- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT In plants, guanosine monophosphate (GMP) is synthesized from adenosine monophosphate via inosine monophosphate and xanthosine monophosphate (XMP) in the cytosol. It has been shown
recently that the catabolic route for adenylate-derived nucleotides bifurcates at XMP from this biosynthetic route. Dephosphorylation of XMP and GMP by as yet unknown phosphatases can
initiate cytosolic purine nucleotide catabolism. Here we show that _Arabidopsis thaliana_ possesses a highly XMP-specific phosphatase (XMPP) which is conserved in vascular plants. We
demonstrate that XMPP catalyzes the irreversible entry reaction of adenylate-derived nucleotides into purine nucleotide catabolism in vivo, whereas the guanylates enter catabolism via an
unidentified GMP phosphatase and guanosine deaminase which are important to maintain purine nucleotide homeostasis. We also present a crystal structure and mutational analysis of XMPP
providing a rationale for its exceptionally high substrate specificity, which is likely required for the efficient catalysis of the very small XMP pool in vivo. SIMILAR CONTENT BEING VIEWED
BY OTHERS MECHANISMS OF FEEDBACK INHIBITION AND SEQUENTIAL FIRING OF ACTIVE SITES IN PLANT ASPARTATE TRANSCARBAMOYLASE Article Open access 11 February 2021 PLASTID ANCESTORS LACKED A
COMPLETE ENTNER-DOUDOROFF PATHWAY, LIMITING PLANTS TO GLYCOLYSIS AND THE PENTOSE PHOSPHATE PATHWAY Article Open access 06 February 2024 PLANT ADENYLATE CYCLASES HAVE COME FULL CIRCLE Article
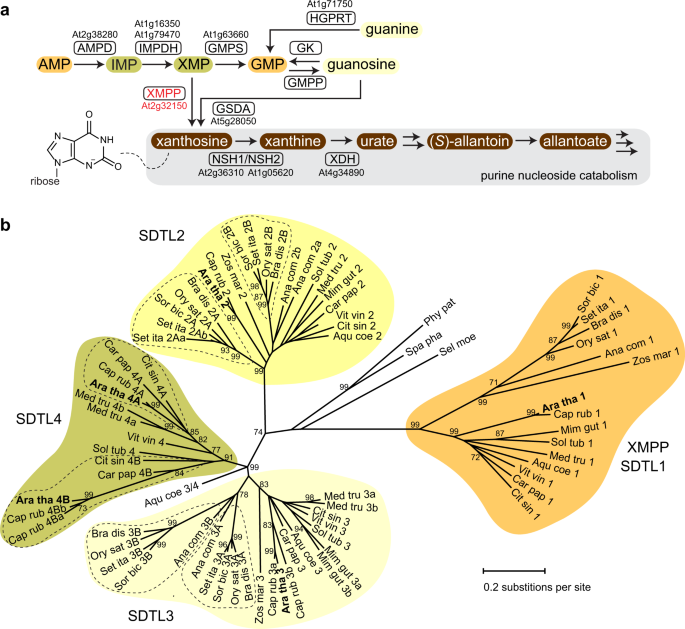
14 September 2023 INTRODUCTION Purine metabolism in plants and other eukaryotes differs in several aspects. For example, in plants, purine nucleotide biosynthesis generating AMP is
localized in chloroplasts, whereas in mammals or in yeasts it is located in the cytosol. However, plant GMP biosynthesis, starting from AMP, occurs in the cytosol (Fig. 1a and Supplementary
Fig. 1)1,2. In contrast to mammals, plants are able to degrade purine nucleotides completely, disintegrating the purine ring via intermediates like urate and allantoate into glyoxylate,
carbon dioxide and ammonium3,4. Recently, it was shown in _Arabidopsis thaliana_ that there are two independent entry reactions into purine nucleotide catabolism5. One is leading from
xanthosine monophosphate (XMP) to xanthosine presumably catalyzed by an XMP phosphatase (XMPP). This reaction may serve to degrade the adenylate-derived purine nucleotides. The other also
generates xanthosine but via the dephosphorylation of GMP to guanosine by a so far unknown GMP phosphatase (GMPP) and the subsequent deamination of guanosine by guanosine deaminase (GSDA),
an enzyme found only in plants (Fig. 1a and Supplementary Fig. 1)6. Because xanthosine cannot be converted into other nucleosides and cannot be re-phosphorylated (salvaged) to XMP in
Arabidopsis7,8, the XMPP and GSDA reactions both lead irreversibly into purine catabolism. Although several enzymes with nucleotide monophosphate (NMP) phosphatase activity have been
described in eukaryotes, there are only few examples where their physiological role could be clearly demonstrated9,10. Why is the functional assignment of NMP phosphatases so difficult?
Possible reasons are (i) that NMPs are central metabolites and their pool sizes are affected by a multitude of reactions so that single mutations hardly change the pools at all; (ii)
eukaryotes have many enzymes with NMP phosphatase activity often with a broad substrate spectrum complicating the assignment of specific physiological roles; (iii) NMP phosphatases may act
partially redundant in vivo. In this work, we were able to identify a plant-specific NMP phosphatase, the XMP phosphatase. We have characterized the enzyme biochemically and report its
crystal structure with bound substrate elucidating its specificity determinants. Most importantly, we demonstrate that XMPP is involved in purine nucleotide catabolism in vivo. RESULTS AND
DISCUSSION IDENTIFICATION AND MOLECULAR CHARACTERIZATION OF XMPP In _Saccharomyces cerevisiae_ two homologous enzymes, Sdt1p (Suppressor of disruption of _TFIIS_) and Phm8p (Phosphate
metabolism protein 8), were shown to hydrolyze NMPs in vitro. Phm8p but not Sdt1p is required in vivo for purine and pyrimidine mononucleotide degradation10. Because Phm8p also shows
activity with XMP and GMP, we searched for genes in the Arabidopsis genome encoding homologous proteins to Phm8p/Sdt1p with the aim to identify XMP- and GMP-specific plant phosphatases (Fig.
1a and Supplementary Fig. 1). Five proteins homologous to Phm8p/Sdt1p were found which we called SDTL for Sdt1p-like as they were more similar to Sdt1p than to Phm8p. Because enzymes of
primary metabolism are usually highly conserved in plants, we assumed that only SDTLs that are present in all vascular plants are good XMP/GMP phosphatase candidates. To identify such SDTLs,
a maximum likelihood tree was constructed using the full complement of SDTL sequences from 15 phylogenetically distant vascular plants and 3 moss species (Fig. 1b and Supplementary Table
1). Four major clades of SDTL proteins were found. The clades SDTL1, SDTL2, and SDTL3 each contain at least one representative sequence from every analyzed vascular plant species, whereas
the SDTL4 clade lacks sequences from several plant species, in particular from the monocots. Each of the mosses only has a single SDTL not associated with any of the clades. The clades
likely represent groups of orthologous proteins with identical or at least similar functions. The SDTL1 clade is more distant from the others and contains only a single protein from each of
the analyzed vascular plants, whereas in other clades also sub-clades are found indicating some functional diversification. We transiently expressed cDNAs coding for C-terminal Strep-tagged
variants of the SDTL proteins from the clades 1, 2, and 3 of Arabidopsis in leaves of _Nicotiana benthamiana_, affinity purified the proteins, and screened their activity with XMP and GMP as
substrates. GMP phosphatase activity was not observed for any of the enzymes but SDTL from clade 1 showed activity with XMP. We extended the substrate survey for this enzyme to other
mononucleotides and found that it is highly specific for XMP (Fig. 2a). Such high specificity is unusual for NMP phosphatases. The enzyme has a _K_M value of 3.9 ± 0.2 μM and a _k_cat value
of 9.2 ± 0.1 s−1 for XMP (Fig. 2b). An N-terminal Strep-tagged variant has similar kinetic constants (Supplementary Fig. 2) indicating that the tags and the tag position do not interfere
with the activity. Based on these data, we named the enzyme XMP phosphatase (XMPP). The subcellular localization of XMPP was investigated by confocal laser scanning microscopy in the leaves
of _N. benthamiana_ transiently expressing the enzyme as C-terminal yellow fluorescent protein (YFP)-tagged variant. XMPP is present in the cytosol and the nucleus (Fig. 2c) as are other
enzymes upstream and downstream of XMPP, i.e. IMP dehydrogenase (IMPDH), GMP synthetase (GMPS)2, guanosine deaminase (GSDA)6, and the nucleoside hydrolase heterocomplex (NSH1/NSH2)11,12,13
(Fig. 1a). THE FUNCTION OF XMPP IN PURINE NUCLEOTIDE CATABOLISM IN VIVO To investigate whether XMPP is involved in degrading XMP in vivo, we used a series of single (_xmpp, nsh1, gsda_),
double (_xmpp nsh1_, _xmpp gsda, nsh1 gsda_), and triple (_xmpp nsh1 gsda_) mutants as well as two complementation lines containing a _TwinStrep-XMPP_ transgene driven from the native
promoter (Supplementary Fig. 3). The pools of XMP, xanthosine, guanosine, and guanine in these genetic variants were quantified in fresh seeds, because it is known that purine catabolism is
active during seed development. The XMP concentration was close to the detection limit in most genotypes but was slightly elevated in the _xmpp_ line (Fig. 2d). In _nsh1_ seeds, more XMP was
detected and in the _xmpp nsh1_ seeds the largest XMP pool was observed, suggesting that XMPP is involved in XMP catalysis in vivo. The absence of NSH1, which plays a central role for
xanthosine hydrolysis5, has a positive effect on the XMP concentration possibly because the strong accumulation of xanthosine in _nsh1_ seeds5,11,13 (Fig. 2d) leads to a partial inhibition
of the two XMP converting enzymes: XMPP and GMPS (Fig. 1a). For XMPP, we confirmed an inhibition by xanthosine (Supplementary Fig. 4). XMP accumulation in _xmpp_ seeds is comparatively low
probably because XMP is easily aminated by GMPS to GMP, which can be degraded by an unknown GMP phosphatase (GMPP) and GSDA via guanosine to xanthosine (Fig. 1a). This hypothesis is
supported by hyperaccumulation of guanosine and guanine in seeds with _xmpp gsda_ background compared to _gsda_ seeds (Supplementary Fig. 5). GMP can also be phosphorylated leading in
tendency to GTP hyperaccumulation in this double mutant background, which is prevented and even over-compensated by the introduction of an XMPP transgene (Supplementary Fig. 6). It has been
noted before that guanosine accumulation in _gsda_ plants results in higher concentrations of guanylates and in consequence adenylates14. The GMPP and GSDA reactions seem to guard guanylate
homeostasis. Xanthosine is the first common product of direct XMP dephosphorylation by XMPP and of GMP degradation by GMPP and GSDA (Fig. 1a). The xanthosine pool size in _nsh1_ background
may therefore serve as a proxy for assessing if both routes are operative. Whereas in _nsh1 gsda_ seeds the xanthosine concentration is reduced strongly compared to the _nsh1_ background,
this is not the case in _xmpp nsh1_ seeds (Fig. 2d), suggesting at first sight that the XMPP route plays no role. However, one needs to bear in mind that XMP can easily be converted to GMP
by GMPS, especially if XMPP is not functional, thus only in _xmpp nsh1 gsda_ seeds one can assess whether XMPP contributes to the xanthosine pool in vivo. This is indeed the case because in
the triple mutant the xanthosine concentration is strongly reduced and this effect is reversed if the triple mutant expresses an _XMPP_ transgene. These data demonstrate that XMPP catalyzes
the dephosphorylation of XMP in seeds representing an entry point of the adenylate-derived nucleotides into purine catabolism in vivo. The XMPP reaction is likely the main entry point for
the adenylates into catabolism, whereas the GMPP reaction serves for the degradation of the guanylates. Note, that xanthosine cannot be salvaged in Arabidopsis whereas guanosine can8—thus
the XMPP reaction leads irreversibly into purine nucleotide degradation. Purine catabolism is enhanced by prolonged darkness5,15. Therefore, we assessed the role of XMPP in seedlings exposed
to a night prolonged by 48 h in comparison to seedlings in a 16 h day/8 h night regime. Dark treatment led to a marked increase of the steady-state pool sizes of urate and allantoate (Fig.
3a), which are intermediates of purine nucleoside catabolism4,16 (Fig. 1a and Supplementary Fig. 1). In _gsda_ seedlings under long-day conditions, the urate pool was already smaller than in
the wild type or in _xmpp_ seedlings, while in _xmpp gsda_ plants, the urate concentration was even more reduced and allantoate could not be detected anymore. In the prolonged night, urate
and allantoate concentrations rose but not in the _xmpp gsda_ seedlings, demonstrating that the degradation of XMP by XMPP and of GMP via GSDA feeds these pools. The dark-exposure experiment
was repeated including a line expressing an _XMPP_ transgene in _xmpp gsda_ background (Supplementary Fig. 3c), which complemented metabolic changes observed in the _xmpp gsda_ line
compared with the _gsda_ line (Supplementary Fig. 7). Together these results show that XMP and GMP dephosphorylation can initiate cytosolic purine nucleotide catabolism, but it is not
possible to quantify the relative contributions of XMP versus GMP degradation from the data. Reasons are that the XMPP reaction can be bypassed in _xmpp_ background via GMP and guanosine
(Fig. 1a), and that in _gsda_ seedlings guanosine accumulates so strongly that it partially inhibits the NSH1/NSH2 complex, which leads to xanthosine buildup in the dark (Fig. 3a and
Supplementary Fig. 7), and consequently to partial inhibition of XMPP (Supplementary Fig. 4). The dark-induced xanthosine accumulation in _gsda_ background is not observed in _xmpp gsda_
seedlings, again showing that the XMPP reaction directly contributes to the xanthosine pool in vivo. Because XMP cannot be channeled into degradation in _xmpp gsda_ plants, these double
mutants accumulate more guanosine than _gsda_ seedlings in the dark (Fig. 3a). In an independent experiment we observed that the _xmpp gsda_ plants also over-accumulate GTP and in
consequence ATP in tendency, which is prevented by the introduction of an _XMPP_ transgene (Fig. 3b and Supplementary Fig. 8). These tendencies were repeatedly observed in the long day and
in the prolonged night. It is clear that GSDA, probably in concert with the unknown GMPP, is necessary to keep guanylate and adenylate homeostasis, and that XMPP prevents adenylate-derived
nucleotides destined for degradation to spill into the guanylate pool. The mutation of _XMPP_ does not create an obvious macroscopic phenotype at any developmental stage (Supplementary Fig.
9) and also not during dark stress. This is not surprising because (1) many purine catabolism mutants of Arabidopsis do not show phenotypes under standard growth conditions except those that
accumulate toxic intermediates like guanosine15 or uric acid16 and because (2) it is possible to bypass the XMPP reaction via GMP and guanosine as shown above. However, this bypass is
energetically costly, because the GMPS reaction requires the conversion of ATP to AMP and uses glutamine for the amination of XMP. Subsequently, this amino group is released as ammonia by
GSDA and must be re-assimilated into glutamine which is energized by the conversion of ATP to ADP. Thus in total the bypass requires the hydrolysis of three phosphoanhydride bonds that are
spared when the XMPP reaction is used. THE MOLECULAR BASIS OF SUBSTRATE RECOGNITION BY XMPP XMPP is highly substrate specific (Fig. 2a). To elucidate the molecular basis for this
specificity, we determined the crystal structure of XMPP with and without XMP bound (Fig. 4, Supplementary Fig. 10a and Supplementary Table 2). Crystals could only be obtained for a
shortened XMPP variant lacking 13 amino acids at the C-terminus (Met1 to Ser250). Consistent with the monomeric state of the enzyme in solution (Supplementary Fig. 10b), crystalized XMPP is
monomeric with a two-domain architecture (Fig. 4a). In the α/β-folded central domain with five repeating β-α units, the central parallel six-stranded β-sheet is flanked by five α-helices on
both sides. A 20-residue-long loop (Pro142 to Ser161) protrudes from the β3–α8 unit, and a cap domain (Leu20 to Ser89) with a four α-helices bundle-like architecture is inserted in the β1–α5
unit. The loop probably stabilizes the orientation of the cap domain (Supplementary Fig. 11). XMPP is a member of the Haloacid Dehalogenase (HAD) protein superfamily with unique
characteristics since HAD enzymes typically have either a protruding loop or a cap structure17, but not both, like XMPP. At the junction of the cap and the central domains, a Mg2+ ion
coordinates six ligands within 2.1 Å in a square bi-pyramidal geometry (Fig. 4b) involving Asp12 and Asp14. These aspartates are conserved (Supplementary Fig. 10a) and important for the
catalytic activity of HAD enzymes. In the complex with XMP, the 5′-monophosphate moiety of XMP provides an equatorial ligand across Asp184 in the Mg2+-coordination shell, and the ribose and
xanthine moieties face toward the cap domain (Fig. 4c, d and Supplementary Fig. 12). Unlike the ribose moiety, xanthine embedded on the concave side of the cap domain interacts extensively
with the enzyme via (i) hydrophobic interactions with Val28 and Ile32 and (ii) by base stacking with Phe55, and (iii) via direct or water-mediated indirect hydrogen bonds of O2 with Thr61,
O6 indirectly with Arg51, and N3 and N7 indirectly with Tyr77 and Lys29, respectively (Fig. 4d). To validate the structure-based functional assignments, enzymatic activities and kinetic
constants of XMPP variants were determined. The specific activities of the XMPP variants closely reflect the assigned functional roles of the respective residues (Fig. 4d, e).
Mg2+-coordination by Asp12, Asp183, and Asp184 is linked to catalysis and is of critical importance since the respective mutants are inactive. The interaction of Asn188 with the phosphate
and of various residues with the xanthine moiety are required for full activity (Fig. 4e). Further kinetic analysis revealed that mutations of xanthine-interacting residues do not affect the
_k_cat but the _K_M values for XMP (Fig. 4f and Supplementary Fig. 13). For variants altered in hydrophobic residues, the _K_M increases 2.2- to 26.9-fold. For those changed in hydrogen
bond-forming residues, the _K_M increase ranges from 3.6- to 9.9-fold. The XMP specificity is thus collectively mediated by the hydrophobic residues Val28, Ile31, and Phe55 that shape the
xanthine-binding pocket in the cap domain and by direct and indirect hydrogen bonds to Thr61, Lys29, and Arg51. These amino acids are largely conserved in putative XMPPs from other plants
(Supplementary Fig. 10a) but there are some slight variations. Thr61, for example, which forms a direct hydrogen bond to the characteristic O2 of xanthine can be replaced by serine in
enzymes from other plants. We were unable to detect XMP in seedling extracts but estimate that its concentration must be at least three orders of magnitude smaller than the GMP
concentration. The high specificity of XMPP together with a _K_M in the low micromolar range and a high _k_cat are therefore of physiological importance, because a less specific enzyme would
constantly dephosphorylate other more abundant NMPs. Additionally, a precise adjustment of the XMPP amount is apparently required because even a moderate overexpression can strongly disturb
GTP homeostasis (Supplementary Fig. 6). It will be interesting to investigate how this enzyme at the crossroad between nucleotide biosynthesis and degradation is regulated and coordinated
with the activity of GMP synthetase to adjust the flux of XMP towards the guanylates or into catabolism. METHODS PLANT MATERIAL AND CULTIVATION _Arabidopsis thaliana_ ecotype Columbia-0 was
used throughout. The T-DNA mutants _xmpp-1_ (SALK067037) and _xmpp-2_ (SALK131244, At2g32150), _nsh1_ (SALK083120, At2g36310)11, and _gsda_ (GK432D08, At5g28050)6 were obtained from the
SALK18 and GABI-Kat19 collections. Seedlings of _xmpp-1_ and _xmpp-2_ were tested for the presence of intact transcript using the primers N188 and N233 (Supplementary Table 3). The presence
of _Actin2_ (At3g18780) transcript was tested with the primers 1033 and 1034. The mutants _xmpp-_1, _nsh1_, and _gsda_ were used to generate double and triple mutants by crossing.
Complementation lines of _xmpp gsda_ and _xmpp nsh1 gsda_ were generated by transformation with construct H453 (Supplementary Fig. 3c,d). Arabidopsis and _N. benthamiana_ were cultivated
under long-day conditions (16 h light of 85 μmol m−2 s−1, 22 °C/8 h dark, 20 °C) at 60% relative humidity. For metabolite analysis of Arabidopsis seeds, six individual plants of each
genotype were grown next to each other in a randomized setup. Xanthosine, guanosine and guanine were quantified directly after seed harvest. Nucleotide analysis was performed with
six-month-old seeds. Seedlings were grown on half-strength Murashige and Skoog medium. For metabolite analysis, each plate contained all genotypes representing one biological replicate. To
equalize germination, plates were stored in the dark at 4 °C for 2 days before transfer to the growth chamber. Five days after germination half of the seedlings were transferred into the
dark while the other half was kept under long-day conditions. After 48 h, whole seedlings were harvested. For the characterization of the complementation lines, seedlings were grown on
separate plates for 10 days after germination and placed into the dark for 2 days. CLONING The coding sequence of _XMPP_ was amplified from cDNA obtained from flower RNA introducing _Nco_I
and _Xma_I sites with the primers N188 and N189 or _Cla_I and _Xma_I sites with the primers N359 and N360. Via _Cla_I and _Xma_I, the coding sequence was introduced into pXCS-HAStrep20
(vector V13, construct X130) to generate a C-terminal hemeaglutinin (HA)- and Strep-tagged protein by transient expression in _N. benthamiana_. Similarly, it was introduced into pXCS-YFP21
(V36, X131) for producing a C-terminal yellow fluorescent protein (YFP)-tagged protein. The coding sequence of YFP in V36 was exchanged for a mTFP1 coding sequence amplified from a wave_1T
vector22 with the primers P243 and P244 introducing _Xma_I and _Sac_I sites. This generated a vector for the expression of a cytosolic cyan fluorescent protein (V108). For production of an
N-terminal Strep-tagged protein, the coding sequence was cloned into pXNS2cpmv-strep5 (V90, construct X144) via _Nco_I and _Xma_I. To obtain the complementation lines _xmpp gsda::C_ and
_xmpp nsh1 gsda::C_, we cloned a genomic fragment of XMPP including 999 bp upstream of the translation start codon with an introduced coding sequence for a TwinStrep tag for N-terminal
tagging (Supplementary Fig. 3d). For this, pXNS2pat-Strep-sl (V28) was generated by cloning the annealed primers 1709 and 1710 into the binary vector pAMPAT-MCS (accession number AY436765)
opened with _Xho_I and _Eco_RI. V28 was used to generate pXNS2patTwinStrep-sI (V163) by opening V28 with _Kas_I and _Nco_I and introducing a second Strep tag with a preceding linker sequence
formed by the annealed primers P1097 and P1098. _XMPP_ genomic DNA was amplified using the primers N188 and N189 and cloned into V163 via _Nco_I and _Xma_I. Into this construct the _XMPP_
promoter, amplified with the primers P1094 and P1103, was cloned via _Asc_I and _Xho_I replacing a 35 S promoter cassette (construct H453). To generate untagged Arabidopsis XMPP in
_Escherichia coli_ strain BL21 (DE3) for antibody production, _XMPP_ amplified with the primers N188 and N189 was cloned into pET30nco-CTH23 (V48) using _Cla_I and _Sma_I for the _XMPP_ cDNA
and _Cla_I and _Eco_RV for the vector (H325). PHYLOGENTIC ANALYSIS Sequences were obtained from Phytozome V12.1 (Supplementary Table 1) using the protein sequence of XMPP from _Arabidopsis
thaliana_ as query. A multiple alignment (Supplementary Data 1) was generated with muscle hosted at the website of the European Bioinformatics Institute. The Maximum Likelihood method and
the JTT matrix-based model24 were employed to infer the evolutionary history within the MEGA X25 software. Bootstrapping with 1000 repeats was performed and the results are shown at the
respective branches. To start the heuristic search, the Neighbor-Join and BioNJ algorithms were applied to a matrix of pairwise distances estimated using the JTT model, and the tree topology
with the best log likelihood was selected. The evolutionary rate differences among sites was modeled with a discrete Gamma (G) distribution of five categories (G = 1.1995). In all, 8.28% of
sites were treated as invariable. In total, 76 amino acid sequences were analyzed excluding positions with more than 5% gaps. The final dataset comprised 234 positions. Evolutionary
analyses were conducted in MEGA X25. SUBCELLULAR LOCALIZATION _Nicotiana benthamiana_ leaves were used for transient Agrobacterium (_Rhizobium radiobacter_)-mediated (co-)-expression of the
constructs X131 for production of XMPP-YFP and V108 for production of mTFP1 as a cyan fluorescent cytosolic marker. Abaxial leaf surfaces were analyzed using a Leica TSC SP8 microscope
equipped with an HC PL APO CS2 ×40 1.10 water immersion objective (Leica, Germany). To prevent crosstalk, images were obtained by sequential scanning with an excitation of 448 nm for mTFP1
(emission 465–495 nm) and 514 nm for YFP (emission 524–539 nm). Images were processed using the Leica Application Suite Advanced Fluorescence software (Leica Microsystems). PROTEIN
PURIFICATION AND DETERMINATION OF KINETIC CONSTANTS Transient expression of Strep-tagged proteins from constructs X130 and X144 were performed by infiltrating _N. benthamiana_ leaves with
_Rhizobium radiobacter_ at an optical density of 0.5 (at 600 nm). Three days after infiltration 0.75 g of leaves were ground in 1.5 mL buffer E containing 100 mM HEPES (pH 8.0), 100 mM NaCl,
5 mM EDTA (pH 8.0), 0.005% Triton X-100, 10 mM dithiothreitol, 1:625 diluted Biolock (IBA Life Sciences), 1:10 diluted protease inhibitor (complete protease inhibitor cocktail, Roche).
After centrifugation, 40 μL of StrepTactin Macroprep (IBA Life Sciences) was added to the supernatant and incubated for 10 min on a rotation wheel at 4 °C. The matrix was washed three times
with 1 mL buffer W1 (100 mM HEPES (pH 8.0), 100 mM NaCl, 0.5 mM EDTA (pH 8.0), 0.005% Triton X-100, 2 mM dithiothreitol), and centrifuged at 700 _g_ for 30 s between washes. Then the matrix
was washed twice with 1 mL buffer W2 (5 mM HEPES (pH 8.0), 100 mM NaCl, 0.5 mM EDTA (pH 8.0), 0.005% Triton X-100, 2 mM dithiothreitol). For batch elution 35 μL of buffer W2 containing 2.5
mM biotin was used. Elution was repeated and fractions were pooled5,26. Phosphatase activity was measured at 22 °C with the Enzchek Phosphate Assay Kit (Thermo Fisher Scientific) according
to the manufacturer’s instructions but in a total volume of 200 μL. In brief, 2-amino-6-mercapto-7-methyl-purine (MESG) and inorganic phosphate produced from the XMPP-dependent reaction
serve as two substrates for purine nucleoside phosphorylase, resulting in a product exhibiting an absorbance at 360 nm. For kinetic measurements, XMPP amounts were adjusted to obtain linear
initial rates at all XMP concentrations. Protein concentrations were determined with bovine serum albumin standards in a Coomassie-stained sodium dodecyl sulfate (SDS) gel using an Odyssey
Fc Imager (LI-COR Biosciences). To determine the kinetic constants, the data were plotted and fitted with the GraphPad Prism 4 software. For the substrate screen and for testing whether XMPP
is inhibited by guanosine or xanthosine, XMPP with a C-terminal HAStrep tag (expressed from X130) was used. CRYSTALLIZATION AND STRUCTURE DETERMINATION Crystallization of XMPP was
successful only after 13 C-terminal residues had been removed, likely because the region is disordered as predicted by the Phyre2 server27 and the XtalPred server28. The corresponding cDNA
(encoding Met1 to Ser250) was amplified with the primers X0001 and X0002 using a codon-optimized synthetic full-length DNA as a template (Supplementary Table 3). The product was cloned via
the In-Fusion method using _Nde_I and _Xho_I into the pET41b expression vector containing a coding sequence for a C-terminal His tag and the construct was transformed into _E. coli_ BL21
(DE3). Protein expression was induced after the cells had reached an optical density of 0.6–0.8 (at 600 nm) by the addition of 0.5 mM IPTG followed by continued culturing for 14–16 h at 20
°C. Collected cells were sonicated and centrifuged in buffer A (50 mM Tris, pH 8.0; 100 mM NaCl) and 1 mM MgCl2. The protein was purified using a HisTrap HP column with buffer A and eluted
using buffer A plus 500 mM imidazole and subsequently subjected to size exclusion chromatography using a Superdex-200 column equilibrated with buffer A plus 5% (v/v) glycerol. For solving
the structure in complex with XMP, a catalytically inactive Asp12 to Ala mutant called XMPP(D12A) was purified the same way. Purified enzymes in buffer A with 5% (v/v) glycerol were
concentrated to 8–15 mg mL−1 and subjected to crystallization using the sitting drop vapor diffusion method at 22 °C. Crystals of XMPP in its ligand-free form were produced using a
crystallization solution of 0.1 M Tris (pH 8.5) and 20% (w/v) PEG6000. For XMPP(D12A) complexed with XMP, crystals which had been grown in solution containing 0.1 M HEPES (pH 7.0) and 18%
(w/v) PEG12000 were further soaked for 1 h in crystallization solution plus 5 mM XMP, 5 mM MgCl2, and 20% (v/v) glycerol. Data collection was performed at 100 K on beamline 7 A at the Pohang
Accelerator Laboratory (Korea) with a 0.5° oscillation angle and 20% (v/v) glycerol as cryoprotectant (Supplementary Table 2). Collected data were processed using the HKL2000 software29 and
the high-resolution cutoff was based on a CC1/2 statistical value of approximately 0.6 (refs. 30,31,32). The space group of both crystals was _P2__1_, with one monomer in the asymmetric
unit. The structure of unliganded XMPP was solved by molecular replacement using the Phaser utility of PHENIX software33. A search model was generated by the PHENIX Ensembler34 and Sculptor
utilities35. Specifically, the structures of the phosphatase Sdt1p from _Saccharomyces cerevisiae_ (PDB: 3NUQ, 3OPX, and 3ONN) were used for the generation and truncation of a search model.
The initial electron density map calculated based on a solution from the Phaser utility did not produce a high-quality map, even with 1.4-Å resolution. Extensive manual model building was
required using the program COOT36 and subsequent refinement using PHENIX33 produced a highly reliable electron density map. During refinement, we observed unequivocal electron density for a
metal ion on the _F_o−_F_c map. Subsequently, the structure of XMPP(D12A) in complex with XMP was determined using the refined structure of unliganded XMPP as a starting model. We clearly
identified electron density for XMP in the vicinity of the metal-binding site of XMPP(D12A). Details of data collection and refinement are listed in Supplementary Table 2. FUNCTIONAL
ANALYSIS OF XMPP VARIANTS As for the enzyme purified from plants, the Enzchek Phosphate Assay Kit (Thermo Fisher Scientific) was used for the activity assays of enzyme purified from _E.
coli_. C-terminal His-tagged full-length XMPP and various mutants were expressed and purified by affinity chromatography using a HisTrap HP column with buffer A and were eluted using buffer
A containing 500 mM imidazole. A Hi-Prep 26/10 column equilibrated with buffer A was further used as final step for desalting and buffer exchange. Genes for mutant XMPPs were constructed by
_Dpn_I-mediated site-directed mutagenesis using mutagenic primers (Supplementary Table 3). Enzymes mutated in Asp14, His78, His82, Thr111, Asn112, and Lys159, respectively, caused problems
in expression or solubility and were excluded from further analyses. Activity and steady-state kinetic analyses were performed according to the manufacturer’s instructions, but at 30 °C. We
validated that all components in the reaction mixture were present at saturating concentrations and were not limiting the measured rates. In the activity assay, 18 nM XMPP was added per
reaction containing 50 mM Tris (pH 7.5), 200 μM MESG, 1.25 mM MgCl2, and 1 U of purine nucleoside phosphorylase. A higher XMPP concentration of 145 nM was employed for the relatively
inactive variants D12A, D183A, and D184A. Activities were determined after 140 s of reaction with 125 μM XMP. For the steady-state kinetic analysis, the reaction was initiated by adding the
indicated concentrations of XMP and the initial linear rate determined for the first 35 s. The _K_M and _V_max values were obtained using the SigmaPlot software (Systat Software).
ELECTROPHORESIS AND IMMUNOBLOTTING Proteins were extracted with buffer E (1:2 tissue to buffer ratio) and 15 μL of supernatants were separated by electrophoresis on a 12% SDS-gel and
transferred by semi-dry blotting to a nitrocellulose membrane (0.45 μm pore size, Thermo Fisher Scientific). The membrane was blocked with 5% milk powder in TBS-T (20 mM TRIS-HCl, pH 7.6;
150 mM NaCl; 0.1% (v/v) Tween 20) and was incubated with custom-made rabbit polyclonal anti-XMPP antibody (1 μg mL−1) in TBS-T with 0.5% (w/v) milk powder over night at 4 °C. The membrane
was washed three times for 10 min with TBS-T and incubated with secondary goat anti-rabbit IgG horseradish peroxidase conjugated antibody (RABHRP1, 1:5000, Sigma-Aldrich) in TBS-T for 1 h at
room temperature and washed as before. For detection, SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) was used. The chemiluminescence was detected by a
Lumi-Imager F1 (Hoffmann-La Roche). For detection of YFP, membranes were incubated with monoclonal anti-GFP antibody from mouse (Roche 11814460001; clones 7.1 and 13.1; 1:15000 diluted).
This antibody does not bind to mTFP1. Anti-mouse IgG conjugated to alkaline phosphatase (Sigma-Aldrich A3562; 1:10,000 diluted) was used as a secondary antibody. PRODUCTION OF CUSTOM-MADE
POLYCLONAL ANTI-XMPP ANTIBODY Untagged Arabidopsis XMPP protein was produced from construct H325 in _E. coli_ strain BL21(DE3) and precipitated in inclusion bodies. Cells were grown in 0.5 L
of Luria-Bertani medium to an optical density of 0.5 (at 600 nm) and then induced by 0.5 mM isopropyl-β-d-thiogalactoside. After 3 h of induction, cells were harvested, the pellet was
resuspended in 27 mL of lysis buffer (50 mM Tris-HCl (pH 8.0), 0.25% (w/v) sucrose, 1 mM EDTA (pH 8.0), 7.5 mg mL−1 lysozyme), vortexed, and incubated on ice for 30 min. Cells were disrupted
by sonication on ice and 70 mL of detergent buffer (20 mM Tris-HCl (pH 7.5), 2 mM EDTA (pH 8.0), 200 mM NaCl, 1% (w/v) deoxycholic acid, and 1% (v/v) Nonidet P-40) was added. The lysate was
centrifuged at 5000 _g_ for 10 min, the supernatant was removed, and the pellet was resuspended in 80 mL washing buffer (0.5% (v/v) Triton X-100 and 1 mM EDTA (pH 8.0)). Centrifugation and
resuspension were repeated until a tight pellet was obtained. The pellet was washed in 80 mL of 70% ethanol (v/v), resuspended in 1 mL of freshly prepared PBS by sonication, and used for
commercial antisera production in rabbit and antibody purification (immunoGlobe Antikörpertechnik GmbH). METABOLITE ANALYSIS The extraction method was adapted from Hauck et al.16. For
metabolite analysis, frozen plant material was homogenized with steel beads using a mixer mill MM 400 (Retsch). Tube racks were pre-cooled in liquid nitrogen. Ten milligrams of seeds were
ground with one 4 mm steel bead and five 2 mm steel beads for 4.5 min at 28 s−1. For seedlings, 50 mg were ground with five 2 mm steel beads for 4.5 min at 28 s−1. For the extraction, 10 mM
ammonium acetate buffer (pH 7.5) pre-warmed to 60 °C was used and samples were immediately transferred to a heat block at 95 °C for shaking at 1000 rpm for 10 min. For seeds and seedlings, 1
mL and 500 μL of extraction buffer were used, respectively. Following the heat treatment, samples were chilled on ice for 5 min. After centrifugation at 20,000 _g_ for 10 min at 4 °C, 80%
of the supernatant was transferred to a new tube to remove beads and cell debris. By centrifugation at 45,000 _g_ for 15 min at 4 °C particles which might block the HPLC system were removed.
Xanthosine, xanthine, guanosine, guanine, urate and allantoate were quantified using an Agilent HPLC 1200 system with a Polaris 5 C18-A 50 ×4.6 mm column (Agilent Technologies) coupled to
an Agilent 6460 C series triple quadrupole mass spectrometer. Except for urate, measurements were performed in the positive ion mode. Ammonium acetate (10 mM, pH 7.5) and 100% methanol
served as solvents A and B to form the following gradient: 0 min, 5% B; 1.5 min, 5% B; 3.5 min, 15% B; 6 min, 100% B; 7 min, 100% B; 7.1 min, 5% B; and 13 min, 5% B5. The flow rate was 0.8
mL min−1 and samples of 20 μL were injected. Seed extracts were diluted 50 fold for the quantification of xanthosine, guanosine and guanine. Seedling extracts were not diluted for the
quantification of xanthosine, urate and allantoate and 100-fold diluted for the quantification of guanosine. External standard dilutions spiked 1:10 into the wild-type matrix were used for
quantification of xanthosine, guanosine and guanine. For the quantification of urate and allantoate, the _xmpp gsda_ matrix was used. The MS parameters are listed in Supplementary Table 4.
Measurements failing the quality criteria for retention time, qualifier to quantifier ratio or with a signal to noise ratio below 10 were called “not detected”. The quantification of
nucleotides was performed as recently described14,37. STATISTICAL ANALYSIS Data for the determination of kinetic constants were plotted and analyzed using the GraphPad Prism 4 software. For
metabolite data, the R software (version 1.2.5042) and CRAN packages multcomp and sandwich were used to perform two-sided Tukey’s pairwise comparisons and to consider heteroscedasticity of
the dataset with the sandwich variance estimator38,39. Using the same packages, two-sided Dunnett’s comparisons to the reference group (XMPP + 10 μM XMP) were performed for the inhibitor
test. The number of replicates, the test values and the multiplicity-adjusted _p_ values are reported in the Source Data file. ACCESSION NUMBERS Sequence data from this article can be found
with the following locus identifiers: _XMPP_, At2g32150; _NSH1_, At2g36310; _GSDA_, At5g28050, _Actin2_ At3g18780. REPORTING SUMMARY Further information on research design is available in
the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY All data generated or analyzed during this study are included in this published article (and its Supplementary
Information file). Mass spectrometry raw data can be supplied upon request. Atomic coordinates and structure factors of XMPP with and without XMP bound have been deposited in the Protein
Data Bank (PDB) under the accession codes 7EF7 and 7EF6, respectively. Source data are provided with this paper. REFERENCES * Smith, P. M. C. & Atkins, C. A. Purine biosynthesis. Big in
cell division, even bigger in nitrogen assimilation. _Plant Physiol._ 128, 793–802 (2002). Article CAS Google Scholar * Witte, C.-P. & Herde, M. Nucleotide metabolism in plants.
_Plant Physiol._ 182, 63–78 (2020). Article CAS Google Scholar * Werner, A. K., Romeis, T. & Witte, C.-P. Ureide catabolism in Arabidopsis thaliana and Escherichia coli. _Nat. Chem.
Biol._ 6, 19–21 (2010). Article CAS Google Scholar * Werner, A. K. & Witte, C.-P. The biochemistry of nitrogen mobilization: purine ring catabolism. _Trends Plant Sci._ 16, `381–387
(2011). Article CAS Google Scholar * Baccolini, C. & Witte, C.-P. AMP and GMP catabolism in Arabidopsis converge on xanthosine, which is degraded by a nucleoside hydrolase
heterocomplex. _Plant Cell_ 31, 734–751 (2019). Article CAS Google Scholar * Dahncke, K. & Witte, C.-P. Plant purine nucleoside catabolism employs a guanosine deaminase required for
the generation of xanthosine in Arabidopsis. _Plant Cell_ 25, 4101–4109 (2013). Article CAS Google Scholar * Yin, Y., Katahira, R. & Ashihara, H. Metabolism of purine nucleosides and
bases in suspension-cultured Arabidopsis thaliana cells. _Eur. Chem. Bull._ 3, 925–934 (2014). Google Scholar * Ashihara, H., Stasolla, C., Fujimura, T. & Crozier, A. Purine salvage in
plants. _Phytochemistry_ 147, 89–124 (2018). Article CAS Google Scholar * Sa, N., Rawat, R., Thornburg, C., Walker, K. D. & Roje, S. Identification and characterization of the missing
phosphatase on the riboflavin biosynthesis pathway in Arabidopsis thaliana. _Plant J_. 88, 705-716 (2016). * Xu, Y.-F. et al. Nucleotide degradation and ribose salvage in yeast. _Mol. Syst.
Biol._ 9, 665 (2013). Article Google Scholar * Jung, B., Hoffmann, C. & Möhlmann, T. Arabidopsis nucleoside hydrolases involved in intracellular and extracellular degradation of
purines. _Plant J._ 65, 703–711 (2011). Article CAS Google Scholar * Jung, B. et al. Uridine-ribohydrolase is a key regulator in the uridine degradation pathway of Arabidopsis. _Plant
Cell_ 21, 876–891 (2009). Article CAS Google Scholar * Riegler, H., Geserick, C. & Zrenner, R. Arabidopsis thaliana nucleosidase mutants provide new insights into nucleoside
degradation. _N. Phytol._ 191, 349–359 (2011). Article CAS Google Scholar * Straube, H., Niehaus, M., Zwittian, S., Witte, C.-P. & Herde, M. Enhanced nucleotide analysis enables the
quantification of deoxynucleotides in plants and algae revealing connections between nucleoside and deoxynucleoside metabolism. _Plant Cell_ 33, 270–289 (2020). Article Google Scholar *
Schroeder, R. Y., Zhu, A., Eubel, H., Dahncke, K. & Witte, C.-P. The ribokinases of Arabidopsis thaliana and Saccharomyces cerevisiae are required for ribose recycling from nucleotide
catabolism, which in plants is not essential to survive prolonged dark stress. _N. Phytol._ 217, 233–244 (2018). Article CAS Google Scholar * Hauck, O. K. et al. Uric acid accumulation in
an Arabidopsis urate oxidase mutant impairs seedling establishment by blocking peroxisome maintenance. _Plant Cell_ 26, 3090–3100 (2014). Article CAS Google Scholar * Seifried, A.,
Schultz, J. & Gohla, A. Human HAD phosphatases: structure, mechanism, and roles in health and disease. _FEBS J._ 280, 549–571 (2013). Article CAS Google Scholar * Alonso, J. M. et al.
Genome-wide insertional mutagenesis of Arabidopsis thaliana. _Science_ 301, 653–657 (2003). Article ADS Google Scholar * Kleinboelting, N., Huep, G., Kloetgen, A., Viehoever, P. &
Weisshaar, B. GABI-Kat simple search. New features of the Arabidopsis thaliana T-DNA mutant database. _Nucleic Acids Res._ 40, D1211–D1215 (2012). Article CAS Google Scholar * Witte,
C.-P., Noël, L. D., Gielbert, J., Parker, J. E. & Romeis, T. Rapid one-step protein purification from plant material using the eight-amino acid StrepII epitope. _Plant Mol. Biol._ 55,
135–147 (2004). Article CAS Google Scholar * Chen, M. & Witte, C.-P. A kinase and a glycosylase catabolize pseudouridine in the peroxisome to prevent toxic pseudouridine monophosphate
accumulation. _Plant Cell_ 32, 722–739 (2020). Article CAS Google Scholar * Geldner, N. et al. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor
marker set. _Plant J._ 59, 169–178 (2009). Article CAS Google Scholar * Myrach, T., Zhu, A. & Witte, C.-P. The assembly of the plant urease activation complex and the essential role
of the urease accessory protein G (UreG) in delivery of nickel to urease. _J. Biol. Chem._ 292, 14556–14565 (2017). Article CAS Google Scholar * Jones, D. T., Taylor, W. R. &
Thornton, J. M. The rapid generation of mutation data matrices from protein sequences. _Comput. Appl. Biosci._ 8, 275–282 (1992). CAS PubMed Google Scholar * Kumar, S., Stecher, G., Li,
M., Knyaz, C. & Tamura, K. MEGA X: molecular evolutionary genetics analysis across computing platforms. _Mol. Biol. Evol._ 35, 1547–1549 (2018). Article CAS Google Scholar * Werner,
A. K., Sparkes, I. A., Romeis, T. & Witte, C.-P. Identification, biochemical characterization, and subcellular localization of allantoate amidohydrolases from Arabidopsis and soybean.
_Plant Physiol._ 146, 418–430 (2008). Article CAS Google Scholar * Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. E. The Phyre2 web portal for protein
modeling, prediction and analysis. _Nat. Protocol_ 10, 845–858 (2015). Article CAS Google Scholar * Slabinski, L. et al. XtalPred: a web server for prediction of protein
crystallizability. _Bioinformatics_ 23, 3403–3405 (2007). Article CAS Google Scholar * Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode.
_Methods Enzymol._ 276, 307–326 (1997). Article CAS Google Scholar * Winn, M. D. et al. Overview of the CCP4 suite and current developments. _Acta Crystallogr. D._ 67, 235–242 (2011).
Article CAS Google Scholar * Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. _Science_ 336, 1030–1033 (2012). * Diederichs, K. & Karplus, P. A.
Better models by discarding data? _Acta Crystallogr. D_ 69, 1215–1222 (2013). Article CAS Google Scholar * Adams, P. D. et al. PHENIX: a comprehensive Python-based system for
macromolecular structure solution. _Acta Crystallogr. D_ 66, 213–221 (2010). Article CAS Google Scholar * Terwilliger, T. C. et al. Interpretation of ensembles created by multiple
iterative rebuilding of macromolecular models. _Acta Crystallogr. D_ 63, 597–610 (2007). Article CAS Google Scholar * Bunkóczi, G. & Read, R. J. Improvement of molecular-replacement
models with Sculptor. _Acta Crystallogr. D_ 67, 303–312 (2011). Article Google Scholar * Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. _Acta
Crystallogr. D_ 66, 486–501 (2010). * Straube, H., Witte, C.-P. & Herde, M. Analysis of nucleosides and nucleotides in plants: an update on sample preparation and LC-MS techniques.
_Cells_ 10, 689 (2021). Article CAS Google Scholar * Herberich, E., Sikorski, J. & Hothorn, T. A robust procedure for comparing multiple means under heteroscedasticity in unbalanced
designs. _PloS ONE_ 5, e9788 (2010). Article ADS Google Scholar * Pallmann, P. & Hothorn, L. A. Analysis of means. A generalized approach using R. _J. Appl. Stat._ 43, 1541–1560
(2016). Article MathSciNet Google Scholar Download references ACKNOWLEDGEMENTS We thank André Specht and Hildegard Thölke for technical assistance and Anting Zhu for generating the clones
X130, X131, and X144; Christel Schmiechen for generating the clone H453 and Mingjia Chen for generating the vector V108 as well as Ludwig Hothorn for advice concerning the statistical
analysis. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grants WI3411/8-1, INST 187/741-1 FUGG, and GRK1798 “Signaling at the Plant-Soil Interface” for C.-P.W., and by
the National Research Foundation of Korea (NRF) grant 2020R1A4A1018890 by the Korea government (MSIT) for S.R. FUNDING Open Access funding enabled and organized by Projekt DEAL. AUTHOR
INFORMATION Author notes * These authors contributed equally: Katharina J. Heinemann, Sun-Young Yang. AUTHORS AND AFFILIATIONS * Leibniz Universität Hannover, Department of Molecular
Nutrition and Biochemistry of Plants, Herrenhäuser Strasse 2, 30419, Hannover, Germany Katharina J. Heinemann, Henryk Straube, Nieves Medina-Escobar, Marina Varbanova-Herde, Marco Herde
& Claus-Peter Witte * Seoul National University, Department of Agricultural Biotechnology, 151-921, Seoul, Republic of Korea Sun-Young Yang & Sangkee Rhee Authors * Katharina J.
Heinemann View author publications You can also search for this author inPubMed Google Scholar * Sun-Young Yang View author publications You can also search for this author inPubMed Google
Scholar * Henryk Straube View author publications You can also search for this author inPubMed Google Scholar * Nieves Medina-Escobar View author publications You can also search for this
author inPubMed Google Scholar * Marina Varbanova-Herde View author publications You can also search for this author inPubMed Google Scholar * Marco Herde View author publications You can
also search for this author inPubMed Google Scholar * Sangkee Rhee View author publications You can also search for this author inPubMed Google Scholar * Claus-Peter Witte View author
publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS C.-P.W. devised the project and supervised the work except for the generation of the crystal structure
and the biochemical analysis of enzyme mutants, which was supervised by S.R. C.-P.W., S.R., K.J.H., and S.-Y.Y. designed the experiments and interpreted the results. K.J.H. performed the
biochemical analyses of XMPP produced in plants, generated the Arabidopsis mutants, and performed the metabolite analyses of nucleosides and purine catabolic products. H.S. performed the
metabolite analysis of nucleotides. N.M.-E. performed the confocal microscopy and the characterization of the T-DNA lines. C.-P.W., M.V.-H., and M.H. made bioinformatic analyses and M.V.-H.
performed preliminary assays leading to the discovery of XMPP. C.-P.W. performed the phylogenetic analysis. S.-Y.Y. determined the crystal structure and made the biochemical analyses of the
XMPP variants. K.J.H. conducted most of the statistical analyses. K.J.H., C.-P.W., and S.-Y.Y. prepared the figures. C.-P.W. and S.R. wrote the manuscript with the contribution of K.J.H. and
S.-Y.Y. The manuscript was revised by all authors. CORRESPONDING AUTHORS Correspondence to Sangkee Rhee or Claus-Peter Witte. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no
competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Communications_ thanks Santiago Ramón-Maiques and the other, anonymous, reviewer(s) for their contribution to the
peer review of this work. Peer reviewer reports are available. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE REPORTING SUMMARY SOURCE DATA SOURCE DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under
a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate
credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article
are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and
your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this
license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Heinemann, K.J., Yang, SY., Straube, H. _et al._ Initiation of
cytosolic plant purine nucleotide catabolism involves a monospecific xanthosine monophosphate phosphatase. _Nat Commun_ 12, 6846 (2021). https://doi.org/10.1038/s41467-021-27152-4 Download
citation * Received: 17 May 2021 * Accepted: 02 November 2021 * Published: 25 November 2021 * DOI: https://doi.org/10.1038/s41467-021-27152-4 SHARE THIS ARTICLE Anyone you share the
following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer
Nature SharedIt content-sharing initiative








