase confer resistance to remdesivir by distinct mechanisms. _Sci. Transl. Med._ 14, eabo0718 (2022). Article CAS PubMed Google Scholar
* Soga, T. et al. Characterization of influenza A(H1N1)pdm09 viruses isolated in the 2018-2019 and 2019-2020 influenza seasons in Japan. _Viruses_ 15, https://doi.org/10.3390/v15020535
(2023). * Herlocher, M. L. et al. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. _J. Infect. Dis._ 190, 1627–1630 (2004). Article CAS
PubMed Google Scholar * Imai, M. et al. Influenza A variants with reduced susceptibility to baloxavir isolated from Japanese patients are fit and transmit through respiratory droplets.
_Nat. Microbiol._ 5, 27–33 (2020). Article CAS PubMed Google Scholar * Farrukee, R. et al. Characterization of influenza B virus variants with reduced neuraminidase inhibitor
susceptibility. _Antimicrob. Agents Chemother._ 62, https://doi.org/10.1128/AAC.01081-18 (2018). * Kwon, J. J. et al. An I436N substitution confers resistance of influenza A(H1N1)pdm09
viruses to multiple neuraminidase inhibitors without affecting viral fitness. _J. Gen. Virol._ 99, 292–302 (2018). Article CAS PubMed Google Scholar * Wang, M. Z., Tai, C. Y. &
Mendel, D. B. Mechanism by which mutations at his274 alter sensitivity of influenza a virus n1 neuraminidase to oseltamivir carboxylate and zanamivir. _Antimicrob. Agents Chemother._ 46,
3809–3816 (2002). Article CAS PubMed PubMed Central Google Scholar * Poland, G. A., Jacobson, R. M. & Ovsyannikova, I. G. Influenza virus resistance to antiviral agents: a plea for
rational use. _Clin. Infect. Dis._ 48, 1254–1256 (2009). Article PubMed Google Scholar * Kumar, D. et al. Combining baloxavir marboxil with standard-of-care neuraminidase inhibitor in
patients hospitalised with severe influenza (FLAGSTONE): a randomised, parallel-group, double-blind, placebo-controlled, superiority trial. _Lancet Infect. Dis._ 22, 718–730 (2022). Article
CAS PubMed Google Scholar * Chen, Q. et al. Deep-sequencing reveals broad subtype-specific HCV resistance mutations associated with treatment failure. _Antiviral Res._ 174, 104694
(2020). Article CAS PubMed Google Scholar * He, X. et al. Generation of SARS-CoV-2 reporter replicon for high-throughput antiviral screening and testing. _Proc. Natl Acad. Sci. USA_ 118,
e2025866118 (2021). Article CAS PubMed PubMed Central Google Scholar * Furusawa, Y., Yamayoshi, S. & Kawaoka, Y. The accuracy of reverse genetics systems for SARS‐CoV‐2: Circular
polymerase extension reaction versus bacterial artificial chromosome. _Influenza Respiratory Viruses_ 17, e13109 (2023). Article CAS Google Scholar * Itokawa, K., Sekizuka, T., Hashino,
M., Tanaka, R. & Kuroda, M. Disentangling primer interactions improves SARS-CoV-2 genome sequencing by multiplex tiling PCR. _PLoS ONE_ 15, e0239403 (2020). Article CAS PubMed PubMed
Central Google Scholar * Vanderheiden, A. et al. Development of a rapid focus reduction neutralization test assay for measuring SARS-CoV-2 neutralizing antibodies. _Curr Protoc Immunol_
131, e116 (2020). CAS PubMed PubMed Central Google Scholar * Imai, M. et al. Characterization of a new SARS-CoV-2 variant that emerged in Brazil. _Proc. Natl Acad. Sci. USA_ 118,
https://doi.org/10.1073/pnas.2106535118 (2021). * Uraki, R. et al. Characterization of SARS-CoV-2 Omicron BA.4 and BA.5 isolates in rodents. _Nature_ 612, 540–545 (2022). Article ADS CAS
PubMed Google Scholar * Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. _Science_ 370, 1464–1468 (2020). Article ADS CAS
PubMed PubMed Central Google Scholar * Iwatsuki-Horimoto, K. et al. Syrian hamster as an animal model for the study of human influenza virus infection. _J. Virol._ 92, e01693–01617
(2018). Article CAS PubMed PubMed Central Google Scholar * Kuroda, T. et al. Efficacy comparison of 3CL protease inhibitors ensitrelvir and nirmatrelvir against SARS-CoV-2 in vitro and
in vivo. _J. Antimicrob. Chemother_, https://doi.org/10.1093/jac/dkad027 (2023). * Abdelnabi, R. et al. The oral protease inhibitor (PF-07321332) protects Syrian hamsters against infection
with SARS-CoV-2 variants of concern. _Nat. Commun._ 13, 719 (2022). Article ADS CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS The authors thank Susan
Watson for editing the manuscript. FUNDING This work was supported by the Japan Agency for Medical Research and Development (JP22wm0125002 and JP223fa627001). AUTHOR INFORMATION AUTHORS AND
AFFILIATIONS * Division of Virology, Institute of Medical Science, University of Tokyo, Tokyo, Japan Maki Kiso, Yuri Furusawa, Ryuta Uraki, Masaki Imai, Seiya Yamayoshi & Yoshihiro
Kawaoka * The Research Center for Global Viral Diseases, National Center for Global Health and Medicine Research Institute, Tokyo, Japan Yuri Furusawa, Ryuta Uraki, Masaki Imai, Seiya
Yamayoshi & Yoshihiro Kawaoka * International Research Center for Infectious Diseases, Institute of Medical Science, University of Tokyo, Tokyo, Japan Masaki Imai & Seiya Yamayoshi *
The University of Tokyo Pandemic Preparedness, Infection and Advanced Research Center, Tokyo, Japan Yoshihiro Kawaoka * Department of Pathobiological Sciences, School of Veterinary
Medicine, University of Wisconsin–Madison, Madison, USA Yoshihiro Kawaoka Authors * Maki Kiso View author publications You can also search for this author inPubMed Google Scholar * Yuri
Furusawa View author publications You can also search for this author inPubMed Google Scholar * Ryuta Uraki View author publications You can also search for this author inPubMed Google
Scholar * Masaki Imai View author publications You can also search for this author inPubMed Google Scholar * Seiya Yamayoshi View author publications You can also search for this author
inPubMed Google Scholar * Yoshihiro Kawaoka View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS M.K., S.Y., and Y.K. designed the study. M.K.,
Y.F., R.U., M.I., and S.Y. performed the experiments. M.K., S.Y., and Y.K. analyzed the data and wrote the manuscript. All authors reviewed the manuscript and approved the final version.
CORRESPONDING AUTHORS Correspondence to Seiya Yamayoshi or Yoshihiro Kawaoka. ETHICS DECLARATIONS COMPETING INTERESTS Y.K. has received unrelated funding support from Daiichi Sankyo
Pharmaceutical, Toyama Chemical, Tauns Laboratories, Inc., Shionogi & Co. LTD, Otsuka Pharmaceutical, and KM Biologics. The other authors have no conflicts of interest. PEER REVIEW PEER
REVIEW INFORMATION _Nature Communications_ thanks Josep Quer, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION
REPORTING SUMMARY PEER REVIEW FILE SOUCE DATA FILE RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits
use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the
Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated
otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds
the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Kiso, M., Furusawa, Y., Uraki, R. _et al._ In vitro and in vivo characterization of SARS-CoV-2 strains resistant to nirmatrelvir. _Nat
Commun_ 14, 3952 (2023). https://doi.org/10.1038/s41467-023-39704-x Download citation * Received: 08 January 2023 * Accepted: 20 June 2023 * Published: 04 July 2023 * DOI:
https://doi.org/10.1038/s41467-023-39704-x SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative