- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Genetic discoveries of Alzheimer’s disease are the drivers of our understanding, and together with polygenetic risk stratification can contribute towards planning of feasible and
efficient preventive and curative clinical trials. We first perform a large genetic association study by merging all available case-control datasets and by-proxy study results (discovery _n_
= 409,435 and validation size _n_ = 58,190). Here, we add six variants associated with Alzheimer’s disease risk (near _APP, CHRNE, PRKD3/NDUFAF7, PLCG2_ and two exonic variants in the
_SHARPIN_ gene). Assessment of the polygenic risk score and stratifying by _APOE_ reveal a 4 to 5.5 years difference in median age at onset of Alzheimer’s disease patients in _APOE_ ɛ4
carriers. Because of this study, the underlying mechanisms of _APP_ can be studied to refine the amyloid cascade and the polygenic risk score provides a tool to select individuals at high
risk of Alzheimer’s disease. SIMILAR CONTENT BEING VIEWED BY OTHERS A GLOBAL VIEW OF THE GENETIC BASIS OF ALZHEIMER DISEASE Article 06 April 2023 STEP BY STEP: TOWARDS A BETTER UNDERSTANDING
OF THE GENETIC ARCHITECTURE OF ALZHEIMER’S DISEASE Article Open access 02 May 2023 IDENTIFYING INDIVIDUALS WITH HIGH RISK OF ALZHEIMER’S DISEASE USING POLYGENIC RISK SCORES Article Open
access 23 July 2021 INTRODUCTION Thus far, multiple loci associated with Alzheimer’s disease (AD) have been described next to causal mutations in two subunits of γ-secretases,
membrane-embedded aspartyl complexes (_PSEN1_, _PSEN2_ genes_)_, and the gene encoding one target protein of these proteases, the amyloid precursor protein gene _(APP)_. The most prominent
locus, _APOE_, was detected almost 30 years ago using linkage techniques1. In addition, genome-wide association studies (GWAS) of AD case-control datasets and by-proxy AD case-control
studies have identified 30 genomic loci that modify the risk of AD2,3,4,5,6,7. These signals account for ~31% of the genetic variance of AD, leaving most of the genetic risk as yet
uncharacterized8. Further disentangling the genetic constellation of common genetic variations underlying AD can drive our biological insights of AD and can point toward novel drug targets.
There are over 50 million people living with dementia and the global cost of dementia is well above 1 trillion US$9. This means there is a medical and economical urgency to efficiently test
interventions that are under development. Therefore, to increase power and reduce duration of trials, pre-symptomatic patients that are at high genetic risk of disease are increasingly
developed10. However, only carriers of causal mutations (_APP, PSEN1,_ and _PSEN2_) and the _APOE_ ɛ4 allele are considered high risk, while other common and rare genetic variants are
ignored11. Despite that, the combined effects of all currently known variants in a polygenic risk score (PRS) is associated with the conversion of mild cognitive impairment to AD12,13, the
neuropathological hallmarks of AD, age at onset (AAO) of disease14,15,16,17 and lifetime risk of AD18. In this work we aim to comprehend and expand the knowledge of the genetic landscape
underlying AD and provide additional evidence that a PRS of variants can be a robust tool to select high risk individuals with an earlier AAO. We first performed a meta-GWAS integrating all
currently published GWAS case-control data, by-proxy case-control data, and the data from the Genome Research at Fundació ACE (GR@ACE) study19. We confirm the observed associations in a
large independent replication study. Then, we construct an update of the PRS and test whether the effects of the PRS are influenced by diagnostic certainty, sex and AAO groups. Lastly, we
test whether the PRS could be used to identify individuals at the highest odds of having AD and we compared AAO of the AD cases. This study describes the identification of six variants
associated with AD risk and provides an extended PRS tool to select individuals at high risk of AD. RESULTS META-GWAS OF AD We combined data from three AD GWASs: the summary statistics
calculated from the GR@ACE19 case-control study (6331 AD cases and 6055 controls), the IGAP20 case-control study (up to 30,344 AD cases and 52,427 controls) and the UKB AD-by-proxy
case-control study21 (27,696 cases of maternal AD with 260,980 controls, and 14,338 cases of paternal AD with 245,941 controls, Fig. 1, Supplementary Data 1). Although we observed inflation
in the resulting summary statistics (_λ_ median = 1.08; see Supplementary Fig. 1d), it was not driven by an un-modeled population structure (LD score regression intercept = 1.036). The full
details of the studies are described in methods. After study-specific variant filtering and quality-control procedures, we performed a fixed effects inverse-variance-weighted meta-analysis22
on the summary statistics of the three studies. Using this strategy, we identified a genome-wide significant (GWS) association (_p_ < 5 × 10−8) for 36 independent genetic variants in 35
genomic regions (the _APOE_ region contains signals for ɛ4 and ɛ2). As a sensitivity analysis, we removed the AD-by-proxy study and compared the resulted effect estimates with and without
this dataset. We found a high correlation between the effect estimates from the case-control and by-proxy approaches for the significant loci (_R_2 = 0.994, _p_ = 8.1 × 10−37; Supplementary
Fig. 1e). Four genomic regions were not previously associated with AD (see Manhattan Plot, Fig. 2a). Next, we aimed at replicating the associated loci in 16 cohorts (19,087 AD cases and
39,101 controls in total), many of them collected and analyzed by the European Alzheimer’s Disease Biobank (JPND-EADB) project. We tested all variants with suggestive association (_p_ <
10−5) located within a 200 kb region from the sentinel SNP. Overall, 384 variants were tested in the replication datasets (Supplementary Data 2). Discovery and replication were combined, and
we identified associations in six variants comprising five genomic loci annotated using FUMA23 (Table 1, Fig. 2b–f, Supplementary Fig. 2 and Supplementary Results). In _APP_, we identified
a common (MAF = 0.46) intronic variant associated with a reduced risk of AD (rs2154481, OR = 0.95 [0.94–0.96], _p_ = 1.39 × 10−11, Fig. 2f). In _SHARPIN_ (SHANK Associated RH Domain
Interactor) gene, we found two missense mutations (rs34173062/p.Ser17Phe and rs34674752/p.Pro294Ser) that are in linkage equilibrium (_R_2 = 1.3 × 10−6, _D_′ = 0.014, _p_ = 0.96). Both
missense variants increased AD risk (p.Ser17Phe, MAF = 0.085, OR = 1.14 [1.10–1.18], _p_ = 9.6 × 10−13 and p.Pro294Ser, MAF = 0.052, OR = 1.13 [1.09–1.18], _p_ = 1.0 × 10−9, Fig. 2b). A
variant close to the genes _PRKD3_ and _NDUFAF7_ (rs876461, MAF = 0.143) emerged as the most significant variant in the region after the combined analysis (OR = 1.07 [1.05–1.09], _p_ = 1.3 ×
10−9, Fig. 2c). In the 3’-UTR region of _CHRNE_ (Cholinergic Receptor Nicotinic Epsilon Subunit), rs72835061 (MAF = 0.085) was associated with a 1.09-fold increased risk of AD (95% CI
[1.06–1.11], _p_ = 1.5 × 10−10, Fig. 2e). Our analysis also strengthened the evidence of association with AD for three additional genomic loci including an association with a variant in
_PLCG2_ (rs3935877, MAF = 0.13, OR = 0.92 [0.90–0.95], _p_ = 6.9 × 10−9, Fig. 2d), and confirmed another common variant in _PLCG2_, a stop gain mutation in _IL-34_ and a variant near
_HS3ST1_ (Table 1, Supplementary Fig. 3 and Supplementary Data 2, 3). We were not able to replicate two loci (_ELK2AP_ and _SPPL2A_ regions) that showed suggestive association with AD (_p_
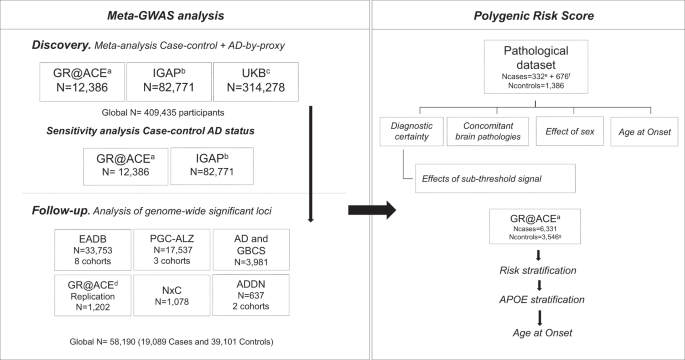
< 1 × 10−7 in discovery). POLYGENIC RISK SCORES In order to assess the robustness and combined effect of the genetic landscape of AD (Fig. 3, Supplementary Data 4), we constructed a
weighted PRS based on the 39 genetic variants (excluding _APOE_ genotypes) that showed GWS evidence of association with AD (see Methods, Fig. 4 and Supplementary Data 5). We tested if the
association of the PRS with AD is independent of clinically important factors that are considered in the selection of individuals for clinical trials. First, we showed that the association
of the PRS with clinically diagnosed AD cases is similar to the association with pathologically confirmed AD (OR = 1.30 vs. 1.38, per 1-SD increase in the PRS). In this setting, adding
variants below the GWS threshold did not lead to a more significant association of the PRS with AD (Fig. 4a). Next, we tested whether the PRS was associated with AD in the presence of
concomitant brain pathologies (besides AD). Among our autopsy-confirmed AD patients (_n_ = 332), 84% had at least one concomitant pathology, and the PRS was associated with AD in the
presence of all tested concomitant pathologies (Fig. 4b). Moreover, the patients often had more than one concomitant pathology (48.8%), but no difference was observed in the effect estimate
of the PRS when more than one pathology was present (Fig. 4b). Last, we investigated the effect of sex and AAO (Fig. 4c). Our analysis revealed that the effect of the PRS was the same in
both sexes (Fig. 4c) and was consistent with both early-onset (onset before 65 years; OR = 1.58, 95% CI [1.22–2.05], _p_ = 5.8 × 10−4) as well as with late-onset AD (onset later than 85
years; OR = 1.29, 95% CI [1.10–1.51], _p_ = 1.5 × 10−3). PRSs has the potential to early identify subjects at risk of complex diseases24. To identify people at the highest genetic risk of AD
based on the PRS, we used the validated 39-variants PRS in the large GR@ACE dataset. The PRS was associated with a 1.27-fold (95% CI [1.23–1.32]) increased risk for every standard deviation
increase in the PRS (_p_ = 7.3 × 10−39) and with a gradual risk increase when we stratified the dataset into 2% percentiles of the PRS (Fig. 5a, Supplementary Data 6). Next, we stratified
the dataset in _APOE_ genotype risk groups. The PRS percentiles were associated with AD within the _APOE_ genotype groups (Fig. 5b, Supplementary Data 7). Finally, we compared the risk
extremes and found a 16.2-fold (95% CI [8.84–29.5], _p_ = 1.5 × 10−19) increased risk for the highest-PRS group (_APOE ɛ4ɛ4_) compared with the lowest-PRS group (_APOE ɛ2ɛ2/ɛ2ɛ3_;
Supplementary Data 8). When we compared the median AAO in AD patients in these extreme risk groups we found a 9-year difference in the median age (_p__Wilcoxon_ = 1.7 × 10−6) (Fig. 5c).
Lastly, we studied the effects on AAO of the PRS in the _APOE_ genotype groups. The PRS differentiated AAO only within _APOE_ ɛ4 carriers. In _APOE_ ɛ4 heterozygotes the PRS determined a
4-year difference in median AAO and in _APOE_ ɛ4 homozygotes (_p__Wilcoxon_ = 6.9 × 10−5), where the PRS determined a median AAO difference of 5.5 years (_p__Wilcoxon_ = 4.6 × 10−5). For the
selection of high-risk individuals, it is important to note that we found no difference in the odds and AAO for AD for _APOE_ ɛ4 heterozygotes with the highest PRS compared to _APOE_ ɛ4
homozygotes with the lowest PRS. The Cox regression also showed an impact of _APOE_ on AAO, mainly on _APOE_ ε4ε4 (significant _APOE_-PRS interaction (_p_ = 0.021), Fig. 5d, Supplementary
Data 9). DISCUSSION This work adds on the ongoing global effort to identify genetic variants associated with AD (Fig. 3). In the present work, we reported on the largest GWAS for AD risk to
date, comprising genetic information of 467,623 individuals of European ancestry. We identified six variants that were not previously associated with the risk of AD and constructed a robust
PRS for AD demonstrating its potential value for selecting subjects at risk of AD, especially within _APOE_ ɛ4 carriers. This PRS was based on European ancestries and may or may not
generalize to other ancestries. Validation in other populations will be required. We also acknowledge that controls included in GR@ACE are younger than cases and some of the controls might
still develop AD later in life. This fact does not invalidate the analysis although reported estimates must be considered conservative. The differences in risk and AAO determined by the PRS
of AD are relevant for design clinical trials that over-represent _APOE ε4_ carriers, as _APOE ε4_ heterozygous with highest-PRS values have a similar risk and AAO to _APOE_ ɛ4 homozygotes
(Fig. 5b). These represents ~1% of our control population, which is the same percentage as all _APOE ε4_ homozygotes. A trial that aims to include _APOE_ ɛ4 homozygotes, could consider
widening the selection criteria and in this way hasten the enrollment process. Also, our PRS could aid at the interpretation of the results of clinical trials, as it determines a relevant
proportion of the AAO, which could either mimic or obscure a treatment effect. The most interesting finding from our GWAS is the discovery of a common protective (MAF (C-allele) = 0.483)
intronic variant in the _APP_ gene. Our results directly support _APP_ production or processing as a causal pathway not only in familial AD but in common sporadic AD. The SNP is in a DNase
hypersensitive area of 295 bp (chr21:27473781-27474075) possibly involved in the transcriptional regulation of the _APP_ gene. rs2154481 is an eQTL for the _APP_ mRNA and an antisense
transcript of the _APP_ gene named AP001439.2 in public eQTL databases25 (Supplementary Fig. 4). Functional evidence supports a modified _APP_ transcription26 as an LD block of 13 SNPs
within the _APP_ locus (including rs2154481) increased the TFCP2 transcription factor avidity to its binding site and increased the enhancer activity of this specific intronic region26.
Based on this evidence, we can postulate that a life-long slightly higher _APP_ gene expression protects the brain from AD insults. Still, this seems counterintuitive as duplications of the
gene lead to early-onset AD27. A U-shaped effect, or hormesis effect of _APP_ might help explain our observations and it might also fit the accelerated cognitive deterioration observed in AD
patients treated with beta-secretase inhibitors28,29 as these reduce beta-amyloid in their brain. An alternative hypothesis is that mechanisms underlying the variant are related to the
overexpression of protective fragments of the APP protein30. Disentangling the molecular mechanism of our finding will help refine and steer the amyloid hypothesis. Additionally, other three
variants identified are altering protein sequence or affecting regulatory motifs. Two independent missense mutations in _SHARPIN_ increased the AD risk. _SHARPIN_ was previously proposed as
an AD candidate gene31,32, and functional analysis of a rare missense variant (NM_030974.3:p.Gly186Arg) resulted in the aberrant cellular localization of the variant protein and attenuated
the activation of NF-κB, a central mediator of inflammatory and immune responses. Functional analysis of the two identified missense variants will show if the effect on immune reaction in AD
is similar. The variant located in the _CHRNE_ which encodes a subunit of the cholinergic receptor (AChR) is a strong modulator of _CHRNE_ expression. The same allele that increases AD risk
increases the expression in the brain and other tissues according to GTEx (_p_ = 2.1 × 10−13) (Supplementary Fig. 5). The detection of a potential hypermorph allele linked to AD risk and
affecting cholinergic function could reintroduce this neurotransmitter pathway into the search for preventative strategies. Further functional studies are needed to consolidate this
hypothesis. Altogether, we described six additional loci associated with sporadic AD. These signals reinforce that AD is a complex disease in which amyloid processing and immune response
play key roles. We add to the growing body of evidence that the polygenic scores of all genetic loci to date, in combination with _APOE_ genotypes, are robust tools that are associated with
AD and its AAO. These properties make PRS promising in selecting individuals at risk to apply preventative therapeutic strategies. METHODS DATA Participants in this study were obtained from
multiple sources, including raw data from case-control samples collected by GR@ACE/DEGESCO, summary statistics data from the case-control samples in the IGAP and the summary statistics of
AD-by-proxy phenotype from the UK Biobank. An additional case-control samples from 16 independent cohorts (19,087 AD cases and 39,101 controls) was used for replication, largely collected
and analyzed by the European Alzheimer’s Disease Biobank (JPND-EADB) project. Full descriptions of the samples and their respective phenotyping and genotyping procedures are provided in the
Supplementary Methods. GR@ACE The GR@ACE study19 recruited AD patients from Fundació ACE, Institut Català de Neurociències Aplicades (Catalonia, Spain), and control individuals from three
centers: Fundació ACE (Barcelona, Spain), Valme University Hospital (Seville, Spain), and the Spanish National DNA Bank–Carlos III (University of Salamanca, Spain) (http://www.bancoadn.org).
Additional cases and controls were obtained from dementia cohorts included in the Dementia Genetics Spanish Consortium (DEGESCO)33. At all sites, AD diagnosis was established by a
multidisciplinary working group—including neurologists, neuropsychologists, and social workers—according to the DSM-IV criteria for dementia and the National Institute on Aging and
Alzheimer’s Association’s (NIA–AA) 2011 guidelines for diagnosing AD. In our study, we considered as AD cases any individuals with dementia diagnosed with probable or possible AD at any
point in their clinical course. For further details on the contribution of the sites, see Supplementary Data 10. Written informed consent was obtained from all the participants. The ethics
and scientific committees have approved this research protocol (Acta 25/2016, Ethics Committee H., Clinic I Provincial, Barcelona, Spain). _Genotyping, quality control, and imputation_. DNA
was extracted from peripheral blood according to standard procedures using the Chemagic system (Perkin Elmer). Samples reaching DNA concentrations of >10 ng/µl and presenting high
integrity were included for genotyping. Cases and controls were randomized across sample plates to avoid batch effects. Genotyping was conducted using the Axiom 815K Spanish biobank array
(Thermo Fisher) at the Spanish National Center for Genotyping (CeGEN, Santiago de Compostela, Spain). The genotyping array not only is an adaptation of the Axiom biobank genotyping array but
also contains rare population-specific variations observed in the Spanish population. The DNA samples were genotyped according to the manufacturer’s instructions (Axiom™ 2.0 Assay Manual
Workflow). The Axiom 2.0 assay interrogates biallelic SNPs and simple indels in a single-assay workflow. Starting with 200 ng of genomic DNA, the samples were processed through a manual
target preparation protocol, followed by automated processing of the array plates in the GeneTitan Multi-Channel (MC) instrument. Target preparation involved DNA amplification,
fragmentation, purification, and resuspension of the target in a hybridization cocktail. The hyb-ready targets were then transferred to the GeneTitan MC instrument for automated, hands-free
processing, including hybridization, staining, washing, and imaging. The CEL files were generated using the GeneTitan MC instrument. Quality control (QC) was performed for samples and plates
using the Affymetrix power tool (APT) 1.15.0 software following the Axiom data analysis workflow. The sample quality was determined based on the resolution of AT and GC channels in a group
of non-polymorphic SNPs (resolution > 0.82). Samples with a call rate greater than 97% and plates with an average call rate above 98.5% were included for final SNP calling. The samples
were jointly called. Markers passing all the QC tests were used in downstream analysis (_N_SNPs = 729,868; 95.4%) using the SNPolisher R package (Thermo Fisher). To assess the sample
genotyping concordance, we intentionally resampled 200 samples and determined a concordance rate of 99.5%. We also conducted previously described standard QC prior to imputation19. In brief,
individual QC includes genotype call rates >97%, sex checks, and no excess heterozygosity; we removed population outliers as well (European cluster of 1000 Genomes). We included variants
with a call rate of >95%, with a minor allele frequency (MAF) of >0.01, in Hardy–Weinberg equilibrium (_p_ < 1 × 10−4 in controls) and without differential missingness between
cases and controls (Supplementary Data 11, Supplementary Fig. 1). Imputation was carried out using the Haplotype reference consortium34 (HRC, full panel) and the 1000 Genomes reference
panel35 (for indels only) on the Michigan Imputation Server (https://imputationserver.sph.umich.edu). Rare variants (MAF < 0.001) and variants with low imputation quality (_R_2 < 0.30)
were excluded. Logistic regression models, adjusted for the first four ancestry principal components19, were fitted using Plink (v2.00a). Population-based controls were used; therefore, age
was not included as a covariate. Age and gender statistically behave like phenotype proxies (for AD status in this case). Therefore, adjusting for co-variation with age and gender could
result in an over-adjustment of GWAS results. After QC steps, we included 6,331 AD cases and 6,055 control individuals and tested 14,542,816 genetic variants for association with AD. IGAP
SUMMARY STATISTICS The GWAS summary results from the IGAP were downloaded from the National Institute on Aging Genetics of Alzheimer’s Disease Data Storage Site (NIAGADS,
https://www.niagads.org/)20. Details on data generation and analyses by the IGAP have been previously described20. In brief, the IGAP is a large study based upon genome-wide association
using individuals of European ancestry. Stage 1 of the IGAP comprises 21,982 AD cases and 41,944 cognitively normal controls from four consortia: the Alzheimer Disease Genetics Consortium
(ADGC), the European Alzheimer’s Disease Initiative (EADI), the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium, and the Genetic and Environmental Risk in
AD/Defining Genetic, Polygenic, and Environmental Risk for Alzheimer’s Disease (GERAD/PERADES) Consortium. Summary statistics are available for 11,480,632 variants, both genotyped and
imputed (1000 Genomes phase 1, v3). In Stage 2, 11,632 SNPs were genotyped in an independent set of 8362 AD cases and 10,483 controls. UK BIOBANK SUMMARY STATISTICS UK Biobank data—including
health, cognitive, and genetic data—was collected on over 500000 individuals aged 37–73 years from across Great Britain (England, Wales, and Scotland) at the study baseline (2006–2010)
(http://www.ukbiobank.ac.uk)36. Several groups have demonstrated the utility of self-report of parental history of AD for case ascertainment in GWAS (proxy–AD approach)21,37,38. For this
study, we used the published summary statistics of Marioni et al.21. They included, after stringent QC, 314,278 unrelated individuals for whom AD information was available on at least one
parent in the UK Biobank (https://datashare.is.ed.ac.uk/handle/10283/3364). In brief, the 27,696 participants whose mothers had dementia (maternal cases) were compared with the 260,980
participants whose mothers did not have dementia. Likewise, the 14,338 participants whose fathers had dementia (paternal cases) were compared with the 245,941 participants whose fathers did
not have dementia21. The phenotype of the parents is independent, and therefore, the estimates could be meta-analyzed. After analysis, the effect estimates were made comparable to a
case-control setting. Further information on the transformation of the effect sizes can be found elsewhere21,39. The data available comprises summary statistics of 7,794,553 SNPs imputed to
the HRC reference panel (full panel). META-GWAS OF AD After study-specific variant filtering and quality-control procedures, we performed a fixed effects inverse-variance–weighted
meta-analysis22 on the discovery and follow-up stages (Supplementary Data 1 and Supplementary Data 12). To determine the lead SNPs (those with the strongest association per genomic region),
we performed clumping on SNPs with a GWS _p_ value (_p_ < 5 × 10−8) (Plink v1.90, maximal linkage disequilibrium (LD) with _R_2 < 0.001 and physical distance 250 Kb). In the _APOE_
region, we only considered the _APOE_ ɛ4 (rs429358) and _APOE_ ɛ2 (rs7412) SNPs40. LD information was calculated using the GR@ACE imputed genotypes as a reference. Polygenicity and
confounding biases, such as cryptic relatedness and population stratification, can yield an inflated distribution of test statistics in GWAS. To distinguish between inflation from a true
polygenic signal and bias we quantified the contribution of each by examining the relationship between test statistics and linkage disequilibrium (LD) using the LD Score regression intercept
(LDSC software41). Chromosomal regions associated with AD in previous studies were excluded from follow-up (Lambert et al.3, Kunkle et al.42, and Jansen et al.38). We tested all variants
with suggestive association (_p_ < 10−5) located in proximity (200 kb) of genomic regions selected for follow-up to allow for the potential refinement of the top associated variant.
Conditional analyses were performed in regions where multiple variants were associated with AD using logistic regression models, adjusting for the genetic variants in the region
(Supplementary Data 13, 14). Regional plots were generated with a mixture of homemade Python (v2.7) and R (v3.6.0) scripts. Briefly, given an input variant, we calculated the LD between the
input variant and all the surrounding variants within a window of length defined by the user. The LD was calculated in the 1000 Genomes samples of European ancestry. We used gene positions
from RefSeq (release 93); in the case of multiple gene models for a given gene, we reported the model with the largest number of exons. We used recombination rates from HapMap II and
chromatin states from ENCODE/Broad (15 states were grouped to highlight the predicted functional elements). As a reference genome, we used GRCh37. Quantile–quantile plots, Manhattan plots,
and the exploration of genomic inflation factors were performed using the R package qqman. POLYGENIC RISK SCORES We calculated a weighted individual PRS based on the 39 genetic variants that
showed GWS evidence of association with AD in the present study, excluding _APOE_ to check the impact of PRS modulating _APOE_ risk (Table 1 and Supplementary Data 3). The selected variants
were directly genotyped or imputed with high quality (median imputation score _R_² = 0.93). The PRSs were generated by multiplying the genotype dosage of each risk allele for each variant
by its respective weight and then summing across all variants. We weighted this by the effect size from previous IGAP studies [Kunkle et al.42 (36 variants), Sims et al.7 (2 variants), Jun
et al.43 (_MAPT_ locus), Supplementary Data 5]. The generated PRS was validated using logistic regression adjusted by four principal components in a sample of 676 AD cases diagnosed based on
clinical criteria and 332 pathologically confirmed AD cases from the European Alzheimer’s Disease Biobank–Fundació ACE/Barcelona Brain Bank dataset (EADB–F.ACE/BBB, Supplementary
Information). This dataset was not used in prior genetic studies. In this dataset, all pathologically confirmed cases were scored for the presence or absence of concomitant pathologies. In
all analyses, we compared the AD patients to the same control dataset (_n_ = 1386). We performed analyses to test the robustness of the PRS. We tested the effect of adding variants below the
genome-wide significance threshold using a pruning and thresholding approach. For this, we used the summary statistics of the IGAP42 study, and we selected independent variants using the
clump_data() function from the TwoSampleMR package (v0.4.25). We used strict settings for clumping (_R_2 = 0.001 and window = 1 MB) and increasing p value thresholds (>1 × 10−7, >1 ×
10−6, >1 × 10−5, >1 × 10−4, >1 × 10−3, and >1 × 10−2). We tested the association of the results with clinically diagnosed and pathologically confirmed AD patients. To evaluate
the effect of diagnostic certainty, we tested whether the PRS was different between the two patient groups. For the PRS with 39 GWS variants, we tested whether the PRS had sex-specific
effects, whether it resulted in different age-of-onset groups of AD, and the effect of the PRS in the presence of concomitant brain pathologies. _Risk stratification of the validated PRSs_.
We searched for the groups at the highest risk of AD in the GR@ACE dataset (6331 AD cases and 6055 controls). We stratified the population into PRS percentiles, taking into account survival
bias anticipated at old age18. To eliminate selection bias, we calculated the boundaries of the percentiles in the control participants aged 55 years and younger (_n_ = 3546). Based on the
boundaries from this population, the rest of the controls and all AD cases were then assigned into their appropriate percentiles. We first explored risk stratification using only the PRSs.
For this, we split the PRSs into 50 groups (2 percentiles) and compared all groups with that which had the lowest PRS. Second, we explored risk stratification considering both the _APOE_
genotypes and the PRSs. The _APOE_ genotypes were pooled in the analyses as _APOE_ ɛ2ɛ2/ɛ2ɛ3 (_n_ = 998, split into 7 PRS groups), _APOE_ ɛ3ɛ3 (_n_ = 7611, split into 25 PRS groups), _APOE_
ɛ2ɛ4/ɛ3ɛ4 (_n_ = 3399, split into 15 PRS groups), and _APOE_ ɛ4ɛ4 (_n_ = 382, split into 3 PRS groups). We studied the effect of PRS across groups of individuals stratified by the _APOE_
genotypes with the lowest-PRS group (_APOE_ as the reference group using logistic regression models adjusted for four population ancestry components). Finally, we compared the median AAO
using a Wilcoxon test. We implemented a Cox regression model on AAO in the GR@ACE/DEGESCO dataset case-only adjusted for covariates as _APOE_ group, the interaction between the PRS and
_APOE_ and four population ancestry components. All analyses were done in R (v3.4.2). FUNCTIONAL ANNOTATION We used Functional Mapping and Annotation of Genome-Wide Association Studies23
(FUMA, v1.3.4c) to interpret SNP-trait associations (see Supplementary Methods and Supplementary Data 15–18). FUMA is an online platform that annotates GWAS findings and prioritizes the most
likely causal SNPs and genes using information from 18 biological data repositories and tools. As input, we used the summary statistics of our meta-GWAS. Gene prioritization is based on a
combination of positional mapping, expression quantitative trait loci (eQTL) mapping, and chromatin interaction mapping. Functional annotation was performed by applying a methodology similar
to that described by Jansen et al.38. We referred to the original publication for details on the methods and repositories of FUMA23. REPORTING SUMMARY Further information on research design
is available in the Nature Research Reporting Summary linked to this article. DATA AVAILABILITY The discovery summary statistics of this study are publicly available in Fundació ACE server
[https://fundacioace-my.sharepoint.com/:u:/g/personal/iderojas_fundacioace_org/EaTwlPg9cRJHn7Kos4h39OUBaxajsjJHL_C110fC89bc8w?e=ZdcEUy]. CHANGE HISTORY * _ 09 FEBRUARY 2023 A Correction to
this paper has been published: https://doi.org/10.1038/s41467-023-36192-x _ REFERENCES * Pericak-Vance, M. A. et al. Linkage studies in familial Alzheimer disease: evidence for chromosome 19
linkage. _Am. J. Hum. Genet._ 48, 1034–1050 (1991). CAS PubMed PubMed Central Google Scholar * Seshadri, S. et al. Genome-wide analysis of genetic loci associated with Alzheimer
disease. _JAMA J. Am. Med. Assoc_. 303, 1832–1840 (2010). * Lambert, J. C. et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. _Nat.
Genet._ 45, 1452–1458 (2013). Article CAS PubMed PubMed Central Google Scholar * Jonsson, T. et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. _N. Engl. J. Med._
368, 107–116 (2013). Article CAS PubMed Google Scholar * Guerreiro, R. et al. TREM2 variants in Alzheimer’s disease. _N. Engl. J. Med_. 368, 117–127 (2013). Article CAS PubMed Google
Scholar * Jonsson, T. et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. _Nature_ 488, 96–99 (2012). Article ADS CAS PubMed Google Scholar
* Sims, R. et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. _Nat. Genet_. 49, 1373–1384 (2017). * Ridge, P. G. et
al. Assessment of the genetic variance of late-onset Alzheimer’s disease. _Neurobiol. Aging_ 41, 200.e13-200.e20 (2016). * 2020 Alzheimer’s disease facts and figures. _Alzheimer’s Dement_.
16 391–460 (2020). * Reiman, E. M. et al. Alzheimer’s prevention initiative: a plan to accelerate the evaluation of presymptomatic treatments. _J. Alzheimer’s Dis_ 26, 321–329 (2011).
Article Google Scholar * Huang, L.-K., Chao, S.-P. & Hu, C.-J. Clinical trials of new drugs for Alzheimer disease. _J. Biomed. Sci._ 27, 18 (2020). Article CAS PubMed PubMed Central
Google Scholar * Rodríguez-Rodríguez, E. et al. Genetic risk score predicting accelerated progression from mild cognitive impairment to Alzheimer’s disease. _J. Neural Transm._ 120,
807–812 (2013). Article PubMed Google Scholar * Adams, H. H. H. et al. Genetic risk of neurodegenerative diseases is associated with mild cognitive impairment and conversion to dementia.
_Alzheimer’s Dement_. https://doi.org/10.1016/j.jalz.2014.12.008 (2015). * Desikan, R. S. et al. Genetic assessment of age-associated Alzheimer disease risk: Development and validation of a
polygenic hazard score. _PLoS Med_. https://doi.org/10.1371/journal.pmed.1002258 (2017). * Lacour, A. et al. Genome-wide significant risk factors for Alzheimer’s disease: role in progression
to dementia due to Alzheimer’s disease among subjects with mild cognitive impairment. _Mol. Psychiatry_ 22, 153–160 (2016). Article PubMed PubMed Central Google Scholar * Sleegers, K.
et al. A 22-single nucleotide polymorphism Alzheimer’s disease risk score correlates with family history, onset age, and cerebrospinal fluid Aβ 42. _Alzheimer’s Dement_ 11, 1452–1460 (2015).
Article Google Scholar * Zhang, Q. et al. Risk prediction of late-onset Alzheimer’s disease implies an oligogenic architecture. _Nat. Commun_. https://doi.org/10.1038/s41467-020-18534-1
(2020). * van der Lee, S. J. et al. The effect of APOE and other common genetic variants on the onset of Alzheimer’s disease and dementia: a community-based cohort study. _Lancet Neurol_.
https://doi.org/10.1016/S1474-4422(18)30053-X (2018). * Moreno-Grau, S. et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with
Alzheimer’s disease and three causality networks: The GR@ACE project. _Alzheimer’s Dement_. 0, 1333–1347 (2019). * Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s disease
identifies new risk loci and implicates AmyloidBeta, tau, immunity and lipid processing. _Nat. Genet._ 51, 414–430 (2019). Article CAS PubMed PubMed Central Google Scholar * Marioni,
R. E. et al. GWAS on family history of Alzheimer’s disease. _Transl. Psychiatry_ https://doi.org/10.1038/s41398-018-0150-6 (2018). * Willer, C. J., Li, Y. & Abecasis, G. R. METAL: Fast
and efficient meta-analysis of genomewide association scans. _Bioinformatics_ https://doi.org/10.1093/bioinformatics/btq340 (2010) . * Watanabe, K., Taskesen, E., van Bochoven, A. &
Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. _Nat. Commun_. https://doi.org/10.1038/s41467-017-01261-5 (2017). * Khera, A. V. et al. Genome-wide
Polygenic Scores for Common Diseases Identify Individuals With Risk Equivalent to Monogenic Mutations. _Nat. Genet_. 50, 1219–1224 (2018). * Lonsdale, J. et al. The genotype-tissue
expression (GTEx) project. _Nat. Genet._ 45, 580–585 (2013). Article CAS Google Scholar * Myrum, C., Nikolaienko, O., Bramham, C. R., Haavik, J. & Zayats, T. Implication of the APP
gene in intellectual abilities. _J. Alzheimer’s Dis_ 59, 723–735 (2017). Article CAS Google Scholar * Cenini, G. et al. Wild type but not mutant APP is involved in protective adaptive
responses against oxidants. _Amino Acids_ 39, 271–283 (2010). Article CAS PubMed Google Scholar * Egan, M. F. et al. Randomized trial of verubecestat for mild-to-moderate Alzheimer’s
disease. _N. Engl. J. Med._ 378, 1691–1703 (2018). Article CAS PubMed PubMed Central Google Scholar * Panza, F., Lozupone, M. & Watling, M. & Imbimbo, B. P. Do BACE inhibitor
failures in Alzheimer patients challenge the amyloid hypothesis of the disease? _Expert Rev. Neurother._ 19, 599–602 (2019). * Hefter, D. & Draguhn, A. APP as a protective factor in
acute neuronal insults. _Front. Mol. Neurosci._ 10, 22 (2017). Article PubMed PubMed Central Google Scholar * Lancour, D. et al. One for all and all for One: Improving replication of
genetic studies through network diffusion. _PLoS Genet_ 14, e1007306 (2018). Article PubMed PubMed Central Google Scholar * Asanomi, Y. et al. A rare functional variant of SHARPIN
attenuates the inflammatory response and associates with increased risk of late-onset Alzheimer’s disease. _Mol. Med._ 25, 20 (2019). Article PubMed PubMed Central Google Scholar * Ruiz,
A. et al. Assessing the role of the TREM2 p.R47H variant as a risk factor for Alzheimer’s disease and frontotemporal dementia. _Neurobiol. Aging_ 35, 444.e1–444.e4 (2014). Article CAS
PubMed Google Scholar * McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. _Nat. Genet._ 48, 1279–1283 (2016). Article CAS PubMed PubMed Central Google
Scholar * 1000 Genomes Project Consortium. et al. A global reference for human genetic variation. _Nature_ 526, 68–74 (2015). Article Google Scholar * Sudlow, C. et al. UK Biobank: an
open access resource for identifying the causes of a wide range of complex diseases of middle and old Age. _PLOS Med._ 12, e1001779 (2015). Article PubMed PubMed Central Google Scholar *
Liu, J. Z., Erlich, Y. & Pickrell, J. K. Case-control association mapping by proxy using family history of disease. _Nat. Genet._ 49, 325–331 (2017). Article CAS PubMed Google
Scholar * Jansen, I. E. et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. _Nat. Genet._ 51, 404–413 (2019). Article CAS
PubMed PubMed Central Google Scholar * Lloyd-Jones, L. R., Robinson, M. R., Yang, J. & Visscher, P. M. Transformation of summary statistics from linear mixed model association on
all-or-none traits to odds ratio. _Genetics_ 208, 1397–1408 (2018). Article CAS PubMed PubMed Central Google Scholar * Jun, G. et al. Comprehensive search for Alzheimer disease
susceptibility loci in the APOE region. _Arch. Neurol._ 69, 1270–1279 (2012). Article PubMed PubMed Central Google Scholar * Bulik-Sullivan, B. et al. LD score regression distinguishes
confounding from polygenicity in genome-wide association studies. _Nat. Genet_. https://doi.org/10.1038/ng.3211 (2015). * Kunkle, B. W. et al. Genetic meta-analysis of diagnosed Alzheimer’s
disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. _Nat. Genet._ 51, 414–430 (2019). Article CAS PubMed PubMed Central Google Scholar * Jun, G. et
al. A novel Alzheimer disease locus located near the gene encoding tau protein. _Mol. Psychiatry_ 21, 108–117 (2016). Article MathSciNet CAS PubMed Google Scholar Download references
ACKNOWLEDGEMENTS We would like to thank patients and controls who participated in this project. The present work has been performed as part of the doctoral program of I. de Rojas at the
Universitat de Barcelona (Barcelona, Spain) supported by national grant from the Instituto de Salud Carlos III FI20/00215. The Genome Research @ Fundació ACE project (GR@ACE) is supported by
Grifols SA, Fundación bancaria “La Caixa”, Fundació ACE, and CIBERNED. A.R. and M.B. receive support from the European Union/EFPIA Innovative Medicines Initiative Joint undertaking ADAPTED
and MOPEAD projects (grant numbers 115975 and 115985, respectively). M.B. and A.R. are also supported by national grants PI13/02434, PI16/01861, PI17/01474, PI19/01240 and PI19/01301. Acción
Estratégica en Salud is integrated into the Spanish National R + D + I Plan and funded by ISCIII (Instituto de Salud Carlos III)—Subdirección General de Evaluación and the Fondo Europeo de
Desarrollo Regional (FEDER—“Una manera de hacer Europa”). Some control samples and data from patients included in this study were provided in part by the National DNA Bank Carlos III
(www.bancoadn.org, University of Salamanca, Spain) and Hospital Universitario Virgen de Valme (Sevilla, Spain); they were processed following standard operating procedures with the
appropriate approval of the Ethical and Scientific Committee. Amsterdam dementia Cohort (ADC): Research of the Alzheimer center Amsterdam is part of the neurodegeneration research program of
Amsterdam Neuroscience. The Alzheimer Center Amsterdam is supported by Stichting Alzheimer Nederland and Stichting VUmc fonds. The clinical database structure was developed with funding
from Stichting Dioraphte. Genotyping of the Dutch case-control samples was performed in the context of EADB (European Alzheimer DNA biobank) funded by the JPco-fuND FP-829-029 (ZonMW project
number 733051061). 100-Plus study: We are grateful for the collaborative efforts of all participating centenarians and their family members and/or relations. This work was supported by
Stichting Alzheimer Nederland (WE09.2014-03), Stichting Diorapthe, horstingstuit foundation, Memorabel (ZonMW project number 733050814, 733050512) and Stichting VUmc Fonds. Genotyping of the
100-Plus Study was performed in the context of EADB (European Alzheimer DNA biobank) funded by the JPco-fuND FP-829-029 (ZonMW project number 733051061). Longitudinal Aging Study Amsterdam
(LASA) is largely supported by a grant from the Netherlands Ministry of Health, Welfare and Sports, Directorate of Long-Term Care. The authors are grateful to all LASA participants, the
fieldwork team and all researchers for their ongoing commitment to the study. This work was supported by a grant (European Alzheimer DNA BioBank, EADB) from the EU Joint
Program—Neurodegenerative Disease Research (JPND) and also funded by Inserm, Institut Pasteur de Lille, the Lille Métropole Communauté Urbaine, the French government’s LABEX DISTALZ program
(development of innovative strategies for a transdisciplinary approach to AD). Genotyping of the German case-control samples was performed in the context of EADB (European Alzheimer DNA
biobank) funded by the JPco-fuND (German Federal Ministry of Education and Research, BMBF: 01ED1619A). Full acknowledgments for the studies that contributed data can be found in the
Supplementary Note. We thank the numerous participants, researchers, and staff from many studies who collected and contributed to the data. We thank the International Genomics of Alzheimer’s
Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not
participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was
funded by the French National Foundation on AD and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm,
Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer’s Research UK (Grant n°
503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420.
CHARGE was partly supported by the NIA/NHLBI grants AG049505, AG058589, HL105756 and AGES contract N01–AG–12100, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus
University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728. This research has been conducted using
the UK Biobank public resource obtained through the University of Edinburg Data Share (https://datashare.is.ed.ac.uk/handle/10283/3364). AUTHOR INFORMATION Author notes * These authors
contributed equally: Itziar de Rojas, Sonia Moreno-Grau, Niccolo Tesi, Benjamin Grenier-Boley, Victor Andrade, Iris E. Jansen. * These authors jointly supervised this work: Jordi Clarimón,
Mercè Boada, Wiesje M. van der Flier, Alfredo Ramirez, Jean-Charles Lambert, Sven J. van der Lee, Agustín Ruiz. AUTHORS AND AFFILIATIONS * Research Center and Memory clinic Fundació ACE,
Institut Català de Neurociències Aplicades, Universitat Internacional de Catalunya, Barcelona, Spain Itziar de Rojas, Sonia Moreno-Grau, Isabel Hernández, Laura Montrreal, Pablo
García-González, Emilio Alarcón-Martín, Sergi Valero, Oscar Sotolongo-Grau, Alba Benaque, Alba Pérez-Cordón, Ana Espinosa, Ángela Sanabria, Carla Abdelnour, Gemma Ortega, Maitée
Rosende-Roca, Montserrat Alegret, Natalia Roberto, Silvia Gil, C. Abdelnour, N. Aguilera, E. Alarcon, M. Alegret, A. Benaque, M. Boada, M. Buendia, P. Cañabate, I. de Rojas, S. Diego, A.
Espinosa, A. Gailhajenet, P. García-González, S. Gil, M. Guitart, I. Hernández, M. Ibarria, A. Lafuente, E. Martín, M. Marquié, A. Mauleón, L. Montrreal, S. Moreno-Grau, M. Moreno, A.
Orellana, G. Ortega, A. Pancho, E. Pelejá, A. Pérez-Cordon, S. Preckler, M. Rosende-Roca, A. Ruiz, A. Sanabria, O. Sotolongo-Grau, L. Tárraga, S. Valero, L. Vargas, E. Alarcón-Martín, M.
Boada, I. de Rojas, I. Hernández, M. Marquié, A. Ruiz, S. Valero, Marta Marquié, Adelina Orellana, Lluís Tárraga, Mercè Boada & Agustín Ruiz * CIBERNED, Network Center for Biomedical
Research in Neurodegenerative Diseases, National Institute of Health Carlos III, Madrid, Spain Itziar de Rojas, Sonia Moreno-Grau, Isabel Hernández, Miguel Calero, Rafael Blesa, Laura
Cervera-Carles, Ana Frank-García, Fermin Moreno, Carmen Lage, Sergi Valero, Alberto Rábano, Ana Espinosa, Angel Martín Montes, Ángela Sanabria, Carla Abdelnour, Dolores Buiza-Rueda, Daniel
Alcolea, Eloy Rodriguez-Rodriguez, Gemma Ortega, Juan Fortea, Maitée Rosende-Roca, Miren Zulaica, Montserrat Alegret, Sara López-García, C. Abdelnour, M. Alegret, M. Boada, P. Cañabate, A.
Espinosa, I. Hernández, S. Moreno-Grau, G. Ortega, A. Ruiz, A. Sanabria, L. Tárraga, S. Valero, A. D. Adarmes-Gómez, R. Blesa, M. Boada, D. Buiza-Rueda, M. J. Bullido, M. Calero, F.
Carrillo, M. Carrión-Claro, J. Clarimón, I. de Rojas, R. Escuela, L. Garrote-Espina, J. Fortea, A. Frank-García, P. Gómez-Garre, I. Hernández, S. Jesús, M. A. Labrador Espinosa, C. Lage, A.
Lleó, S. López-García, D. Macias-García, M. Marquié, A. Martín Montes, M. Medina, P. Mir, J. Pérez Tur, T. Periñán-Tocino, R. Pineda-Sanchez, G. Piñol-Ripoll, A. Rábano, E.
Rodríguez-Rodríguez, A. Ruiz, P. Sánchez-Juan, I. Sastre, S. Valero, R. Vigo-Ortega, Pascual Sánchez-Juan, Jordi Pérez-Tur, Adolfo Lopez de Munain, Jose María García-Alberca, María J.
Bullido, Alberto Lleó, Pablo Mir, Miguel Medina, Marta Marquié, Adelina Orellana, Lluís Tárraga, Jordi Clarimón, Mercè Boada & Agustín Ruiz * Alzheimer Center Amsterdam, Department of
Neurology, Amsterdam Neuroscience, Vrije Universiteit Amsterdam, Amsterdam UMC, Amsterdam, The Netherlands Niccolo Tesi, Iris E. Jansen, Afina W. Lemstra, Marc Hulsman, Yolande A. L.
Pijnenburg, Iris E. Jansen, Philip Scheltens, Henne Holstege, Wiesje M. van der Flier & Sven J. van der Lee * Section Genomics of Neurodegenerative Diseases and Aging, Department of
Clinical Genetics, Vrije Universiteit Amsterdam, Amsterdam UMC, Amsterdam, The Netherlands Niccolo Tesi, Marc Hulsman, Henne Holstege & Sven J. van der Lee * Delft Bioinformatics Lab,
Delft Univeristy of Technology, Delft, The Netherlands Niccolo Tesi * Univ. Lille, Inserm, Institut Pasteur de Lille, CHU Lille, U1167-Labex DISTALZ-RID-AGE-Risk Factors and Molecular
Determinants of Aging-Related Diseases, Lille, France Benjamin Grenier-Boley, Céline Bellenguez, A. Corbatón-Anchuelo, Philippe Amouyel & Jean-Charles Lambert * Division of Neurogenetics
and Molecular Psychiatry, Department of Psychiatry and Psychotherapy, University of Cologne, Medical Faculty, Cologne, Germany Victor Andrade, Luca Kleineidam, Maria Carolina Dalmasso &
Alfredo Ramirez * Department of Neurodegenerative diseases and Geriatric Psychiatry, University Clinic Bonn, Bonn, Germany Victor Andrade, Anja Schneider, Klaus Fließbach, Luca Kleineidam,
Michael T. Heneka, Michael Wagner, Wolfgang Maier, L. M. Real & Alfredo Ramirez * Department of Complex Trait Genetics, Center for Neurogenomics and Cognitive Research, Amsterdam
Neuroscience, VU University, Amsterdam, The Netherlands Iris E. Jansen, Iris E. Jansen & Danielle Posthuma * Department of Medical Epidemiology and Biostatistics, Karolinska Institutet,
Stockholm, Sweden Nancy L. Pedersen, Ida K. Karlsson & Sara Hägg * Amsterdam UMC-Vrije Universiteit Amsterdam, Department of Epidemiology and Data Science, Amsterdam Public Health
Research Institute, Amsterdam, The Netherlands Najada Stringa, Almar A. L. Kok, Natasja M. van Schoor & Martijn Huisman * Neuropsychiatric Epidemiology Unit, Department of Psychiatry and
Neurochemistry, Institute of Neuroscience and Physiology, Sahlgrenska Academy, Centre for Ageing and Health (AgeCap), University of Gothenburg, Gothenburg, Sweden Anna Zettergren, Ingmar
Skoog, Nazib M. Seidu & Silke Kern * Unidad de Demencias, Hospital Clínico Universitario Virgen de la Arrixaca, Murcia, Spain Carmen Antúnez, M. Antequera, C. Antúnez, A. Legaz, J.
Marín-Muñoz, B. Martínez, V. Martínez, M. P. Vicente & L. Vivancos * Alzheimer’s disease and other cognitive disorders unit. Service of Neurology, Hospital Clínic of Barcelona. Institut
d’Investigacions Biomèdiques August Pi i Sunyer, University of Barcelona, Barcelona, Spain Anna Antonell, Laura Molina-Porcel, R. Sanchez del Valle Díaz & Raquel Sánchez-Valle *
Mathematics and Statistics, Murdoch University, Perth, WA, Australia Rick M. Tankard & Nicola J. Armstrong * Cardiovascular Health Research Unit, Department of Medicine, University of
Washington, Seattle, WA, USA Joshua C. Bis * Division of Psychological Medicine and Clinial Neurosciences, MRC Centre for Neuropsychiatric Genetics and Genomics, Cardiff University, Cardiff,
UK Rebecca Sims, Peter A. Holmans, Rachel Marshall, Valentina Escott-Price, Catherine Bresner, Detelina Grozeva, Eftychia Bellou, Ewen W. Sommerville, Ganna Leonenko, Janet Harwood, Lauren
Luckuck, Salha Saad & Julie Williams * Grupo de Medicina Xenómica, Centro Nacional de Genotipado (CEGEN-PRB3-ISCIII), Universidade de Santiago de Compostela, Santiago de Compostela,
Spain Inés Quintela, A. Carracedo, O. Maroñas & Ángel Carracedo * CAEBI, Centro Andaluz de Estudios Bioinformáticos, Sevilla, Spain Antonio González-Perez, A. González-Pérez, M. E. Sáez
& María Eugenia Sáez * UFIEC, Instituto de Salud Carlos III, Madrid, Spain Miguel Calero * CIEN Foundation/Queen Sofia Foundation Alzheimer Center, Madrid, Spain Miguel Calero, Alberto
Rábano, Ana Belén Pastor, M. Calero, A. Rábano & Miguel Medina * Unidad de Demencias, Servicio de Neurología y Neurofisiología, Instituto de Biomedicina de Sevilla (IBiS), Hospital
Universitario Virgen del Rocío/CSIC/Universidad de Sevilla, Sevilla, Spain Emilio Franco-Macías, María Bernal Sánchez-Arjona, A. D. Adarmes-Gómez, M. Bernal, D. Buiza-Rueda, F. Carrillo, M.
Carrión-Claro, R. Escuela, L. Garrote-Espina, E. Franco-Macías, P. Gómez-Garre, S. Jesús, M. A. Labrador Espinosa, D. Macias-García, M. Marín, P. Mir, T. Periñán-Tocino, R. Pineda-Sanchez,
S. Rodrigo & R. Vigo-Ortega * Unidad Clínica de Enfermedades Infecciosas y Microbiología, Hospital Universitario de Valme, Sevilla, Spain Juan Macías, Anaïs Corma-Gómez, Juan A. Pineda,
Marta Fernández-Fuertes, J. Macias, J. A. Pineda & Luis M. Real * Department of Neurology, II B Sant Pau, Hospital de la Santa Creu i Sant Pau, Universitat Autònoma de Barcelona,
Barcelona, Spain Rafael Blesa, Laura Cervera-Carles, Daniel Alcolea, Juan Fortea, R. Blesa, J. Clarimón, J. Fortea, A. Lleó, Alberto Lleó & Jordi Clarimón * Servicio de Neurología,
Hospital Universitario Central de Asturias, Oviedo, Spain Manuel Menéndez-González & M. Menéndez-González * Instituto de Investigación Sanitaria del Principado de Asturias (ISPA),
Oviedo, Spain Manuel Menéndez-González, Carmen Martínez Rodríguez, Irene Rosas Allende, Carmen Martínez, V. Álvarez, C. Martínez, M. Menéndez-González & Victoria Álvarez * Departamento
de Medicina, Universidad de Oviedo, Oviedo, Spain Manuel Menéndez-González * Department of Neurology, La Paz University Hospital, Instituto de Investigación Sanitaria del Hospital
Universitario La Paz, IdiPAZ, Madrid, Spain Ana Frank-García * Hospital La Paz Institute for Health Research, IdiPAZ, Madrid, Spain Ana Frank-García, Angel Martín Montes, A. Frank-García
& A. Martín Montes * Universidad Autónoma de Madrid, Madrid, Spain Ana Frank-García & María J. Bullido * Departamento de Especialidades Quirúrgicas, Bioquímicas e Inmunología, School
of Medicine, University of Málaga, Málaga, Spain Jose Luís Royo, Emilio Alarcón-Martín, E. Alarcon, I. Quintela, L. M. Real, E. Alarcón-Martín, J. M. Cruz-Gamero & J. L. Royo *
Department of Neurology, Hospital Universitario Donostia, San Sebastian, Spain Fermin Moreno, A. Lopez de Munain & Adolfo Lopez de Munain * Neurosciences Area, Instituto Biodonostia, San
Sebastian, Spain Fermin Moreno, Miren Zulaica, A. Lopez de Munain & Adolfo Lopez de Munain * Unitat Trastorns Cognitius, Hospital Universitari Santa Maria de Lleida, Lleida, Spain
Raquel Huerto Vilas, Alfonso Arias Pastor, G. Piñol-Ripoll & Gerard Piñol-Ripoll * Institut de Recerca Biomedica de Lleida (IRBLLeida), Lleida, Spain Raquel Huerto Vilas, Alfonso Arias
Pastor, G. Piñol-Ripoll & Gerard Piñol-Ripoll * Servei de Neurologia, Hospital Universitari i Politècnic La Fe, Valencia, Spain Miquel Baquero, M. Baquero & J. A. Burguera * Fundació
Docència i Recerca MútuaTerrassa, Terrassa, Barcelona, Spain Mónica Diez-Fairen, Ignacio Alvarez, I. Álvarez, M. Diez-Fairen, P. Pastor & Pau Pastor * Memory Disorders Unit, Department
of Neurology, Hospital Universitari Mutua de Terrassa, Terrassa, Barcelona, Spain Mónica Diez-Fairen, Ignacio Alvarez, I. Álvarez, M. Diez-Fairen, P. Pastor & Pau Pastor * Neurology
Service, Marqués de Valdecilla University Hospital (University of Cantabria and IDIVAL), Santander, Spain Carmen Lage, Eloy Rodriguez-Rodriguez, Sara López-García, C. Lage, S. López-García,
E. Rodríguez-Rodríguez, P. Sánchez-Juan & Pascual Sánchez-Juan * Hospital Universitario Ramon y Cajal, IRYCIS, Madrid, Spain Sebastián García-Madrona, M. Calero, S. Garcia Madrona, G.
Garcia-Ribas & Guillermo Garcia-Ribas * Karolinska Institutet, Center for Alzheimer Research, Department NVS, Division of Neurogeriatrics, Stockholm, Sweden Abbe Ullgren, Anne Kinhult
Ståhlbom, Caroline Graff, Charlotte Johansson & Håkan Thonberg * Unit for Hereditary Dementias, Theme Aging, Karolinska University Hospital-Solna, Stockholm, Sweden Abbe Ullgren, Anne
Kinhult Ståhlbom, Caroline Graff, Charlotte Johansson & Håkan Thonberg * Department of Biostatistics, Epidemiology and Informatics, University of Pennsylvania Perelman School of
Medicine, Philadelphia, PA, USA Adam C. Naj * Penn Neurodegeneration Genomics Center, Department of Pathology and Laboratory Medicine, University of Pennsylvania Perelman School of Medicine,
Philadelphia, PA, USA Adam C. Naj, Li-San Wang & Gerard D. Schellenberg * Centre for Neurodegenerative Disorders, Department of Clinical and Experimental Sciences, University of
Brescia, Brescia, Italy Alberto Benussi, Alessandro Padovani & Barbara Borroni * BT-CIEN, Madrid, Spain Alberto Rábano, Ana Belén Pastor & A. Rábano * Department of Biomedical
Sciences, Section of Neuroscience and Clinical Pharmacology, University of Cagliari, Cagliari, Italy Alessio Squassina, Claudia Pisanu & Maria Del Zompo * Faculty of Medicine, University
of Lisbon, Lisbon, Portugal Alexandre de Mendonça * Amsterdam UMC, Vrije Universiteit Amsterdam, Department of Psychiatry, Amsterdam Public Health Research Institute, Amsterdam, The
Netherlands Almar A. L. Kok * UK Dementia Research Institute at Cardiff, Cardiff University, Cardiff, UK Alun Meggy, Georgina Menzies & Julie Williams * Department of Neurology, La Paz
University Hospital, Madrid, Spain Angel Martín Montes * Department of Neurology, Boston University School of Medicine, Boston, MA, USA Anita L. DeStefano, Claudia L. Satizabal & Sudha
Seshadri * Department of Biostatistics, Boston University School of Public Health, Boston, MA, USA Anita L. DeStefano & Qiong Yang * German Center for Neurodegenerative Diseases (DZNE),
Bonn, Germany Anja Schneider, Annika Spottke, Frank Jessen, Matthias Schmid, Michael T. Heneka, Michael Wagner, Wolfgang Maier, Henning Boecker & Alfredo Ramirez * A.I Virtanen Institute
for Molecular Sciences, University of Eastern Finland, Kuopio, Finland Annakaisa Haapasalo * Department of Clinical Biochemistry, Rigshospitalet, Copenhagen, Denmark Anne Tybjærg-Hansen,
Jesper Qvist Thomassen & Ruth Frikke-Schmidt * Department of Clinical Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark Anne Tybjærg-Hansen,
Børge G. Nordestgaard & Ruth Frikke-Schmidt * Martin-Luther-University Halle-Wittenberg, University Clinic and Outpatient Clinic for Psychiatry, Psychotherapy and Psychosomatics, Halle
(Saale), Germany Annette M. Hartmann, Dan Rujescu & Ina Giegling * Department of Neurology, University of Bonn, Bonn, Germany Annika Spottke * Instituto de Investigación Sanitaria,
Hospital Clínico San Carlos (IdISSC), Madrid, Spain Arturo Corbatón-Anchuelo, María Teresa Martínez-Larrad & Manuel Serrano-Ríos * Spanish Biomedical Research Centre in Diabetes and
Associated Metabolic Disorders (CIBERDEM), Madrid, Spain Arturo Corbatón-Anchuelo, María Teresa Martínez-Larrad, M. Serrano-Rios & Manuel Serrano-Ríos * Haugesund Hospital, Helse Fonna,
Department of Research and Innovation, Haugesund, Norway Arvid Rongve, M. T. Martínez & Arvid Rongve * University of Bergen, Institute of Clinical Medicine (K1), Bergen, Norway Arvid
Rongve * Department of Clinical Sciences and Community Health, University of Milan, Milan, Italy Beatrice Arosio * Geriatic Unit, Fondazione Cà Granda, IRCCS Ospedale Maggiore Policlinico,
Milan, Italy Beatrice Arosio, Evelyn Ferri, Paolo Dionigi Rossi & Simona Ciccone * Department of Neuroscience, Psychology, Drug Research and Child Health University of Florence,
Florence, Italy Benedetta Nacmias, Sandro Sorbi, Valentina Bessi, Camilla Ferrari, Irene Piaceri & Silvia Bagnoli * IRCCS Fondazione Don Carlo Gnocchi, Florence, Italy Benedetta Nacmias
& Sandro Sorbi * Department of Clinical Biochemistry, Herlev Gentofte Hospital, Herlev, Denmark Børge G. Nordestgaard * Dr. John T. Macdonald Foundation Department of Human Genetics,
University of Miami Miller School of Medicine, Miami, FL, USA Brian W. Kunkle * John P. Hussman Institute for Human Genomics, University of Miami Miller School of Medicine, Miami, FL, USA
Brian W. Kunkle & Margaret A. Pericak-Vance * Normandie Univ, UNIROUEN, Inserm U1245, CHU Rouen, Department of Genetics and CNR-MAJ, FHU G4 Génomique, F-76000 Rouen, France Camille
Charbonnier & Olivier Quenez * Institute of Neurology, Catholic University of the Sacred Heart, School of Medicine, Milan, Italy Carlo Masullo * Hospital de Cabueñes, Gijón, Spain Carmen
Martínez Rodríguez & Gaël Nicolas * Servicio de Neurología, Hospital Universitario de Gran Canaria Dr.Negrín, Las Palmas, Spain Carmen Muñoz-Fernandez * Inserm, Bordeaux Population
Health Research Center, UMR 1219, Univ. Bordeaux, ISPED, CIC 1401-EC, Univ Bordeaux, Bordeaux, France Carole Dufouil & Geneviève Chêne * CHU de Bordeaux, Pole de Santé Publique,
Bordeaux, France Carole Dufouil & Geneviève Chêne * Instituto de Medicina Molecular João lobo Antunes, Faculdade de Medicina, Universidade de Lisboa, Lisboa, Portugal Catarina B.
Ferreira * Unit of Clinical Pharmacology, University Hospital of Cagliari, Cagliari, Italy Caterina Chillotti * Department of Psychology, University of California—Riverside, Riverside, CA,
USA Chandra A. Reynolds * University of Milan, Dino Ferrari Center, Milan, Italy Chiara Fenoglio, Daniela Galimberti, Elio Scarpini & Maria Serpente * VIB Center for Molecular Neurology,
Antwerp, Belgium Christine Van Broeckhoven, Fahri Küçükali, Jasper Van Dongen & Kristel Sleegers * Laboratory of Neurogenetics, Institute Born-Bunge, Antwerp, Belgium Christine Van
Broeckhoven, Fahri Küçükali, Jasper Van Dongen & Kristel Sleegers * Department of Biomedical Sciences, University of Antwerp., Antwerp, Belgium Christine Van Broeckhoven, Fahri Küçükali
& Kristel Sleegers * Insititute for Regenerative Medicine, University of Zürich, Zürich, Switzerland Christopher Clark * Glenn Biggs Institute for Alzheimer’s and Neurodegenerative
Diseases, San Antonio, TX, USA Claudia L. Satizabal, Xueqiu Jian & Sudha Seshadri * Department of Population Health Sciences, UT Health San Antonio, San Antonio, TX, USA Claudia L.
Satizabal * Division of Clinical Neurosciences, School of Medicine, University of Southampton, Southampton, UK Clive Holmes * Unidad de Trastornos del Movimiento, Servicio de Neurología y
Neurofisiología, Instituto de Biomedicina de Sevilla (IBiS), Hospital Universitario Virgen del Rocío/CSIC/Universidad de Sevilla, Sevilla, Spain Dolores Buiza-Rueda & Pablo Mir *
Department of Old Age Psychiatry, Institute of Psychiatry, Psychology & Neuroscience, King’s College London, London, UK Dag Aarsland * Centre of Age-Related Medicine, Stavanger
University Hospital, Stavanger, Norway Dag Aarsland * Fondazione IRCCS Ca’ Granda, Ospedale Policlinico, Milan, Italy Daniela Galimberti, Elio Scarpini & Marina Arcaro * Normandie Univ,
UNIROUEN, Inserm U1245, CHU Rouen, Department of Neurology and CNR-MAJ, FHU G4 Génomique, F-76000 Rouen, France David Wallon * Complex Structure of Geriatrics, Department of Medical Sciences
Fondazione IRCCS Casa Sollievo della Sofferenza, San Giovanni Rotondo (FG), Italy Davide Seripa * Department of Child and Adolescent Psychiatry and Psychotherapy, Psychiatric University
Hospital Zurich (PUK), University of Zurich, Zurich, Switzerland Edna Grünblatt * Neuroscience Center Zurich, University of Zurich and ETH Zurich, Zurich, Switzerland Edna Grünblatt * Zurich
Center for Integrative Human Physiology, University of Zurich, Zurich, Switzerland Edna Grünblatt * School of Medicine, University of Thessaly, Larissa, Greece Efthimios Dardiotis * German
Center for Neurodegenerative Diseases (DZNE), Magdeburg, Germany Emrah Düzel * Institute of Cognitive Neurology and Dementia Research (IKND), Otto-von-Guericke University, Magdeburg, Germany
Emrah Düzel * School of Medicine and Surgery, University of Milano-Bicocca and Milan Center for Neuroscience, Milan, Italy Elisa Conti, Ildebrando Appollonio, Lucio Tremolizzo, Carlo
Ferrarese, Chiara Paola Zoia, Gessica Sala & Simona Andreoni * Department of Neuroscience and Mental Health, AOU Città della Salute e della Scienza di Torino, Torino, Italy Elisa Rubino
* Neurological Tissue Bank of the Biobanc-Hospital Clinic-IDIBAPS, Institut d’Investigacions Biomèdiques August Pi i Sunyer, Barcelona, Spain Ellen Gelpi & Laura Molina-Porcel * Division
of Neuropathology and Neurochemistry, Department of Neurology, Medical University of Vienna, Vienna, Austria Ellen Gelpi * APHP, Hôpital Brousse, equipe INSERM 1178, MOODS, Villejuif,
France Emmanuelle Duron * Université Paris-Saclay, UVSQ, Inserm, CESP, Team MOODS, Le Kremlin-Bicêtre, Paris, France Emmanuelle Duron * APHP, Hôpital Broca, Paris, France Emmanuelle Duron,
Henri Bailly, Jean-Sebastien Vidal & Oliver Hanon * School of Public Health, Human Genetics Center, University of Texas Health Science Center at Houston, Houston, TX, USA Eric Boerwinkle
* Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX, USA Eric Boerwinkle * Fondazione IRCCS Istituto Neurologico Carlo Besta, Milan, Italy Fabrizio Tagliavini,
Giacomina Rossi & Giorgio Giaccone * Inserm U1172, CHU, DISTAlz, LiCEND, Univ Lille, Lille, France Florence Pasquier, Thibaud Lebouvier & Vincent Deramecourt * CHU CNR-MAJ, Lille,
France Florence Pasquier, Thibaud Lebouvier & Vincent Deramecourt * Servicio de Inmunología, Hospital Universitario de Gran Canaria Dr. Negrín, Las Palmas de Gran Canaria, Spain
Florentino Sanchez-Garcia & M. Candida Deniz-Naranjo * Division of Clinical Geriatrics, Center for Alzheimer Research, Department of Neurobiology, Care Sciences and Society (NVS),
Karolinska Institutet, Stockholm, Sweden Francesca Mangialasche, Miia Kivipelto, Tiia Ngandu & Alina Solomon * Department of Psychiatry and Psychotherapy, University of Cologne, Medical
Faculty, Cologne, Germany Frank Jessen * Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Cologne, Germany Frank Jessen *
Norwegian National Advisory Unit on Ageing and Health, Vestfold Hospital Trust, Tønsberg, Norway Geir Selbæk * Department of Geriatric Medicine, Oslo University Hospital, Oslo, Norway Geir
Selbæk * Institute of Clinical Medicine, University of Oslo, Oslo, Norway Geir Selbæk * Department of Neurology, Medical School, University of Cyprus, Nicosia, Cyprus Georgios Hadjigeorgiou
* Laboratory of Neuropsychiatry, IRCCS Santa Lucia Foundation, Rome, Italy Gianfranco Spalletta, Nerisa Banaj, Fabrizio Piras, Federica Piras, Francesca Assogna & Valentina Ciullo * Beth
K. and Stuart C. Yudofsky Division of Neuropsychiatry, Department of Psychiatry and Behavioral Sciences, Baylor College of Medicine, Houston, TX, USA Gianfranco Spalletta * Aging Research
Center, Department of Neurobiology, Care Sciences and Society, Karolinska Institutet and Stockholm University, Stockholm, Sweden Giulia Grande & Goran Papenberg * MAC—Memory Clinic,
IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy Giuliano Binetti * Molecular Markers Laboratory, IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli,
Brescia, Italy Giuliano Binetti, Luisa Benussi, Roberta Ghidoni & Silvia Fostinelli * Sorbonne University, GRC n° 21, Alzheimer Precision Medicine (APM), AP-HP, Pitié-Salpêtrière
Hospital, Paris, France Harald Hampel * EA 4468, Sorbonne Paris Cité, Université Paris Descartes, Paris, France Henri Bailly, Jean-Sebastien Vidal & Oliver Hanon * Clinical
Neurochemistry Laboratory, Sahlgrenska University Hospital, Mölndal, Sweden Henrik Zetterberg & Kaj Blennow * Department of Psychiatry and Neurochemistry, Institute of Neuroscience and
Physiology, Sahlgrenska Academy at the University of Gothenburg, Gothenburg, Sweden Henrik Zetterberg & Kaj Blennow * Department of Neurodegenerative Disease, UCL Institute of Neurology,
London, UK Henrik Zetterberg * UK Dementia Research Institute at UCL, London, UK Henrik Zetterberg * Institute of Clinical Medicine Neurology, University of Eastern Finland, Kuopio, Finland
Hilkka Soininen, Jenni Lehtisalo & Alina Solomon * Neurocenter, neurology, Kuopio University Hospital, Kuopio, Finland Hilkka Soininen * Institute for Gerontology and Aging Research
Network—Jönköping (ARN-J), School of Health and Welfare, Jönköping University, Jönköping, Sweden Ida K. Karlsson * Neurology Unit, ‘San Gerardo’ hospital, Monza, Italy Ildebrando Appollonio,
Lucio Tremolizzo & Carlo Ferrarese * Department of Geriatrics, Clinic of Medicine, St Olavs Hospital, University Hospital of Trondheim, Trondheim, Norway Ingvild Saltvedt * Department
of Neuromedicine and Movement Science, Norwegian University of Science and Technhology (NTNU), Trondheim, Norway Ingvild Saltvedt * Department of Neuroscience “Rita Levi Montalcini”,
University of Torino, Torino, Italy Innocenzo Rainero, Silvia Boschi, Alessandro Vacca & Fausto Roveta * Laboratorio de Genética, Hospital Universitario Central de Asturias, Oviedo,
Spain Irene Rosas Allende, V. Álvarez & Victoria Álvarez * Memory Clinic, Department of Neurology, 2nd Faculty of Medicine and Motol University Hospital, Charles University, Prague,
Czech Republic Jakub Hort, Martin Vyhnalek, Ondrej Lerch & Jan Laczo * International Clinical Research Center, St. Anne’s University Hospital Brno, Brno, Czech Republic Jakub Hort,
Martin Vyhnalek, Ondrej Lerch & Jan Laczo * Department of Psychiatry and Psychotherapy, School of Medicine Klinikum rechts der Isar, Technical University of Munich, Munich, Germany
Janine Diehl-Schmid, Oliver Goldhardt & Timo Grimmer * Population Health Unit, Finnish Institute for Health and Welfare, Helsinki, Finland Jenni Lehtisalo, Tiia Ngandu, Jaakko
Tuomilehto, Jaana Lindström & Markku Peltonen * Department of Psychiatry and Psychotherapy, University Medical Center Goettingen, Goettingen, Germany Jens Wiltfang & Jonathan
Vogelgsang * German Center for Neurodegenerative Diseases (DZNE), Goettingen, Germany Jens Wiltfang * Neurosciences and Signaling Group, Institute of Biomedicine (iBiMED), Department of
Medical Sciences, University of Aveiro, Aveiro, Portugal Jens Wiltfang * Department of Psychiatry and Psychotherapy, Universitätsklinikum Erlangen, Friedrich-Alexander Universität
Erlangen-Nürnberg, Erlangen, Germany Johannes Kornhuber * Department of Population & Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH, USA Jonathan L. Haines
* Cleveland Institute for Computational Biology, Case Western Reserve University, Cleveland, OH, USA Jonathan L. Haines * Translational Neuroscience Laboratory, McLean Hospital, Harvard
Medical School, Belmont, MA, USA Jonathan Vogelgsang * Department of Geriatric Psychiatry, University Hospital of Psychiatry Zürich, Zürich, Switzerland Julius Popp * University of Zürich,
Zürich, Switzerland Julius Popp * Old age Psychiatry, University Hospital of Lausanne, Lausanne, Switzerland Julius Popp * Department of Psychiatry, Psychosomatics and Psychotherapy, Center
of Mental Health, University Hospital, Wuerzburg, Germany Jürgen Deckert, Martin J. Herrmann, Thomas Polak, Deckert Jürgen, Heike Weber & Martin J. Herrman * Institute for Stroke and
Dementia Research, Klinikum der Universität München, Ludwig-Maximilians-Universität LMU, Munich, Germany Katharina Buerger, Martin Dichgans & Michael Ewers * German Center for
Neurodegenerative Diseases (DZNE), Munich, Germany Katharina Buerger, Martin Dichgans & Michael Ewers * Schools of Life Sciences and Medicine, University of Nottingham, Nottingham, UK
Kevin Morgan * Department of Public Health and Caring Sciences/Geriatrics, Uppsala, Sweden Lena Kilander, Malin Löwenmark, Martin Ingelsson & Vilmantas Giedraitis * Institute of Medical
Biometry, Informatics and Epidemiology, University Hospital of Bonn, Bonn, Germany Leonie Weinhold & Matthias Schmid * Departments of Medicine (Biomedical Genetics), Neurology,
Ophthalmology, Epidemiology, and Biostatistics, Boston University Schools of Medicine and Public Health, Boston, MA, USA Lindsay A. Farrer * Centre for Memory Disturbances, Lab of Clinical
Neurochemistry, Section of Neurology, University of Perugia, Perugia, Italy Lucia Farotti, Lucilla Parnetti, Elena Chipi, Nicola Salvadori & Viviana Lisetti * Department of Geriatric
Psychiatry, Central Institute for Mental Health Mannheim, Medical Faculty Mannheim, University of Heidelberg, Heidelberg, Germany Lucrezia Hausner & Lutz Froelich * Department of
Epidemiology, Erasmus Medical Center, Rotterdam, The Netherlands M. Arfan Ikram & Cornelia M. van Duijn * 1st Department of Neurology Aristotle University of Thessaloniki, Thessaloniki,
Greece Magda Tsolaki, Anna Anastasiou, Arnaoutoglou Marianthi, Effrosyni Koutsouraki & Thomas Tegos * Azienda Ospedaliero-Universitaria, Parma, Italy Marco Spallazzi * Nuffield
Department of Clinical Neurosciences, Oxford, UK Margaret Esiri * Institute of Human Genetics, University of Bonn, School of Medicine & University Hospital Bonn, Bonn, Germany Markus M.
Nöthen, Per Hoffmann & Stefanie Heilmann-Heimbach * Munich Cluster for Systems Neurology (SyNergy), Munich, Germany Martin Dichgans & Robert Perneczky * Department of Primary Medical
Care, University Medical Centre Hamburg-Eppendorf, Hamburg, Germany Martin Scherer * Laboratory of Cognitive Neuroscience, School of Psychology, Aristotle University of Thessaloniki,
Thessaloniki, Greece Mary H. Kosmidis & Mary H. Kosmidis * Department of Nutrition and Dietetics, Harokopio University, Athens, Greece Mary Yannakoulia & Costas Anastasiou *
Institute of Gerontology and Geriatrics, Department of Medicine, University of Perugia, Perugia, Italy Michela Scamosci, Patrizia Mecocci, Roberta Cecchetti & Patrizia Bastiani *
Institute of Public Health and Clinical Nutrition, University of Eastern Finland, Kuopio, Finland Miia Kivipelto & Tiina Laatikainen * Neuroepidemiology and Ageing Research Unit, School
of Public Health, Imperial College London, London, UK Miia Kivipelto * Stockholms Sjukhem, Research & Development Unit, Stockholm, Sweden Miia Kivipelto * Institute of Biomedicine,
University of Eastern Finland, Kuopio, Finland Mikko Hiltunen, Teemu Kuulasmaa, Mari Takalo & Teemu Natunen * Brown Foundation Institute of Molecular Medicine, University of Texas Health
Sciences Center at Houston, Houston, TX, USA Myriam Fornage * 1st Department of Neurology, Aiginition Hospital, National and Kapodistrian University of Athens, Medical School, Athens,
Greece Nikolaos Scarmeas * Taub Institute for Research in Alzheimer’s Disease and the Aging Brain, The Gertrude H. Sergievsky Center, Depatment of Neurology, Columbia University, New York,
NY, USA Nikolaos Scarmeas * LVR-Hospital Essen, Department of Psychiatry and Psychotherapy, Medical Faculty, University of Duisburg-Essen, Essen, Germany Norbert Scherbaum * Department of
Psychiatry and Psychotherapy and Experimental and Clinical Research Center (ECRC), Charité-Universitätsmedizin Berlin, Berlin, Germany Oliver Peters * German Center for Neurodegenerative
Diseases (DZNE), Berlin, Germany Oliver Peters & Josef Priller * Bristol Medical School (THS), University of Bristol, Southmead Hospital, Bristol, UK Olivia Anna Skrobot, Seth Love &
Patrick Gavin Kehoe * Experimental Neuro-psychobiology Laboratory, Department of Clinical and Behavioral Neurology, IRCCS Santa Lucia Foundation, Rome, Italy Paola Bossù & Francesca
Salani * Unit of Neuroscience, DIMEC, University of Parma, Parma, Italy Paolo Caffarra * Athens Association of Alzheimer’s disease and Related Disorders, Athens, Greece Paraskevi Sakka *
Institute of Medical Genetics and Pathology, University Hospital Basel, Basel, Switzerland Per Hoffmann * Department of Psychiatry, Social Medicine Center East- Donauspital, Vienna, Austria
Peter Fischer * Center of Mental Health, Clinic and Policlinic of Psychiatry, Psychosomatics and Psychotherapy, University Hospital of Würzburg, Würzburg, Germany Peter Riederer *
Translational and Clincial Research Institute, Newcastle University, Newcastle upon Tyne, UK Rajesh N. Kalaria & Tuomo M. Polvikoski * Campus for Ageing anf Vitality, Newcastle upon
Tyne, UK Rajesh N. Kalaria & Tuomo M. Polvikoski * Taub Institute on Alzheimer’s Disease and the Aging Brain, Department of Neurology, Columbia University, New York, NY, USA Richard
Mayeux * Gertrude H. Sergievsky Center, Columbia University, New York, NY, USA Richard Mayeux * Department of Neurology, Columbia University, New York, NY, USA Richard Mayeux * Laboratory
for Cognitive Neurology, Department of Neurosciences, University of Leuven, Leuven, Belgium Rik Vandenberghe * Neurology Department, University Hospitals Leuven, Leuven, Belgium Rik
Vandenberghe * Center for Neurosciences, Vrije Universiteit Brussel (VUB), Brussels, Belgium Sebastiaan Engelborghs, Ellen De Roeck & Maria Bjerke * Reference Center for Biological
Markers of Dementia (BIODEM), University of Antwerp, Antwerp, Belgium Sebastiaan Engelborghs * Institute Born-Bunge, University of Antwerp, Antwerp, Belgium Sebastiaan Engelborghs *
Department of Neurology, VUB University Hospital Brussels (UZ Brussel), Brussels, Belgium Sebastiaan Engelborghs * Institute of Clinical Medicine, Internal Medicine, University of Eastern
Finland, Kuopio, Finland Seppo Helisalmi * Department of Neurology and Clinical Neurophysiology, University Hospital of Trondheim, Trondheim, Norway Sigrid Botne Sando * Department of
Neuromedicine and Movement Science, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Trondheim, Norway Sigrid Botne Sando * Department of Laboratory
Diagnostics, III Laboratory of Analysis, Brescia Hospital, Brescia, Italy Silvana Archetti * Alzheimer Research Center & Memory Clinic, Andalusian Institute for Neuroscience, Málaga,
Spain Silvia Mendoza, J. M. García-Alberca, S. Hevilla, T. Marín & Jose María García-Alberca * MRC Prion Unit at UCL, Institute of Prion Diseases, London, UK Simon Mead * Department of
Medical Genetics, Oslo University Hospital, Oslo, Norway Srdjan Djurovic * NORMENT, Department of Clinical Science, University of Bergen, Bergen, Norway Srdjan Djurovic * Institute of Social
Medicine, Occupational Health and Public Health, University of Leipzig, Leipzig, Germany Steffi Riedel-Heller * Department of Neurology/CIEN Foundation/Queen Sofia Foundation Alzheimer
Center, Madrid, Spain Teodoro del Ser * Azienda Ospedaliero-Universitaria Careggi Largo Brambilla, Florence, Italy Valentina Bessi * UKDRI Cardiff, Cardiff University, Cardiff, UK Valentina
Escott-Price * Unitat de Genètica Molecular, Institut de Biomedicina de València-CSIC, Valencia, Spain J. Pérez Tur & Jordi Pérez-Tur * Unidad Mixta de Neurologia Genètica, Instituto de
Investigación Sanitaria La Fe, Valencia, Spain J. Pérez Tur & Jordi Pérez-Tur * Department of Neurosciences, Faculty of Medicine and Nursery, University of the Basque Country, San
Sebastián, Spain Adolfo Lopez de Munain * Centro de Biología Molecular Severo Ochoa (UAM-CSIC), Madrid, Spain M. J. Bullido, A. Martín Montes, I. Sastre & María J. Bullido * Instituto de
Investigacion Sanitaria ‘Hospital la Paz’ (IdIPaz), Madrid, Spain M. J. Bullido, T. del Ser, A. Frank-García, M. Medina & María J. Bullido * Departamento de Especialidades Quirúrgicas,
Bioquímica e Inmunología. Facultad de Medicina, Universidad de Málaga, Málaga, Spain Luis M. Real * Fundación Pública Galega de Medicina Xenómica-CIBERER-IDIS, Santiago de Compostela, Spain
A. Carracedo & Ángel Carracedo * Framingham Heart Study, Framingham, MA, USA Sudha Seshadri * Nuffield Department of Population Health, University of Oxford, Oxford, UK Cornelia M. van
Duijn * Centre for Healthy Brain Ageing (CHeBA), School of Psychiatry, Faculty of Medicine, University of New South Wales, Sydney, NSW, Australia Karen A. Mather * Neuroscience Research
Australia, Sydney, NSW, Australia Karen A. Mather * Department of Sociology, VU University, Amsterdam, The Netherlands Martijn Huisman * NORMENT Centre, Institute of Clinical Medicine,
University of Oslo, Oslo, Norway Ole A. Andreassen * Division of Mental Health and Addiction, Oslo University Hospital, Oslo, Norway Ole A. Andreassen * Department of Psychiatry, Glenn Biggs
Institute for Alzheimer’s and Neurodegenerative Diseases, San Antonio, TX, USA Alfredo Ramirez * University of Oxford (OPTIMA), Oxford, UK A. David Smith * OPTIMA, Department of
Pharmacology, University of Oxford, Oxford, UK A. David Smith * Dirección de Atención de Adultos Mayores del Min., Salud Desarrollo Social y Deportes de la Pcia. de Mendoza, Mendoza,
Argentina Aldo Saenz & Mario Rios * Geriatrics Unit Fondazione Policlinico A. Gemelli IRCCS, Rome, Italy Alessandra Bizzarro & Alessandra Lauria * Cerebral Function Unit, Greater
Manchester Neurosciences Centre, Salford Royal Hospital, Salford, UK Anna Richardson, David Neary, Jenny Harris, Julie Snowden & Matthew Jones * Division of Neuroscience and Experimental
Psychology, School of Biological Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester, UK Anna Richardson, Julie Snowden & Nigel M. Hooper * Centre
National de Recherche en Génomique Humaine (CNRGH), Institut de Biologie François Jacob, CEA, Université Paris-Saclay, F-91057, Evry, France Anne Boland, Bertrand Fin, Céline Besse, Delphine
Daian, Jean-François Deleuze & Robert Olaso * Department of Neurology, Kuopio University Hospital, Kuopio, Finland Anne Koivisto * Insitute of Clinical Medicine-Neurology, University of
Eastenr Finland, Kuopio, Finland Anne Koivisto * Institute of Neurology, Catholic University of Sacred Heart, Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy Antonio
Daniele, Emanuele Costantini, Giordano Lacidogna & Michela Orsini * Research Laboratory, Complex Structure of Geriatrics, San Giovanni Rotondo, Fondazione IRCCS Casa Sollievo della
Sofferenza, Foggia, Italy Antonio Greco * Centre for Public Health, School of Medicine, Queen’s University Belfast, Belfast, UK Bernadette McGuinness, David Craig, Janet Johnston, Peter
Passmore & Stephen Todd * Clinica Medica “Frugoni” and Geriatric Medicine-Memory Unit, University of Bari Aldo Moro, Bari, Italy Carlo Custodero & Vincenzo Solfrizzi *
CENECON-FMED-UBA, Buenos Aires, Argentina Carlos Ingino, Carlos Mangone, Carlos Reyes Toso, Carolina Muchnik, Luis Ignacio Brusco, Matias Rojo, Natividad Olivar & Sandra Germani * ENERI,
Vienna, Austria Carlos Ingino & Luis Ignacio Brusco * Htal Santojani, Buenos Aires, Argentina Carlos Mangone * Servicio de Neurología, Hospital de Cabueñes, Gijón, Spain Carmen Martínez
& C. Martínez * UBA, Buenos Aires, Argentina Carolina Cuesta, Daniel G. Politis, Pablo Galeano & Valeria Cores * HIGA Eva Perón, Billinghurst, Argentina Carolina Cuesta, Daniel G.
Politis, Federico Ariel Prestia & Valeria Cores * University of Oxford, Oxford, UK Catharine Joachim * Neurología Clinica, Madrid, Spain Cecilia Ortiz & Juan Ignacio Bacha * German
Center for Neurodegenerative Diseases (DZNE), Tübingen, Germany Christoph Laske * Section for Dementia Research, Hertie Institute for Clinical Brain Research and Department of Psychiatry and
Psychotherapy, University of Tübingen, Tübingen, Germany Christoph Laske * Hospital Dr. Lucio Molas, Santa Rosa, Argentina Dana Lis Palacio, Ignacio Etchepareborda & Matias Kohler *
Fundacion Ayuda Enfermo Renal y Alta Complejidad (FERNAC), Santa Rosa, Argentina Dana Lis Palacio & Ignacio Etchepareborda * CONICET, Buenos Aires, Argentina Daniel G. Politis &
Valeria Cores * Institute for Stroke and Dementia Research (ISD), University Hospital, Ludwig-Maximilian University Munich, Munich, Germany Daniel Janowitz & Malik Rainer * Faculty of
Medical and Human Sciences, Institute of Brain, Behaviour and Mental Health, Manchester, UK David M. Mann & Stuart Pickering-Brown * Clinic of Neurology UH ‘Alexandrovska’, Medical
University—Sofia, Sofia, Bulgaria Diyana Belezhanska, Latchezar Traykov & Shima Mehrabian * Fundacion Sinapsis, Santa Rosa, Argentina Eduardo Kohler & Matias Kohler * Laboratory of
Brain Aging and Neurodegeneration- FIL, Buenos Aires, Argentina Eduardo M. Castaño, Federico Ariel Prestia, Gisela Novack, Laura Morelli & Pablo Galeano * IIBBA (CONICET), Buenos Aires,
Argentina Eduardo M. Castaño, Gisela Novack, Laura Morelli & Pablo Galeano * Salford Royal NHS Foundation Trust, Salford, England Emma R. L. C. Vardy * Nuffield Department of Clinical
Neurosciences, University of Oxford, Oxford, UK Gordon Wilcock * Department of Psychiatry, University of Bonn, Bonn, Germany Heike Kölsch & Reinhard Heun * Department of Radiology,
University Hospital Bonn, Bonn, Germany Henning Boecker * Department of Public Health, University of Helsinki, Helsinki, Finland Jaakko Tuomilehto * National School of Public Health, Madrid,
Spain Jaakko Tuomilehto * South Ostrobothnia Central Hospital, Seinäjoki, Finland Jaakko Tuomilehto * Dementia Research Centre, Department of Neurodegenerative Disease, UCL Queen Square
Institute of Neurology, University College London, London, UK Jonathan M. Schott & Nick C. Fox * Department of Neuropsychiatry, Charité, Berlin, Germany Josef Priller * ENYS (Estudio en
Neurociencias y Sistemas Complejos) CONICET-Hospital El Cruce ‘Nestor Kirchner’-UNAJ, Buenos Aires, Argentina Julieta Lisso, Nancy Medel, Patricia Solis, Silvia Kochen & Zulma Sevillano
* Molecular Medicine Center, Department of Medical chemistry and biochemistry, Medical University of Sofia, Sofia, Bulgaria Kalina Yonkova Mihova * Laboratory of Neurochemistry, UZ Brussel,
Brussel, Belgium Maria Bjerke * Unit of Clinical Pharmacology, Teaching Hospital of Cagliari, AOUCA, Cagliari, Italy Maria Del Zompo * Programa del Adulto Mayor-Min, Salud de la Pcia. de
Jujuy, San Salvador de Jujuy, Argentina Mariana Sanchez Abalos * Division of Neuroscience and Experimental Psychology, University of Manchester, Manchester, UK Matthew Jones * UK Dementia
Research Institute at UCL, UCL Queen Square Institute of Neurology, University College London, London, UK Nick C. Fox * Center for Life Course Health Research, University of Oulu, Oulu,
Finland Riitta Antikainen & Timo Strandberg * Medical Research Center Oulu, Oulu University Hospital, Oulu, Finland Riitta Antikainen * Oulu City Hospital, Oulu, Finland Riitta
Antikainen * German Center for Neurodegenerative Diseases (DZNE, Munich), Munich, Germany Robert Perneczky * Department of Psychiatry and Psychotherapy, University Hospital LMU Munich,
Munich, Germany Robert Perneczky * Ageing Epidemiology Research Unit (AGE), School of Public Health, Imperial College London, London, UK Robert Perneczky * University of Bristol Institute of
Clinical Neurosciences, School of Clinical Sciences, Frenchay Hospital, Bristol, UK Seth Love * German Center for Neurodegenerative Diseases (DZNE), Rostock, Germany Stefan Teipel *
Department of Psychosomatic Medicine, Rostock University Medical Center, Rostock, Germany Stefan Teipel * Finnish Institute for Health and Welfare, Helsinki, Finland Tiina Laatikainen *
Joint Municipal Authority for North Karelia Social and Health Services (Siun Sote), Joensuu, Finland Tiina Laatikainen * Helsinki University Hospital, University of Helsinki, Helsinki,
Finland Timo Strandberg * Department of Clinical Biochemistry, Hematology and Immunology, Na Homolce Hospital, Prague, Czech Republic Vaclav Matoska * Servei de Neurologia, Hospital Clínic
Universitari de València, València, Spain M. D. Alonso * Department of Neurology, Hospital Universitario Son Espases, Palma, Spain G. Amer-Ferrer & S. Manzanares * Hospital Universitario
Ramón y Cajal, Madrid, Spain M. J. Casajeros * BIOMICs, País Vasco, Centro de Investigación Lascaray, Universidad del País Vasco UPV/EHU, Leioa, Spain M. M. de Pancorbo * Fundación para la
Formación e Investigación Sanitarias de la Región de Murcia, El Palmar, Spain S. Manzanares * Fundación CITA-alzheimer, Centro de Investigacio´n y Terapias Avanzadas, San Sebastián, Spain P.
Martínez-Lage Álvarez * Navarrabiomed, Pamplona, Spain M. Mendioroz Iriarte * Barcelona beta Brain Research Center–Fundació Pasqual Maragall, Barcelona, Spain J. L. Molinuevo * Hospital
Universitario La Princesa, Madrid, Spain D. Real de Asúa * School of Health Sciences, Bangor University, Bangor, UK C. Macleod & Gill Windle * Institute of Psychology, Health and
Society, University of Liverpool, Liverpool, England C. McCracken * School of Clinical Medicine, Cambridge Institute of Public Health, University of Cambridge, Cambridge, England Carol
Brayne * Faculty of Medicine, Institute of Health and Society, University of Newcastle, Callaghan, NSW, Australia F. Matthews * Dementia Studies, University of Stirling, Stirling, Scotland
Judith Phillips * School of Psychology, University of Liverpool, Liverpool, England K. Bennett * School of Psychology, Centre for Research in Ageing and Cognitive Health (REACH), University
of Exeter, Exeter, England Linda Clare * Dementia Services Development Centre, Bangor University, Wales, UK Robert Woods * Swansea University, School of Human Sciences, Centre for Innovative
Ageing, Swansea, UK Vanessa Burholt Authors * Itziar de Rojas View author publications You can also search for this author inPubMed Google Scholar * Sonia Moreno-Grau View author
publications You can also search for this author inPubMed Google Scholar * Niccolo Tesi View author publications You can also search for this author inPubMed Google Scholar * Benjamin
Grenier-Boley View author publications You can also search for this author inPubMed Google Scholar * Victor Andrade View author publications You can also search for this author inPubMed
Google Scholar * Iris E. Jansen View author publications You can also search for this author inPubMed Google Scholar * Nancy L. Pedersen View author publications You can also search for this
author inPubMed Google Scholar * Najada Stringa View author publications You can also search for this author inPubMed Google Scholar * Anna Zettergren View author publications You can also
search for this author inPubMed Google Scholar * Isabel Hernández View author publications You can also search for this author inPubMed Google Scholar * Laura Montrreal View author
publications You can also search for this author inPubMed Google Scholar * Carmen Antúnez View author publications You can also search for this author inPubMed Google Scholar * Anna Antonell
View author publications You can also search for this author inPubMed Google Scholar * Rick M. Tankard View author publications You can also search for this author inPubMed Google Scholar *
Joshua C. Bis View author publications You can also search for this author inPubMed Google Scholar * Rebecca Sims View author publications You can also search for this author inPubMed
Google Scholar * Céline Bellenguez View author publications You can also search for this author inPubMed Google Scholar * Inés Quintela View author publications You can also search for this
author inPubMed Google Scholar * Antonio González-Perez View author publications You can also search for this author inPubMed Google Scholar * Miguel Calero View author publications You can
also search for this author inPubMed Google Scholar * Emilio Franco-Macías View author publications You can also search for this author inPubMed Google Scholar * Juan Macías View author
publications You can also search for this author inPubMed Google Scholar * Rafael Blesa View author publications You can also search for this author inPubMed Google Scholar * Laura
Cervera-Carles View author publications You can also search for this author inPubMed Google Scholar * Manuel Menéndez-González View author publications You can also search for this author
inPubMed Google Scholar * Ana Frank-García View author publications You can also search for this author inPubMed Google Scholar * Jose Luís Royo View author publications You can also search
for this author inPubMed Google Scholar * Fermin Moreno View author publications You can also search for this author inPubMed Google Scholar * Raquel Huerto Vilas View author publications
You can also search for this author inPubMed Google Scholar * Miquel Baquero View author publications You can also search for this author inPubMed Google Scholar * Mónica Diez-Fairen View
author publications You can also search for this author inPubMed Google Scholar * Carmen Lage View author publications You can also search for this author inPubMed Google Scholar * Sebastián
García-Madrona View author publications You can also search for this author inPubMed Google Scholar * Pablo García-González View author publications You can also search for this author
inPubMed Google Scholar * Emilio Alarcón-Martín View author publications You can also search for this author inPubMed Google Scholar * Sergi Valero View author publications You can also
search for this author inPubMed Google Scholar * Oscar Sotolongo-Grau View author publications You can also search for this author inPubMed Google Scholar * Abbe Ullgren View author
publications You can also search for this author inPubMed Google Scholar * Adam C. Naj View author publications You can also search for this author inPubMed Google Scholar * Afina W. Lemstra
View author publications You can also search for this author inPubMed Google Scholar * Alba Benaque View author publications You can also search for this author inPubMed Google Scholar *
Alba Pérez-Cordón View author publications You can also search for this author inPubMed Google Scholar * Alberto Benussi View author publications You can also search for this author inPubMed
Google Scholar * Alberto Rábano View author publications You can also search for this author inPubMed Google Scholar * Alessandro Padovani View author publications You can also search for
this author inPubMed Google Scholar * Alessio Squassina View author publications You can also search for this author inPubMed Google Scholar * Alexandre de Mendonça View author publications
You can also search for this author inPubMed Google Scholar * Alfonso Arias Pastor View author publications You can also search for this author inPubMed Google Scholar * Almar A. L. Kok View
author publications You can also search for this author inPubMed Google Scholar * Alun Meggy View author publications You can also search for this author inPubMed Google Scholar * Ana Belén
Pastor View author publications You can also search for this author inPubMed Google Scholar * Ana Espinosa View author publications You can also search for this author inPubMed Google
Scholar * Anaïs Corma-Gómez View author publications You can also search for this author inPubMed Google Scholar * Angel Martín Montes View author publications You can also search for this
author inPubMed Google Scholar * Ángela Sanabria View author publications You can also search for this author inPubMed Google Scholar * Anita L. DeStefano View author publications You can
also search for this author inPubMed Google Scholar * Anja Schneider View author publications You can also search for this author inPubMed Google Scholar * Annakaisa Haapasalo View author
publications You can also search for this author inPubMed Google Scholar * Anne Kinhult Ståhlbom View author publications You can also search for this author inPubMed Google Scholar * Anne
Tybjærg-Hansen View author publications You can also search for this author inPubMed Google Scholar * Annette M. Hartmann View author publications You can also search for this author
inPubMed Google Scholar * Annika Spottke View author publications You can also search for this author inPubMed Google Scholar * Arturo Corbatón-Anchuelo View author publications You can also
search for this author inPubMed Google Scholar * Arvid Rongve View author publications You can also search for this author inPubMed Google Scholar * Barbara Borroni View author publications
You can also search for this author inPubMed Google Scholar * Beatrice Arosio View author publications You can also search for this author inPubMed Google Scholar * Benedetta Nacmias View
author publications You can also search for this author inPubMed Google Scholar * Børge G. Nordestgaard View author publications You can also search for this author inPubMed Google Scholar *
Brian W. Kunkle View author publications You can also search for this author inPubMed Google Scholar * Camille Charbonnier View author publications You can also search for this author
inPubMed Google Scholar * Carla Abdelnour View author publications You can also search for this author inPubMed Google Scholar * Carlo Masullo View author publications You can also search
for this author inPubMed Google Scholar * Carmen Martínez Rodríguez View author publications You can also search for this author inPubMed Google Scholar * Carmen Muñoz-Fernandez View author
publications You can also search for this author inPubMed Google Scholar * Carole Dufouil View author publications You can also search for this author inPubMed Google Scholar * Caroline
Graff View author publications You can also search for this author inPubMed Google Scholar * Catarina B. Ferreira View author publications You can also search for this author inPubMed Google
Scholar * Caterina Chillotti View author publications You can also search for this author inPubMed Google Scholar * Chandra A. Reynolds View author publications You can also search for this
author inPubMed Google Scholar * Chiara Fenoglio View author publications You can also search for this author inPubMed Google Scholar * Christine Van Broeckhoven View author publications
You can also search for this author inPubMed Google Scholar * Christopher Clark View author publications You can also search for this author inPubMed Google Scholar * Claudia Pisanu View
author publications You can also search for this author inPubMed Google Scholar * Claudia L. Satizabal View author publications You can also search for this author inPubMed Google Scholar *
Clive Holmes View author publications You can also search for this author inPubMed Google Scholar * Dolores Buiza-Rueda View author publications You can also search for this author inPubMed
Google Scholar * Dag Aarsland View author publications You can also search for this author inPubMed Google Scholar * Dan Rujescu View author publications You can also search for this author
inPubMed Google Scholar * Daniel Alcolea View author publications You can also search for this author inPubMed Google Scholar * Daniela Galimberti View author publications You can also
search for this author inPubMed Google Scholar * David Wallon View author publications You can also search for this author inPubMed Google Scholar * Davide Seripa View author publications
You can also search for this author inPubMed Google Scholar * Edna Grünblatt View author publications You can also search for this author inPubMed Google Scholar * Efthimios Dardiotis View
author publications You can also search for this author inPubMed Google Scholar * Emrah Düzel View author publications You can also search for this author inPubMed Google Scholar * Elio
Scarpini View author publications You can also search for this author inPubMed Google Scholar * Elisa Conti View author publications You can also search for this author inPubMed Google
Scholar * Elisa Rubino View author publications You can also search for this author inPubMed Google Scholar * Ellen Gelpi View author publications You can also search for this author
inPubMed Google Scholar * Eloy Rodriguez-Rodriguez View author publications You can also search for this author inPubMed Google Scholar * Emmanuelle Duron View author publications You can
also search for this author inPubMed Google Scholar * Eric Boerwinkle View author publications You can also search for this author inPubMed Google Scholar * Evelyn Ferri View author
publications You can also search for this author inPubMed Google Scholar * Fabrizio Tagliavini View author publications You can also search for this author inPubMed Google Scholar * Fahri
Küçükali View author publications You can also search for this author inPubMed Google Scholar * Florence Pasquier View author publications You can also search for this author inPubMed Google
Scholar * Florentino Sanchez-Garcia View author publications You can also search for this author inPubMed Google Scholar * Francesca Mangialasche View author publications You can also
search for this author inPubMed Google Scholar * Frank Jessen View author publications You can also search for this author inPubMed Google Scholar * Gaël Nicolas View author publications You
can also search for this author inPubMed Google Scholar * Geir Selbæk View author publications You can also search for this author inPubMed Google Scholar * Gemma Ortega View author
publications You can also search for this author inPubMed Google Scholar * Geneviève Chêne View author publications You can also search for this author inPubMed Google Scholar * Georgios
Hadjigeorgiou View author publications You can also search for this author inPubMed Google Scholar * Giacomina Rossi View author publications You can also search for this author inPubMed
Google Scholar * Gianfranco Spalletta View author publications You can also search for this author inPubMed Google Scholar * Giorgio Giaccone View author publications You can also search for
this author inPubMed Google Scholar * Giulia Grande View author publications You can also search for this author inPubMed Google Scholar * Giuliano Binetti View author publications You can
also search for this author inPubMed Google Scholar * Goran Papenberg View author publications You can also search for this author inPubMed Google Scholar * Harald Hampel View author
publications You can also search for this author inPubMed Google Scholar * Henri Bailly View author publications You can also search for this author inPubMed Google Scholar * Henrik
Zetterberg View author publications You can also search for this author inPubMed Google Scholar * Hilkka Soininen View author publications You can also search for this author inPubMed Google
Scholar * Ida K. Karlsson View author publications You can also search for this author inPubMed Google Scholar * Ignacio Alvarez View author publications You can also search for this author
inPubMed Google Scholar * Ildebrando Appollonio View author publications You can also search for this author inPubMed Google Scholar * Ina Giegling View author publications You can also
search for this author inPubMed Google Scholar * Ingmar Skoog View author publications You can also search for this author inPubMed Google Scholar * Ingvild Saltvedt View author publications
You can also search for this author inPubMed Google Scholar * Innocenzo Rainero View author publications You can also search for this author inPubMed Google Scholar * Irene Rosas Allende
View author publications You can also search for this author inPubMed Google Scholar * Jakub Hort View author publications You can also search for this author inPubMed Google Scholar *
Janine Diehl-Schmid View author publications You can also search for this author inPubMed Google Scholar * Jasper Van Dongen View author publications You can also search for this author
inPubMed Google Scholar * Jean-Sebastien Vidal View author publications You can also search for this author inPubMed Google Scholar * Jenni Lehtisalo View author publications You can also
search for this author inPubMed Google Scholar * Jens Wiltfang View author publications You can also search for this author inPubMed Google Scholar * Jesper Qvist Thomassen View author
publications You can also search for this author inPubMed Google Scholar * Johannes Kornhuber View author publications You can also search for this author inPubMed Google Scholar * Jonathan
L. Haines View author publications You can also search for this author inPubMed Google Scholar * Jonathan Vogelgsang View author publications You can also search for this author inPubMed
Google Scholar * Juan A. Pineda View author publications You can also search for this author inPubMed Google Scholar * Juan Fortea View author publications You can also search for this
author inPubMed Google Scholar * Julius Popp View author publications You can also search for this author inPubMed Google Scholar * Jürgen Deckert View author publications You can also
search for this author inPubMed Google Scholar * Katharina Buerger View author publications You can also search for this author inPubMed Google Scholar * Kevin Morgan View author
publications You can also search for this author inPubMed Google Scholar * Klaus Fließbach View author publications You can also search for this author inPubMed Google Scholar * Kristel
Sleegers View author publications You can also search for this author inPubMed Google Scholar * Laura Molina-Porcel View author publications You can also search for this author inPubMed
Google Scholar * Lena Kilander View author publications You can also search for this author inPubMed Google Scholar * Leonie Weinhold View author publications You can also search for this
author inPubMed Google Scholar * Lindsay A. Farrer View author publications You can also search for this author inPubMed Google Scholar * Li-San Wang View author publications You can also
search for this author inPubMed Google Scholar * Luca Kleineidam View author publications You can also search for this author inPubMed Google Scholar * Lucia Farotti View author publications
You can also search for this author inPubMed Google Scholar * Lucilla Parnetti View author publications You can also search for this author inPubMed Google Scholar * Lucio Tremolizzo View
author publications You can also search for this author inPubMed Google Scholar * Lucrezia Hausner View author publications You can also search for this author inPubMed Google Scholar *
Luisa Benussi View author publications You can also search for this author inPubMed Google Scholar * Lutz Froelich View author publications You can also search for this author inPubMed
Google Scholar * M. Arfan Ikram View author publications You can also search for this author inPubMed Google Scholar * M. Candida Deniz-Naranjo View author publications You can also search
for this author inPubMed Google Scholar * Magda Tsolaki View author publications You can also search for this author inPubMed Google Scholar * Maitée Rosende-Roca View author publications
You can also search for this author inPubMed Google Scholar * Malin Löwenmark View author publications You can also search for this author inPubMed Google Scholar * Marc Hulsman View author
publications You can also search for this author inPubMed Google Scholar * Marco Spallazzi View author publications You can also search for this author inPubMed Google Scholar * Margaret A.
Pericak-Vance View author publications You can also search for this author inPubMed Google Scholar * Margaret Esiri View author publications You can also search for this author inPubMed
Google Scholar * María Bernal Sánchez-Arjona View author publications You can also search for this author inPubMed Google Scholar * Maria Carolina Dalmasso View author publications You can
also search for this author inPubMed Google Scholar * María Teresa Martínez-Larrad View author publications You can also search for this author inPubMed Google Scholar * Marina Arcaro View
author publications You can also search for this author inPubMed Google Scholar * Markus M. Nöthen View author publications You can also search for this author inPubMed Google Scholar *
Marta Fernández-Fuertes View author publications You can also search for this author inPubMed Google Scholar * Martin Dichgans View author publications You can also search for this author
inPubMed Google Scholar * Martin Ingelsson View author publications You can also search for this author inPubMed Google Scholar * Martin J. Herrmann View author publications You can also
search for this author inPubMed Google Scholar * Martin Scherer View author publications You can also search for this author inPubMed Google Scholar * Martin Vyhnalek View author
publications You can also search for this author inPubMed Google Scholar * Mary H. Kosmidis View author publications You can also search for this author inPubMed Google Scholar * Mary
Yannakoulia View author publications You can also search for this author inPubMed Google Scholar * Matthias Schmid View author publications You can also search for this author inPubMed
Google Scholar * Michael Ewers View author publications You can also search for this author inPubMed Google Scholar * Michael T. Heneka View author publications You can also search for this
author inPubMed Google Scholar * Michael Wagner View author publications You can also search for this author inPubMed Google Scholar * Michela Scamosci View author publications You can also
search for this author inPubMed Google Scholar * Miia Kivipelto View author publications You can also search for this author inPubMed Google Scholar * Mikko Hiltunen View author publications
You can also search for this author inPubMed Google Scholar * Miren Zulaica View author publications You can also search for this author inPubMed Google Scholar * Montserrat Alegret View
author publications You can also search for this author inPubMed Google Scholar * Myriam Fornage View author publications You can also search for this author inPubMed Google Scholar *
Natalia Roberto View author publications You can also search for this author inPubMed Google Scholar * Natasja M. van Schoor View author publications You can also search for this author
inPubMed Google Scholar * Nazib M. Seidu View author publications You can also search for this author inPubMed Google Scholar * Nerisa Banaj View author publications You can also search for
this author inPubMed Google Scholar * Nicola J. Armstrong View author publications You can also search for this author inPubMed Google Scholar * Nikolaos Scarmeas View author publications
You can also search for this author inPubMed Google Scholar * Norbert Scherbaum View author publications You can also search for this author inPubMed Google Scholar * Oliver Goldhardt View
author publications You can also search for this author inPubMed Google Scholar * Oliver Hanon View author publications You can also search for this author inPubMed Google Scholar * Oliver
Peters View author publications You can also search for this author inPubMed Google Scholar * Olivia Anna Skrobot View author publications You can also search for this author inPubMed Google
Scholar * Olivier Quenez View author publications You can also search for this author inPubMed Google Scholar * Ondrej Lerch View author publications You can also search for this author
inPubMed Google Scholar * Paola Bossù View author publications You can also search for this author inPubMed Google Scholar * Paolo Caffarra View author publications You can also search for
this author inPubMed Google Scholar * Paolo Dionigi Rossi View author publications You can also search for this author inPubMed Google Scholar * Paraskevi Sakka View author publications You
can also search for this author inPubMed Google Scholar * Patrizia Mecocci View author publications You can also search for this author inPubMed Google Scholar * Per Hoffmann View author
publications You can also search for this author inPubMed Google Scholar * Peter A. Holmans View author publications You can also search for this author inPubMed Google Scholar * Peter
Fischer View author publications You can also search for this author inPubMed Google Scholar * Peter Riederer View author publications You can also search for this author inPubMed Google
Scholar * Qiong Yang View author publications You can also search for this author inPubMed Google Scholar * Rachel Marshall View author publications You can also search for this author
inPubMed Google Scholar * Rajesh N. Kalaria View author publications You can also search for this author inPubMed Google Scholar * Richard Mayeux View author publications You can also search
for this author inPubMed Google Scholar * Rik Vandenberghe View author publications You can also search for this author inPubMed Google Scholar * Roberta Cecchetti View author publications
You can also search for this author inPubMed Google Scholar * Roberta Ghidoni View author publications You can also search for this author inPubMed Google Scholar * Ruth Frikke-Schmidt View
author publications You can also search for this author inPubMed Google Scholar * Sandro Sorbi View author publications You can also search for this author inPubMed Google Scholar * Sara
Hägg View author publications You can also search for this author inPubMed Google Scholar * Sebastiaan Engelborghs View author publications You can also search for this author inPubMed
Google Scholar * Seppo Helisalmi View author publications You can also search for this author inPubMed Google Scholar * Sigrid Botne Sando View author publications You can also search for
this author inPubMed Google Scholar * Silke Kern View author publications You can also search for this author inPubMed Google Scholar * Silvana Archetti View author publications You can also
search for this author inPubMed Google Scholar * Silvia Boschi View author publications You can also search for this author inPubMed Google Scholar * Silvia Fostinelli View author
publications You can also search for this author inPubMed Google Scholar * Silvia Gil View author publications You can also search for this author inPubMed Google Scholar * Silvia Mendoza
View author publications You can also search for this author inPubMed Google Scholar * Simon Mead View author publications You can also search for this author inPubMed Google Scholar *
Simona Ciccone View author publications You can also search for this author inPubMed Google Scholar * Srdjan Djurovic View author publications You can also search for this author inPubMed
Google Scholar * Stefanie Heilmann-Heimbach View author publications You can also search for this author inPubMed Google Scholar * Steffi Riedel-Heller View author publications You can also
search for this author inPubMed Google Scholar * Teemu Kuulasmaa View author publications You can also search for this author inPubMed Google Scholar * Teodoro del Ser View author
publications You can also search for this author inPubMed Google Scholar * Thibaud Lebouvier View author publications You can also search for this author inPubMed Google Scholar * Thomas
Polak View author publications You can also search for this author inPubMed Google Scholar * Tiia Ngandu View author publications You can also search for this author inPubMed Google Scholar
* Timo Grimmer View author publications You can also search for this author inPubMed Google Scholar * Valentina Bessi View author publications You can also search for this author inPubMed
Google Scholar * Valentina Escott-Price View author publications You can also search for this author inPubMed Google Scholar * Vilmantas Giedraitis View author publications You can also
search for this author inPubMed Google Scholar * Vincent Deramecourt View author publications You can also search for this author inPubMed Google Scholar * Wolfgang Maier View author
publications You can also search for this author inPubMed Google Scholar * Xueqiu Jian View author publications You can also search for this author inPubMed Google Scholar * Yolande A. L.
Pijnenburg View author publications You can also search for this author inPubMed Google Scholar * Patrick Gavin Kehoe View author publications You can also search for this author inPubMed
Google Scholar * Guillermo Garcia-Ribas View author publications You can also search for this author inPubMed Google Scholar * Pascual Sánchez-Juan View author publications You can also
search for this author inPubMed Google Scholar * Pau Pastor View author publications You can also search for this author inPubMed Google Scholar * Jordi Pérez-Tur View author publications
You can also search for this author inPubMed Google Scholar * Gerard Piñol-Ripoll View author publications You can also search for this author inPubMed Google Scholar * Adolfo Lopez de
Munain View author publications You can also search for this author inPubMed Google Scholar * Jose María García-Alberca View author publications You can also search for this author inPubMed
Google Scholar * María J. Bullido View author publications You can also search for this author inPubMed Google Scholar * Victoria Álvarez View author publications You can also search for
this author inPubMed Google Scholar * Alberto Lleó View author publications You can also search for this author inPubMed Google Scholar * Luis M. Real View author publications You can also
search for this author inPubMed Google Scholar * Pablo Mir View author publications You can also search for this author inPubMed Google Scholar * Miguel Medina View author publications You
can also search for this author inPubMed Google Scholar * Philip Scheltens View author publications You can also search for this author inPubMed Google Scholar * Henne Holstege View author
publications You can also search for this author inPubMed Google Scholar * Marta Marquié View author publications You can also search for this author inPubMed Google Scholar * María Eugenia
Sáez View author publications You can also search for this author inPubMed Google Scholar * Ángel Carracedo View author publications You can also search for this author inPubMed Google
Scholar * Philippe Amouyel View author publications You can also search for this author inPubMed Google Scholar * Gerard D. Schellenberg View author publications You can also search for this
author inPubMed Google Scholar * Julie Williams View author publications You can also search for this author inPubMed Google Scholar * Sudha Seshadri View author publications You can also
search for this author inPubMed Google Scholar * Cornelia M. van Duijn View author publications You can also search for this author inPubMed Google Scholar * Karen A. Mather View author
publications You can also search for this author inPubMed Google Scholar * Raquel Sánchez-Valle View author publications You can also search for this author inPubMed Google Scholar * Manuel
Serrano-Ríos View author publications You can also search for this author inPubMed Google Scholar * Adelina Orellana View author publications You can also search for this author inPubMed
Google Scholar * Lluís Tárraga View author publications You can also search for this author inPubMed Google Scholar * Kaj Blennow View author publications You can also search for this author
inPubMed Google Scholar * Martijn Huisman View author publications You can also search for this author inPubMed Google Scholar * Ole A. Andreassen View author publications You can also
search for this author inPubMed Google Scholar * Danielle Posthuma View author publications You can also search for this author inPubMed Google Scholar * Jordi Clarimón View author
publications You can also search for this author inPubMed Google Scholar * Mercè Boada View author publications You can also search for this author inPubMed Google Scholar * Wiesje M. van
der Flier View author publications You can also search for this author inPubMed Google Scholar * Alfredo Ramirez View author publications You can also search for this author inPubMed Google
Scholar * Jean-Charles Lambert View author publications You can also search for this author inPubMed Google Scholar * Sven J. van der Lee View author publications You can also search for
this author inPubMed Google Scholar * Agustín Ruiz View author publications You can also search for this author inPubMed Google Scholar CONSORTIA EADB CONTRIBUTORS * A. David Smith * , Aldo
Saenz * , Alessandra Bizzarro * , Alessandra Lauria * , Alessandro Vacca * , Alina Solomon * , Anna Anastasiou * , Anna Richardson * , Anne Boland * , Anne Koivisto * , Antonio Daniele * ,
Antonio Greco * , Arnaoutoglou Marianthi * , Bernadette McGuinness * , Bertrand Fin * , Camilla Ferrari * , Carlo Custodero * , Carlo Ferrarese * , Carlos Ingino * , Carlos Mangone * ,
Carlos Reyes Toso * , Carmen Martínez * , Carolina Cuesta * , Carolina Muchnik * , Catharine Joachim * , Cecilia Ortiz * , Céline Besse * , Charlotte Johansson * , Chiara Paola Zoia * ,
Christoph Laske * , Costas Anastasiou * , Dana Lis Palacio * , Daniel G. Politis * , Daniel Janowitz * , David Craig * , David M. Mann * , David Neary * , Deckert Jürgen * , Delphine Daian *
, Diyana Belezhanska * , Eduardo Kohler * , Eduardo M. Castaño * , Effrosyni Koutsouraki * , Elena Chipi * , Ellen De Roeck * , Emanuele Costantini * , Emma R. L. C. Vardy * , Fabrizio
Piras * , Fausto Roveta * , Federica Piras * , Federico Ariel Prestia * , Francesca Assogna * , Francesca Salani * , Gessica Sala * , Giordano Lacidogna * , Gisela Novack * , Gordon Wilcock
* , Håkan Thonberg * , Heike Kölsch * , Heike Weber * , Henning Boecker * , Ignacio Etchepareborda * , Irene Piaceri * , Jaakko Tuomilehto * , Jaana Lindström * , Jan Laczo * , Janet
Johnston * , Jean-François Deleuze * , Jenny Harris * , Jonathan M. Schott * , Josef Priller * , Juan Ignacio Bacha * , Julie Snowden * , Julieta Lisso * , Kalina Yonkova Mihova * ,
Latchezar Traykov * , Laura Morelli * , Luis Ignacio Brusco * , Malik Rainer * , Mari Takalo * , Maria Bjerke * , Maria Del Zompo * , Maria Serpente * , Mariana Sanchez Abalos * , Mario Rios
* , Markku Peltonen * , Martin J. Herrman * , Mary H. Kosmidis * , Matias Kohler * , Matias Rojo * , Matthew Jones * , Michela Orsini * , Nancy Medel * , Natividad Olivar * , Nick C. Fox *
, Nicola Salvadori * , Nigel M. Hooper * , Pablo Galeano * , Patricia Solis * , Patrizia Bastiani * , Patrizia Mecocci * , Peter Passmore * , Reinhard Heun * , Riitta Antikainen * , Robert
Olaso * , Robert Perneczky * , Sandra Germani * , Sara López-García * , Seth Love * , Shima Mehrabian * , Silvia Bagnoli * , Silvia Kochen * , Simona Andreoni * , Stefan Teipel * , Stephen
Todd * , Stuart Pickering-Brown * , Teemu Natunen * , Thomas Tegos * , Tiina Laatikainen * , Timo Strandberg * , Tuomo M. Polvikoski * , Vaclav Matoska * , Valentina Ciullo * , Valeria Cores
* , Vincenzo Solfrizzi * , Viviana Lisetti * & Zulma Sevillano THE GR@ACE STUDY GROUP * C. Abdelnour * , N. Aguilera * , E. Alarcon * , M. Alegret * , A. Benaque * , M. Boada * , M.
Buendia * , P. Cañabate * , A. Carracedo * , A. Corbatón-Anchuelo * , I. de Rojas * , S. Diego * , A. Espinosa * , A. Gailhajenet * , P. García-González * , S. Gil * , M. Guitart * , A.
González-Pérez * , I. Hernández * , M. Ibarria * , A. Lafuente * , J. Macias * , O. Maroñas * , E. Martín * , M. T. Martínez * , M. Marquié * , A. Mauleón * , L. Montrreal * , S. Moreno-Grau
* , M. Moreno * , A. Orellana * , G. Ortega * , A. Pancho * , E. Pelejá * , A. Pérez-Cordon * , J. A. Pineda * , S. Preckler * , I. Quintela * , L. M. Real * , M. Rosende-Roca * , A. Ruiz *
, M. E. Sáez * , A. Sanabria * , M. Serrano-Rios * , O. Sotolongo-Grau * , L. Tárraga * , S. Valero * & L. Vargas DEGESCO CONSORTIUM * A. D. Adarmes-Gómez * , E. Alarcón-Martín * , M.
D. Alonso * , I. Álvarez * , V. Álvarez * , G. Amer-Ferrer * , M. Antequera * , C. Antúnez * , M. Baquero * , M. Bernal * , R. Blesa * , M. Boada * , D. Buiza-Rueda * , M. J. Bullido * , J.
A. Burguera * , M. Calero * , F. Carrillo * , M. Carrión-Claro * , M. J. Casajeros * , J. Clarimón * , J. M. Cruz-Gamero * , M. M. de Pancorbo * , I. de Rojas * , T. del Ser * , M.
Diez-Fairen * , R. Escuela * , L. Garrote-Espina * , J. Fortea * , E. Franco-Macías * , A. Frank-García * , J. M. García-Alberca * , S. Garcia Madrona * , G. Garcia-Ribas * , P. Gómez-Garre
* , I. Hernández * , S. Hevilla * , S. Jesús * , M. A. Labrador Espinosa * , C. Lage * , A. Legaz * , A. Lleó * , A. Lopez de Munain * , S. López-García * , D. Macias-García * , S.
Manzanares * , M. Marín * , J. Marín-Muñoz * , T. Marín * , M. Marquié * , A. Martín Montes * , B. Martínez * , C. Martínez * , V. Martínez * , P. Martínez-Lage Álvarez * , M. Medina * , M.
Mendioroz Iriarte * , M. Menéndez-González * , P. Mir * , J. L. Molinuevo * , L. Montrreal * , A. Orellana * , P. Pastor * , J. Pérez Tur * , T. Periñán-Tocino * , R. Pineda-Sanchez * , G.
Piñol-Ripoll * , A. Rábano * , D. Real de Asúa * , S. Rodrigo * , E. Rodríguez-Rodríguez * , J. L. Royo * , A. Ruiz * , R. Sanchez del Valle Díaz * , P. Sánchez-Juan * , I. Sastre * , O.
Sotolongo-Grau * , S. Valero * , M. P. Vicente * , R. Vigo-Ortega * & L. Vivancos IGAP (ADGC, CHARGE, EADI, GERAD) * C. Macleod * , C. McCracken * , Carol Brayne * , Catherine Bresner *
, Detelina Grozeva * , Eftychia Bellou * , Ewen W. Sommerville * , F. Matthews * , Ganna Leonenko * , Georgina Menzies * , Gill Windle * , Janet Harwood * , Judith Phillips * , K. Bennett *
, Lauren Luckuck * , Linda Clare * , Robert Woods * , Salha Saad * & Vanessa Burholt PGC-ALZ CONSORTIA * Iris E. Jansen * , Nancy L. Pedersen * , Arvid Rongve * , Chandra A. Reynolds *
, Dag Aarsland * , Geir Selbæk * , Ida K. Karlsson * , Ingvild Saltvedt * , Sandro Sorbi * , Seppo Helisalmi * , Simona Ciccone * , Ole A. Andreassen * & Danielle Posthuma
CONTRIBUTIONS A.Ru and S.v.d.L. designed and conceptualized the study, interpreted the data and drafted the paper. I.d.R. contributed to data acquisition, the analysis, interpreted the data,
and co-wrote the paper. S.M.G. and N.T. contributed to the analysis and interpreted the data. S.J.v.d.L. and I.d.R. performed polygenic score analyses. L.C.C. and J.C. conducted the
functional analysis of _APP_. H.Ho, W.v.d.F., S.J.v.d.L., and A.Ru supervised the study. All authors critically revised the paper for important intellectual content and approved the final
paper. GR@ACE/DEGESCO: _Study design or conception_: M.Me, J.C., and A.Ru. _Data generation_: L.M., L.C.C., A.G.P., M.E.S., S.M.G., I.d.R., I.Q., and A.C. _Sample contribution_: M.Bo,
J.M.G.A., M.Me, M.Ma, M.C., T.S., S.G.M., G.G.R., A.L.M., J.M., L.M.R., G.P.R., M.M.G., C.M.R., I.R.A., V.A.l., C.L., E.R.R., P.S.J., D.A.l., P.P., M.D.F., I.A.l., J.P.T., A.C., L.Ta,
A.M.M., M.J.B., A.F.G., I.Q., I.H., L.M., P.G.G., E.A.M., S.V., O.S.G., A.Bena, A.P.C., A.E., A.Sa, C.Ab, G.O., M.R.R., M.A.l., N.R., S.G., A.O., A.Rab, A.Bel, F.Mo, M.Z., A.C.G., J.A.P.,
M.F.F., E.F.M., D.B.R., M.B.S.A., P.M., R.H.V., A.A.P., A.A., L.M.P., R.S.V., E.Ge, A.L., R.B., J.F., J.L.R., S.Men, M.Ba, I.d.R., and S.M.G. _Analysis_: S.M.G. and I.d.R. _Study
supervision/management_: A.C., L.Ta, M.Bo, M.Me, J.C., and A.Ru. IGAP. _Critical revision_: A.C.N., B.W.K., L.A.F., J.L.H., L.S.W., M.A.P.V., R.May, M.A.I., J.C.B., A.L.D.S., C.L.S., E.B.,
M.F., Q.Y., X.J., R.S., C.H., K.M., S.Mea, V.E.P., A.Meg, P.A.H., R.Mar, P.A., G.Sc, J.Will, and S.Se. EADB: _Sample contribution_. N.T., I.E.J., N.St, K.A.M., C.D., G.N., G.C., G.Sp, K.S.,
M.I., M.K., R.F.S., J.C.L., A.Ra, D.G., J.S.V., D.R., E.Gr, H.Ha, I.G., J.K., L.Fa, L.Fr, A.M.H., J.V., L.H., G.H., N.Sca, M.H.K., M.Y., H.Ho, W.M.v.d..F, M.H.u.l., N.M.v.S., A.T.H., B.G.N.,
C.V.B., E.S., R.V., S.E., T.N., F.K., J.V.D., V.G., A.U., A.Benu, A.K.S., B.B., C.Mas, C.F., E.C., F.Ma, G.B., I.Ap, J.Q.T., L.Ki, L.K.l., L.P., L.Tr, L.B., M.L., M.Ar, R.G., S.F., F.J.,
J.D.S., O.G., T.G., M.J.H., T.P., K.Bu, M.E.si, S.R.H., E.Dur, A.Ru, I.H., S.M.G., I.d.R., Y.A.L..P, A.d.M., C.C.l., J.P., S.J.v.d.L., C.G., N.B., O.H., P.B., A.H., T.K., M.E.w., O.A.S.,
R.N.K., J.Wilf, P.F., P.R., P.Sc, P.Sa, N.Sch, D.W., E.R., G.R., H.S., I.R., A.Sc, A.Sp, A.Sq, C.Cha, C.Chi, C.P., A.P., B.A., B.N., C.M.F., D.S., E.Da, E.düz, E.F., F.T., F.P., F.S.G.,
G.Gi, G.Gra, G.P., H.B., J.H., J.L., M.C.D., M.T., M.T.H., M.Schm, M.W., M.S., O.Q., O.L., P.C., P.D., P.M., R.C., S.So, S.He, S.A., S.B., S.C., T.L., V.B., V.D., P.G.K., M.M.N., M.C.D.N.,
O.P., W.M., A.W.L., I.Ap, C.B.F., A.A.L.K., G.B., M.Sca, M.Sp, M.V., M.Hi, K.F., L.W., M.D., P.H., and A.Ra. _Analysis_: N.J.A., R.M.T., V.An, N.T., I.E.J., N.St, S.J.v.d.L., I.d.R., S.M.G.,
B.G.B., and C.B. _Studies supervision/management_: S.H.H., K.A.M., C.D., G.N., G.C., G.S., K.S., M.I., M.K., R.F.S., D.G., J.S.V., D.R., E.Gr, H.Ha, I.G., J.K., L.Fr, A.M.H., J.V., L.H.,
G.H., N.Sca, M.H.K., M.Y., H.Ho, W.M.v.d.F., M.Hui, N.M.v.S., N.J.A., J.D., M.Sche, A.K.S., C.G., N.B., O.H., P.B., A.H., T.K., J.Wilf, P.F., P.R., P.Sa, P.Sc, M.Hul, N.T., I.E.J., A.T.H.,
B.G.N., C.V.B., E.S., R.V., S.E., P.A., A.Ru, and J.C.L. PGC-ALZ: _Sample contribution_. I.E.J., A.Ro, I.Sa, D.Aa, G.Se, S.B.S., S.D., D.P., S.H., I.K.K., N.L.P., C.A.R., and O.A.A.
_Analysis_: S.Ha, I.K.K., and I.E.J. _Study supervision/management_: O.A.A., C.M.v.D., and D.P. AD AND GBCS: _Sample contribution._ H.Z., S.K., I.S., and K.B. _Analysis_: N.M.S. and A.Z.
_Study supervision/management_: I.S.k., A.Z., and K.B.l. NXC: _Sample contribution._ A.C.A., M.T.M., M.S.R., and C.An. CORRESPONDING AUTHORS Correspondence to Sven J. van der Lee or Agustín
Ruiz. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Communications_ thanks the anonymous
reviewers for their contribution to the peer review of this work. Peer reviewer reports are available. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims
in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION PEER REVIEW FILE DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY DATA 1–8
REPORTING SUMMARY RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation,
distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and
indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to
the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will
need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE
CITE THIS ARTICLE de Rojas, I., Moreno-Grau, S., Tesi, N. _et al._ Common variants in Alzheimer’s disease and risk stratification by polygenic risk scores. _Nat Commun_ 12, 3417 (2021).
https://doi.org/10.1038/s41467-021-22491-8 Download citation * Received: 17 July 2020 * Accepted: 17 March 2021 * Published: 07 June 2021 * DOI: https://doi.org/10.1038/s41467-021-22491-8
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to
clipboard Provided by the Springer Nature SharedIt content-sharing initiative