- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Pancreatic cancer is the fourth leading cause of cancer-related deaths in Japan. To identify risk loci, we perform a meta-analysis of three genome-wide association studies
comprising 2,039 pancreatic cancer patients and 32,592 controls in the Japanese population. Here, we identify 3 (13q12.2, 13q22.1, and 16p12.3) genome-wide significant loci (_P_ < 5.0 ×
10−8), of which 16p12.3 has not been reported in the Western population. The lead single nucleotide polymorphism (SNP) at 16p12.3 is rs78193826 (odds ratio = 1.46, 95% confidence interval =
1.29-1.66, _P_ = 4.28 × 10−9), an Asian-specific, nonsynonymous _glycoprotein__ 2_ (_GP2_) gene variant. Associations between selected _GP2_ gene variants and pancreatic cancer are
replicated in 10,822 additional cases and controls of East Asian origin. Functional analyses using cell lines provide supporting evidence of the effect of rs78193826 on KRAS activity. These
findings suggest that _GP2_ gene variants are probably associated with pancreatic cancer susceptibility in populations of East Asian ancestry. SIMILAR CONTENT BEING VIEWED BY OTHERS ALLELIC
EFFECTS ON _KLHL17_ EXPRESSION UNDERLIE A PANCREATIC CANCER GENOME-WIDE ASSOCIATION SIGNAL AT CHR1P36.33 Article Open access 30 April 2025 PAN-CANCER AND CROSS-POPULATION GENOME-WIDE
ASSOCIATION STUDIES DISSECT SHARED GENETIC BACKGROUNDS UNDERLYING CARCINOGENESIS Article Open access 20 June 2023 PATHOGENIC GERMLINE VARIANTS IN CHINESE PANCREATIC ADENOCARCINOMA PATIENTS
Article Open access 05 March 2025 INTRODUCTION With 35,390 related deaths in 2018, pancreatic cancer is the fourth leading cause of cancer deaths in Japan, after lung, colorectal, and
stomach cancers1. The incidence and mortality rates of pancreatic cancer have increased steadily over the past decades, while those of other gastrointestinal cancers have shown decreasing
trends1. Despite the increasing burden levied by pancreatic cancer, few modifiable risk factors other than smoking and type 2 diabetes mellitus (T2D) have been identified, and the 5-year
survival proportions remain the worst (<10%) among major malignancies. Genome-wide association studies (GWASs) have increasingly revealed associations of pancreatic cancer susceptibility
with inherited genetic variations. Since the first GWAS, conducted by the PanScan consortium, identified common variants in the gene coding for the ABO blood group system in 20092, at least
23 genome-wide significant susceptibility loci have been linked to the risk of pancreatic cancer3. However, owing to the smaller sample sizes of the relevant GWASs, fewer loci have been
identified for pancreatic cancer than for other common cancers, including breast and colorectal cancers4,5. Furthermore, the risk variants identified to date explain approximately 13% of the
total heritability on the basis of GWAS-identified single-nucleotide polymorphisms (SNPs) in individuals of European ancestry6. These observations suggest that additional risk loci can be
identified by increasing the sample size, as evidenced by the trend in the numbers of variants reported by the PanScan and PanC4 consortia. It is also important to expand the GWASs to
populations of non-European ancestry because of differences in minor allele frequencies (MAFs) and patterns of linkage disequilibrium (LD) across diverse populations7. In fact, previous
GWASs focusing exclusively on populations of Eastern Asian ancestry led to the identification of additional susceptibility loci for breast and colorectal cancers8,9. The majority of the risk
loci for pancreatic cancer were discovered in the PanScan and PanC4 GWASs, which included populations of European ancestry. Only two GWASs have been conducted in East Asian populations: one
in China10 and one in Japan11. A total of eight risk loci (five genome-wide significant loci and three loci with suggestive evidence of association) have been identified for pancreatic
cancer, but these loci were not replicated in a previous study using samples from European populations12. Therefore, the role of common susceptibility loci in East Asian populations remains
uncertain and needs further exploration. To detect additional susceptibility loci for pancreatic cancer, we perform a meta-analysis combining all published and unpublished GWAS data in
Japan, followed by a replication study involving a Japanese population as well as other populations of East Asian origin. We identify 3 (13q12.2, 13q22.1, and 16p12.3) genome-wide
significant loci (_P_ < 5.0 × 10−8) and 4 suggestive loci (_P_ < 1.0 × 10−6) for the risk of pancreatic cancer. We replicate the associations between selected _GP2_ gene variants at
16p12.3 in 10,822 additional cases and controls of East Asian origin and further explore the functional impact of the top SNP rs78193826 of the _GP2_ gene. We also demonstrate pleiotropic
effects of the _GP2_ variants. Together, these findings indicate that _GP2_ gene variants are probably associated with pancreatic cancer susceptibility in populations of East Asian ancestry.
RESULTS GWAS META-ANALYSIS AND REPLICATION After imputation and quality control of individual subject genotype data, we performed a meta-analysis of 3 Japanese GWASs comprising data from
2039 pancreatic cancer patients and 32,592 controls for 7,914,378 SNPs (Supplementary Table 1 and Supplementary Data 1). Genomic control adjustment was not applied because there was little
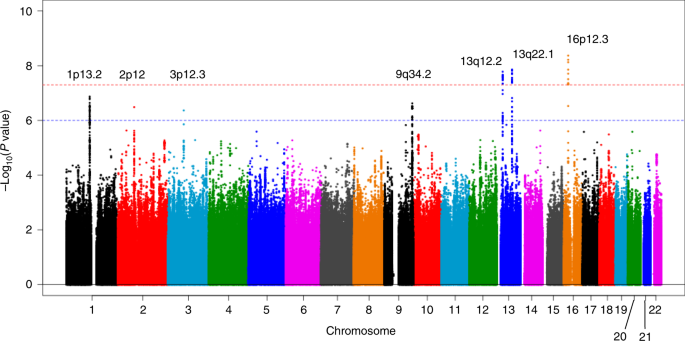
evidence of genomic inflation (lambda = 1.02, Supplementary Fig. 1). We observed genome-wide significant _(P_ < 5.0 × 10−8) association signals at 3 loci (13q12.2, 13q22.1, and 16p12.3;
Fig. 1 and Table 1), for which the genes nearest the lead SNP were _PLUT_ (_PDX1-AS1_) and _PDX1_ on 13q12.2, _KLF5_ and _KLF12_ on 13q22.2, and _GP2_ on 16p12.3. In addition, 4 loci (1p13.2
(_WNT2B_), 2p12 (_CTNNA2_), 3p12.3 (_ROBO2_), and 9q34.2 (_ABO_)) showed suggestive evidence of associations (_P_ < 1.0 × 10−6, Supplementary Table 2). Among these risk loci, genome-wide
significant associations were observed for 10 SNPs at 16p12.3 (rs78193826, rs117267808, rs73541251, rs4609857, rs4544248, rs4632135, rs4420538, rs73541271, rs4383154, and rs4383153;
Supplementary Data 2). The odds ratio (ORs) for these variants ranged from 1.43 to 1.47, indicating generally stronger associations in this region than for GWAS associations for variants
identified in previously published GWASs. The lead SNP here is rs78193826, a nonsynonymous variant of the _GP2_ gene (Fig. 2). The risk increased by 46% per copy of the minor T allele (OR =
1.46, 95% confidence interval (CI) = 1.29–1.66, _P_ = 4.28 × 10−9; Table 1). Regional association plots for the other loci are shown in Supplementary Fig. 2. According to the 1000 Genomes
Project Phase 3 database, rs78193826 is polymorphic, with the MAF ranging from 3.9% to 6.6% in Asian populations, compared with much lower MAFs (<0.1%) in other human populations
(Supplementary Data 2). LD maps of these 10 SNPs at 16p12.3 are shown in Supplementary Fig. 3. Complete LD between nine of these SNPs (all except rs4420538) was observed in the Japanese
population. Among the ten SNPs in this region, only rs4383153 had available association summary statistics in the previous PanScan and Panc4 consortia publications2,13, but this SNP was
negligible and not significantly associated with pancreatic cancer risk (Supplementary Table 4). The functional annotation results for the ten SNPs at 16p12.3 are shown in Supplementary Data
2. The lead SNP rs78193826 was classified as “damaging” according to the Sifting Intolerant from Tolerant (SIFT) algorithm and as “possibly damaging” by Polymorphism Phenotyping v2
(PolyPhen-2). Moreover, the estimated Combined Annotation-Dependent Depletion (CADD) score was 20.3. For replication, we selected four SNPs (rs78193826, rs73541251, rs117267808, rs4632135)
that met either of the following criteria: (1) exonic SNP or (2) intronic SNP with a score of ≤3 according to the RegulomeDB database. We sought the replication of associations between these
4 SNPs and pancreatic cancer in populations of East Asian ancestry, including Japanese, Japanese American, and Chinese subjects, using an additional sample of 1926 cases and 8896 controls
drawn from 6 independent studies. All 4 SNPs were significantly associated with pancreatic cancer risk (_P_ < 0.05) in the combined replication analysis (Supplementary Table 3), with the
ORs for the lead SNP rs78193826 shown for each study cohort in Fig. 3. FUNCTIONAL CHARACTERIZATION OF THE _GP2_-CODING VARIANT We explored the functional impact of the identified coding
variant rs78193826 in the GP2-expressing pancreatic cancer cell line PaTu 8988s. The single-nucleotide change from G to A at codon 282 of the GP2 gene was induced using clustered regularly
interspaced short palindromic repeats (CRISPR)-Cas9-mediated homologous recombination, which enabled the generation of genome-edited PaTu 8988s cells (GP2_V282M PaTu 8988s, Fig. 4a). With
RNA-sequencing (RNA-seq) analysis of two separate clones established from wild-type (GP2_WT, serving as a control group) and genome-edited (GP2_V282M) PaTu 8988s cells, we demonstrated
consistently different gene expression patterns between GP2_WT and GP2_V282M cells (Fig. 4b, Supplementary Data 3). Gene set enrichment analysis (GSEA) using the collections of hallmark (H)
and oncogenic gene sets (C6) in MSigDB v6.214 yielded numerous significantly enriched gene sets for GP2_WT cells. Among them, gene sets related to _KRAS_-activating mutations appeared in
both the H and C6 collections (Supplementary Data 4 and 5). Because _KRAS_ mutation is the most frequently observed mutation and a critical initiating event in pancreatic cancer15, we
focused on these gene sets. Genes in HALLMARK_KRAS_SIGNALING_DN and KRAS.50_UP. V1_DN were significantly expressed at low levels in GP2_V282M PaTu 8988s cells (Fig. 4c). We recapitulated the
downregulation of three genes (_KLK7_, _BMPR1B_, and _KLK8_) in HALLMARK_KRAS_SIGNALLING_DN (Fig. 4d) in three independent GP2_V282M clones compared to GP2_WT clones by quantitative
real-time (qRT)-PCR, suggesting that the monoclonal findings were not due to selection bias (Fig. 4e). We also excluded the possibility of vector transfection–induced off-target effects or
non-specific effects on the expression of genes in specific pathways (Supplementary Figs. 9 and 10, Supplementary Note 1). PLEIOTROPIC EFFECTS OF _GP2_ VARIANTS Epidemiological studies have
consistently shown that longstanding T2D is associated with a mildly increased risk of pancreatic cancer16. Recently, 88 genetic variants were reported in a GWAS meta-analysis of T2D in the
Japanese population17. We found that the top 3 SNPs at 16p12.3 (rs78193826, rs117267808, and rs73541251) were also genome wide significantly associated with the risk of T2D in the latest
GWAS comprising 191,764 Japanese subjects (Supplementary Table 5). In addition, these 3 SNPs were significantly associated with hemoglobin A1c (HbA1c; _P_ < 1 × 10−4) and blood glucose
levels (_P_ < 0.01), which were included in other GWASs of quantitative traits in 42,790 and 93,146 Japanese subjects, respectively18. Among 82 T2D-related SNPs, rs117267808 at 16p12.3
(_GP2)_ and rs2290203 at 15q26.1 (_PRC1-AS1_) were significant after Bonferroni correction in our GWAS meta-analysis (_P_ < 0.0006, Supplementary Data 6). Among 15 blood glucose-related
SNPs, rs4581570 at 13q12.2 (_PDX1_) was significant after Bonferroni correction (_P_ < 0.0033, Supplementary Data 7). In addition, none of the 25 HbA1c-associated (Supplementary Data 8)
and 76 body mass index (BMI)-associated SNPs (Supplementary Data 9) were significant in our GWAS meta-analysis. MENDELIAN RANDOMIZATION (MR) ANALYSIS To examine whether the associations of
metabolic traits with pancreatic cancer are consistent with a systematic association, we performed a MR analysis with the inverse variance-weighted (IVW) and MR–Egger methods. Inconsistent
results were observed for T2D, with no significant association between the SNP index T2D and pancreatic cancer risk based on the IVW method (Fig. 5a). Although the MR–Egger analysis yielded
a significant association, the intercept differed significantly from zero (_P_ < 0.05, Supplementary Fig. 4a), suggesting the presence of horizontal pleiotropy. Applying the MR Pleiotropy
RESidual Sum and Outlier (MR-PRESSO) test enabled the detection of two outlying SNPs (rs117267808 at 16p12.3 (_GP2_) and rs2290203 at 15q26.1 (_PRC1-AS1_); Supplementary Data 6). After
correction for horizontal pleiotropy via outlier removal, the association remained nonsignificant in the IVW analysis (_β_ ± SE = 0.08 ± 0.06, _P_ = 0.16). In contrast, the HbA1c genetic
index level appeared to be related to an increased risk of pancreatic cancer, on the basis of significant results with both the IVW and MR–Egger methods (Fig. 5b and Supplementary Fig. 4b).
In addition, we found no significant associations between genetic index levels for the other two metabolic factors (blood glucose and BMI) and pancreatic cancer risk (Supplementary Fig. 5).
GENE-BASED GWAS To complement the SNP-based GWAS, we performed a gene-based GWAS using MAGMA19 (Supplementary Fig. 6). Among 17,581 genes, we confirmed the significant associations for _GP2_
and _WNT2B_ identified by the SNP-based GWAS. In addition, a significant association (Bonferroni-corrected _P_ = 2.84 × 10−6) for the gene _KRT8_ was observed (Supplementary Table 6 and
Supplementary Figs. 6 and 7), and this association was replicated in the combined PanScan I and PanScan II datasets (_P_ = 0.024)13. REPLICATION OF PANSCAN AND PANC4 CONSORTIA RISK LOCI We
also examined the previously published pancreatic cancer risk loci from the PanScan and PanC4 consortia20, noting that 5 of those 19 SNPs were statistically significant after Bonferroni
correction (_P_ < 0.0026, rs13303010 at 1p36.33, rs505922 at 9q34.2, rs9581943 at 13q12.2, rs7214041 at 17q24.3, and rs9543325 at 13q22.1) in our GWAS meta-analysis (Supplementary Data
10). Notably, we confirmed the significant association (_P_ = 3.84 × 10−5) between rs505922 of the _ABO_ locus and pancreatic cancer risk. DISCUSSION The role of inherited common genetic
variations in pancreatic cancer susceptibility remains incompletely understood. We identified and replicated a risk locus at 16p12.3 by combining three GWAS datasets of East Asian
populations. Furthermore, we provided evidence that the identification of this locus can be attributed to the observed differences in the MAF of the lead SNP (rs78193826) at 16p12.3 and the
LD structure in this region across ethnic populations. Little overlap has been observed when risk loci reported from previous Chinese or Japanese GWASs are compared with those reported in
the PanScan GWASs2. By including more than twice the number of case patients than were included in previous Japanese or Chinese GWASs, as well as by using imputed SNP data, we replicated the
majority of significant loci discovered for pancreatic cancer in the PanScan and PanC4 consortia GWASs (Supplementary Data 10), including the well-established _ABO_ locus. Moreover, for
most variants, the direction and magnitude of the associations in our GWAS meta-analysis of Japanese subjects were consistent with those in populations of European ancestry. These findings
suggest that GWAS-identified variants at many loci are shared across ancestral groups and that lack of replication may be due to insufficient sample sizes in previous Chinese or Japanese
GWASs. The MAF of rs78193826 in cases varies considerably across populations in the replication cohorts when compared with that in control subjects. The main reason may be due to random
variation caused by a small sample size. As shown in Supplementary Table 3, the MAF was higher in controls than in cases in the Japan Public Health Center-based Prospective Study (JPHC),
generating an opposite direction in effect size from that observed in other replication cohorts. However, the overall positive association for rs78193826 was replicated in the analysis
combining all replication cohorts. Another possible reason is population stratification, but the Multiethnic Cohort Study (MEC) results were adjusted for the principal component. After
excluding the JPHC and MEC studies from the replication cohorts, we found no large variations in MAF in the cases. Several lines of evidence indicate that rs78193826, which lies within the
_GP2_ gene on 16p12.3, may be associated with pancreatic cancer risk. First, this variant is nonsynonymous; the nucleotide mutation from C to T causes an amino acid change from valine to
methionine, which could affect protein structure and function. Second, functional annotations in several databases consistently indicate that this variant is likely pathogenic. Third, the
observed differences in the MAF of rs78193826 as well as the LD structure across different ethnic populations provide indirect evidence supporting its role as a significant variant in the
Japanese population. The frequency of the minor T allele of rs78193826 is 0.1% in populations of European ancestry but 7% in the Japanese population. Given this apparent difference in the
MAF, rs78193826 could not have been identified in the PanScan GWASs, although the PanScan GWASs included a much larger sample size than our GWAS. Of the 10 SNPs in LD in this region, only
rs4383153 has association summary statistics available in PanScan publications; however, no significant associations were observed between this SNP and pancreatic cancer risk (Supplementary
Table 4). While complete LD between rs4383153 and rs78193826 was evident in the Japanese population, no LD data were available for these two SNPs in the European ancestry populations (1000
Genomes Project Phase 3 CEU). Genetic variations in the _GP2_ gene have been linked to several phenotypes in addition to pancreatic cancer. The SNP rs12597579, located ~60 kb downstream of
_GP2_, has been associated with BMI in a GWAS including East Asians21. However, rs12597579 was not in LD with rs78193826 (_r_2 = 0.003, calculated from Japanese samples in the 1000 Genomes
Project Phase 3), suggesting that rs12597579 may have functions different from those of rs78193826. Perhaps coincidently, the lead variant (rs117267808) in the _GP2_ gene identified in the
latest GWAS meta-analysis of T2D in the Japanese population was also identified in our GWAS meta-analysis of pancreatic cancer (Supplementary Table 5). Of the 82 T2D-related SNPs, 2 showed
significant associations with pancreatic cancer, suggesting that pancreatic cancer and T2D may share specific genetic susceptibility factors. The identified lead SNP (rs78193826) is in the
coding sequence for the GP2 protein, which is present on the inner surface of zymogen granules in pancreatic acinar cells22. GP2 is a glycosylated protein of ∼90 kDa that contains multiple
sites, such as an asparagine-linked glycosylation site, a zona pellucida domain, and a glycosylphosphatidylinositol linkage to the membrane22. During the secretory process, GP2 is cleaved
from the membrane and secreted into the pancreatic duct along with other digestive enzymes. The expression of GP2 is extremely high in normal pancreatic tissues compared with that in other
tissues (Supplementary Fig. 8)23. Furthermore, a previous RNA transcriptome analysis revealed that pancreatic tumor tissues have a decreased level of GP2 expression compared with adjacent
benign pancreatic tissues24. The functional characterization of GWAS-identified SNPs remains a challenge. We conducted a series of experiments to examine the possible functional impact of
the nonsynonymous lead SNP rs78193826 on global gene expression. RNA sequencing (RNA-seq) analysis revealed consistently differential gene expression patterns between genome-edited GP2_V282M
cells and control GP2_WT cells (Supplementary Data 3). Among many significantly enriched gene sets identified for GP2_WT cells in the GSEA, _KRAS_-related gene sets stood out because of the
well-known role of _KRAS_ mutation in pancreatic carcinogenesis (Supplementary Data 4 and 5)15. Interestingly, gene signatures related to _KRAS_-activating mutations
(HALLMARK_KRAS_SIGNALING_DN and KRAS.50_UP. V1_DN) in two distinct collections (H and C6) showed significant enrichment in GP2_WT cells. Collectively, these experimental findings suggest
that the functional relevance of rs78193826 may involve modulation of KRAS activity. Given that a very high frequency (>93%) of _KRAS_ mutations has been observed in pancreatic cancer
patients15, elucidating interactions among _GP2_ variants, _KRAS_ oncogenic mutations, and other potential effector genes would provide insights into pancreatic carcinogenesis. Previous
epidemiological studies have suggested that HbA1c levels, even in nondiabetic ranges, or changes in HbA1c levels in new-onset T2D are associated with pancreatic cancer risk25,26. Our MR
analysis of selected metabolic factors provided corroborating evidence that HbA1c genetic index levels may be associated with pancreatic cancer risk. This result was also partially
consistent with a previous MR analysis, in which T2D was not implicated but the genetic indices of BMI and fasting insulin were associated with pancreatic cancer27. However, it should be
noted that the MR results may be influenced by a few SNPs with relatively large effect sizes. To address this possibility, we further applied MR-PRESSO and detected two outlying SNPs
(Supplementary Data 6), one of which was the _GP2_ SNP. This finding indicated that the _GP2_ SNP may differ from other T2D-related SNPs in terms of the effect on pancreatic cancer risk. It
is likely that the null findings for the MR association for T2D reflect the phenotypic and genetic heterogeneity of T2D, but T2D may also be both a cause and consequence of pancreatic
cancer28. Additional studies are needed to further explore the associations of T2D and metabolic factors with pancreatic cancer using the best available genetic instruments in the Japanese
population. Three genome-wide significant genes (_GP2_, _WNT2B_, and _KRT8_) emerged in the gene-based GWAS. Among these genes, _WNT2B_ (1p13.1) showed suggestive evidence of association in
the previous PanScan and PanC4 GWAS6. KRT8 belongs to a group of intermediate-filament cytoskeletal proteins involved in maintaining epithelial structural integrity29. KRT8 is expressed in
both ductal and acinar single-layer epithelia, and mutations in the _KRT8_ gene have been linked to exocrine pancreatic disorders and liver disease30,31. In conclusion, our GWAS
meta-analysis identified a risk locus at chromosome 16p12.3 within the _GP2_ gene for pancreatic cancer in populations of East Asian ancestry. Functional analyses using cell lines provided
supporting evidence of the effect of the lead SNP rs78193826 of the _GP2_ gene on KRAS activity. Further fine mapping and functional characterization are needed to elucidate the associations
of common _GP2_ gene variants with pancreatic cancer susceptibility. Our findings also highlight genetic susceptibility factors shared between T2D and pancreatic cancer. METHODS STUDY
SAMPLES We performed a GWAS meta-analysis based on three Japanese studies: the Japan Pancreatic Cancer Research (JaPAN) consortium GWAS, the National Cancer Center (NCC) GWAS, and the
BioBank Japan (BBJ) GWAS. An overview of the characteristics of the study populations is provided in Supplementary Table 1. JAPAN CONSORTIUM GWAS Participants in this GWAS were drawn from
the JaPAN consortium32. Two case–control datasets were combined, resulting in data from a total of 945 pancreatic cancer patients and 3134 controls. The vast majority of patients were
diagnosed with primary adenocarcinoma of the exocrine pancreas (ICD-O-3 codes C250–C259). The first dataset included 622 pancreatic cancer patients who were recruited from January 2010 to
July 2014 at 5 participating hospitals in the Central Japan, Kanto, and Hokkaido regions. This multi-institutional case–control study collected questionnaire data on demographic and
lifestyle factors and 7-ml blood samples from the study participants. The second dataset included 323 patients with newly diagnosed pancreatic cancer and 3134 control subjects recruited for
an epidemiological research program at Aichi Cancer Center (HERPACC) between 2005 and 2012. All outpatients on their first visit to Aichi Cancer Center were invited to participate in
HERPACC. Those who agreed to participate completed a self-administered questionnaire and provided a 7-ml blood sample. After quality control, 943 cases and 3057 controls remained for the
subsequent analysis (Supplementary Data 1). None of the control subjects had a diagnosis of cancer by the time of recruitment. Written informed consent was obtained from all study
participants, and the study protocol was approved by the Ethical Review Board of Aichi Medical University, the Institutional Ethics Committee of Aichi Cancer Center, the Human Genome and
Gene Analysis Research Ethics Committee of Nagoya University, and the ethics committees of all participating hospitals. BIOBANK JAPAN GWAS Pancreatic cancer patient data were obtained from
the BBJ GWAS, which was launched in 2003 and collected DNA and clinical information from approximately 200,000 patients, including those with pancreatic cancer33. Overall, 422 pancreatic
cancer patients with available genotype data were recruited from 2003 to 2008. Clinical information was collected using a standardized questionnaire. This study was approved by the ethics
committees of the RIKEN Center for Integrative Medical Sciences. Controls were drawn from the participants in four population-based cohort studies in Japan: the Japan Multi-Institutional
Collaborative Cohort Study, the JPHC, the Tohoku Medical Megabank Project Organization, and the Iwate Tohoku Medical Megabank Organization (Supplementary Note 2). In total, 28,870 controls
who passed genotype data quality-control assessments were included in the study. In all participating cohort studies, informed consent was obtained from the participants by following the
protocols approved by the corresponding institutional ethics committees. Detailed descriptions of the BBJ and each cohort study are provided in Supplementary Note 2. NATIONAL CANCER CENTER
GWAS The case and control samples were derived from a previous pancreatic cancer GWAS11. Case subjects were 677 patients diagnosed with invasive pancreatic ductal adenocarcinoma at the NCC
Hospital, Tokyo, Japan. Controls consisted of 677 Japanese volunteers who participated in a health check-up program in Tokyo. After preimputation quality control, data from 674 cases and 674
controls remained for the subsequent analysis (Supplementary Data 1). This project was approved by the ethics committee of the NCC. QUALITY CONTROL AND GENOTYPE IMPUTATION Quality control
for samples and SNPs was performed based on the study-specific criteria. For the study that included data genotyped on two different platforms, we performed imputation using those SNPs that
were available from both genotyping platforms. Genotype data in each study were imputed separately based on the 1000 Genomes Project reference panel (Phase 3, all ethnicities). The phasing
was performed with the use of SHAPEIT (v2)34 or Eagle2 (v2)35, and the imputation was performed using minimac336 or IMPUTE (v2)37. Information on the study-specific genotyping, imputation,
and analysis tools is provided in Supplementary Data 1. After genotype imputation, quality control was applied to each study. SNPs with an imputation quality of _r_2 < 0.5 or an MAF of
<0.01 were excluded. SNPs that passed quality control in at least two cohorts were included in the meta-analysis. ASSOCIATION ANALYSIS FOR SNPS AND PANCREATIC CANCER The association of
pancreatic cancer with the SNP allele dose was estimated using logistic regression analysis with adjustment for the first two principal components. The association magnitudes and standard
errors were used in the subsequent meta-analysis. GWAS META-ANALYSIS We performed a meta-analysis of three pancreatic cancer GWASs (JaPAN, BBJ, and NCC). The association results for each SNP
across the studies were combined with the METAL software (v2011-03-25) in a fixed effects IVW meta-analysis. Heterogeneity in allelic associations was assessed using the _I_2 index. The
meta-analysis included 7,914,378 SNPs with genotype data available from at least two cohorts. A _P_ value threshold of 5 × 10−8 was used to establish a threshold for genome-wide
significance. We assessed the inflation of test statistics using the genomic control lambda. REPLICATION ANALYSIS We sought replication of the SNP associations in six additional studies
involving populations of East Asian origin, including Japanese, Japanese American, and Chinese individuals. In total, we assembled genotype data from 1926 cases and 8896 controls for the
replication analysis. Detailed information on the study descriptions, quality-control thresholds, and exclusion criteria for each replication cohort is provided in Supplementary Data 1 and
the Supplementary Note 3. The association between the SNP allele dose and pancreatic cancer risk in each replication cohort was estimated using logistic regression analysis with the
adjustment for study-specific covariates shown in Supplementary Data 1. For the lead SNP rs78193862, we also performed a fixed-effects IVW meta-analysis of SNP associations by combining all
six study sample sets included in the replication analysis. FUNCTIONAL ANNOTATION To prioritize the associated SNPs of the identified loci, we adopted a series of bioinformatic approaches to
collate functional annotations. We first used ANNOVAR38 to obtain an aggregate set of functional annotations—including gene locations and impacts of amino acid substitutions based on
prediction tools, such as SIFT, PolyPhen-2, and CADD—for SNPs with _P_ values <5 × 10–8 for pancreatic cancer. We also explored potential effects on gene regulation by annotating these
SNPs with information from the RegulomeDB database39. FUNCTIONAL CHARACTERIZATION We chose PaTu 8988s for the functional study, because it is the only pancreatic adenocarcinoma cell line
expressing _GP2_ according to the Cancer Cell Line Encyclopedia. PaTu 8988s cells were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany)
and maintained with RPMI 1640 (Invitrogen) plus 10% fetal bovine serum at 37 °C in a 5% CO2 cell culture incubator. PaTu 8988s cells were genotyped for identity at BEX CO., LTD. and tested
routinely for mycoplasma contamination. GENERATION OF THE GP2_V282M PATU 8988S CELL LINE Mutation was induced through CRISPR-Cas9-mediated homologous recombination. The plasmid
pSpCas9(BB)-2A-GFP (PX458) (Addgene plasmid # 48138) was purchased from Addgene (Cambridge, MA)40. To generate a nucleotide mutation from G to A at codon 282 (synonymous with 285, 429, and
432; Supplementary Data 2) of the _GP2_ gene, we selected an single-guide RNA target with CHOPCHOP (https://chopchop.cbu.uib.no)41 and designed a DNA repair template with 45 bp homology
arms. Oligonucleotide pairs were annealed and ligated into the BbsI-linearized PX458 plasmid. Cells were transfected with the vector and the repair template using Lipofectamine 3000 (Life
Technologies) according to the manufacturer’s instructions. To obtain monoclonal clones, GFP-positive cells were sorted as single cells into 96-well plates using a BD FACSAria III cell
sorter (BD Biosciences) 48 h post-transfection. After 3 weeks of culture, cells were distributed into two 24-well plates, followed by Sanger sequencing-based genotyping. Mutation was also
confirmed by RNA-seq. A clone harboring the precise mutation was used for further analysis. All oligonucleotide primers were obtained from FASMAC (Kanagawa, Japan; Supplementary Table 7).
Control cell lines were obtained from clones without induced mutations at codon 282 by the same plasmid transfection. TRANSIENT TRANSFECTION EXPERIMENTS In all, 2 × 105 Patu-S cells plated
in 6-well plate were transfected with the empty vector alone or the sgGP2-cloned plasmid and the repair template using Lipofectamine 3000 (Life Technologies) according to the manufacturer’s
instructions. Total RNA was isolated 72 h post-transfection followed by cDNA synthesis and qRT-PCR. RNA ISOLATION AND CDNA SYNTHESIS Total RNA was isolated using the miRNeasy Mini Kit
(Qiagen) with DNase I (Qiagen) digestion according to the manufacturer’s instructions. cDNA was synthesized from total RNA using Superscript III (Invitrogen) and random primers (Invitrogen).
QUANTITATIVE REAL-TIME PCR qRT-PCR was performed using SYBR® Premix Ex Taq TM (TaKaRa) on an Applied Biosystems 7900HT Real-Time PCR System. All oligonucleotide primers were obtained from
FASMAC (Kanagawa, Japan; Supplementary Table 7). RNA-SEQUENCING RNA-seq was performed by GeneWiz Inc. (Saitama, Japan) in paired-end mode. RNA-seq reads were mapped to NCBI37 with TopHat2
and quantified to the human transcriptome (refGene) with GFOLD42. Gene expression was quantified in the form of RPKM (reads per kilobase of transcript, per million mapped reads). The
reference sequence and refGene GTF files were obtained from iGenomes. DIFFERENTIAL EXPRESSION ANALYSIS AND GSEA Differentially expressed genes were identified using GFOLD (v1.1)42 based on
comparisons between the GP2_V282M and GP2_WT groups. Significantly differentially expressed genes were defined as genes with empirical false discovery rates (FDR) < 0.10. The GSEA was
performed using the MSigDB v6.2 collections—H and C614. Genes with coding lengths <200 bp or <10 mean reads in either the GP2_V282M or GP2_WT groups were excluded. A rank list was
generated by ordering each gene according to the log2-fold-change value (log2fdc from GFOLD), which was calculated from the expression level ratio of GP2_V282M/GP2_WT. These rank lists were
used in a weighted, preranked GSEA. Sets of 1000 permutations of the genes were applied in the preranked GSEA performed with the above-described collections of gene sets. An FDR < 0.10
was considered significant for the GSEA analysis. MR ANALYSIS We performed MR analyses for the associations of pancreatic cancer with selected metabolic traits, including T2D, HbA1c, blood
glucose, and BMI. As genetic instruments for each trait, we selected genome-wide significant SNPs that had been reported in the three previously published GWAS meta-analyses involving
Japanese subjects17,18,43. For the two-sample MR analysis of T2D and pancreatic cancer, we did not exclude duplicate samples (15.5% found only in the controls) because retaining these
samples was unlikely to introduce substantial bias44. Data from 106 pancreatic cancer cases were excluded from the HbA1c GWAS, and the association magnitudes for the HbA1c-associated SNPs
were re-estimated. After the exclusion of SNPs on the X chromosome or SNPs without genotype data (Supplementary Data 6–9), the summary data were available for 82 T2D-related SNPs, 25
HbA1c-related SNPs, 15 blood glucose-related SNPs, and 76 BMI-related SNPs. The associations of these SNPs with pancreatic cancer risk were analyzed using the IVW and MR–Egger regression
methods. MR analysis was performed with the MendelianRandomization package45. Given that the presence of horizontal pleiotropy may violate MR assumptions, leading to invalid results, we
further applied the MR-PRESSO test to detect and correct for horizontal pleiotropic outliers46. GENE-BASED ANALYSIS SNP-based _P_ values for 7,914,378 SNPs were combined into gene-based _P_
values for 17,581 genes using the MAGMA software version 1.0618. SNP summary statistics (_P_ values) from the meta-analysis were used as input for MAGMA. In gene-based association
statistics, LD between SNPs was accounted for, and the _P_ value threshold for genome-wide significant associations was set at 2.84 × 10−6 (=0.05/17,581). The 1000 Genomes reference panel
(Phase 3, East Asian) was used to control for LD. We did not include any upstream/downstream regions around the genes in this analysis; only variants located between the first exon and the
last exon of a gene were used to calculate the gene-based _P_ values. The NCBI Gene database was used to define genomic intervals for protein-coding genes. To replicate the association
between _KRT8_ and pancreatic cancer, we applied SNP summary statistics from PanScan 1 and PanScan 2 (pha002889.1)13 to MAGMA. The MAF of the Haplotype Map (HapMap) project Phase 2 CEU
samples for each SNP was added to the summary statistics, because the pha002889.1 data did not include the MAFs. We excluded variants with call fractions <95% in either the case or
control data, Hardy–Weinberg equilibrium _P_ value <10−6 in the controls, or MAF < 0.01. The 1000 Genomes reference panel (Phase 3, European) was used to control for LD. The
significance level was set at _α_ = 0.05. REPORTING SUMMARY Further information on research design is available in the Nature Research Reporting Summary linked to this article. DATA
AVAILABILITY The summary GWAS statistics for this analysis are available on the website of the JaPAN consortium [http://www.aichi-med-u.ac.jp/JaPAN/current_initiatives-e.html]. The RNA-seq
data are available at the Gene Expression Omnibus under the following accession number: GSE147368. The other datasets generated during this study are available from the corresponding author
upon reasonable request. The source data underlying Fig. 4e are provided as a Source data file. REFERENCES * Ministry of Health, Labour and Welfare, Japan. Vital statistics (1958-2016).
https://ganjoho.jp/reg_stat/statistics/dl/index.html (2017). * Amundadottir, L. et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to
pancreatic cancer. _Nat. Genet._ 41, 986–990 (2009). Article CAS PubMed PubMed Central Google Scholar * Amundadottir, L. T. Pancreatic cancer genetics. _Int. J. Biol. Sci._ 2, 314–325
(2016). Article CAS Google Scholar * Fachal, L. & Dunning, A. M. From candidate gene studies to GWAS and post-GWAS analyses in breast cancer. _Curr. Opin. Genet. Dev._ 30, 32–41
(2015). Article CAS PubMed Google Scholar * Schmit, S. L. et al. Common genetic susceptibility loci for colorectal cancer. _J. Natl Cancer Inst._ 111, 146–157 (2019). Article PubMed
CAS Google Scholar * Childs, E. J. et al. Common variation at 2p13.3, 3q29, 7p13 and 17q25.1 associated with susceptibility to pancreatic cancer. _Nat. Genet._ 47, 911–916 (2015). Article
CAS PubMed PubMed Central Google Scholar * Park, S., Cheng, I. & Haiman, C. A. Genome-wide association studies of cancer in diverse populations. _Cancer Epidemiol. Prev. Biomark._
27, 405–417 (2018). Article CAS Google Scholar * Zhang, B. et al. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. _Nat. Genet._
46, 533–542 (2014). Article CAS PubMed PubMed Central Google Scholar * Cai, Q. et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at
1q32.1, 5q14.3 and 15q26.1. _Nat. Genet._ 46, 886–890 (2014). Article CAS PubMed PubMed Central Google Scholar * Wu, C. et al. Genome-wide association study identifies five loci
associated with susceptibility to pancreatic cancer in Chinese populations. _Nat. Genet._ 44, 62–66 (2012). Article CAS Google Scholar * Low., S. K. et al. Genome-wide association study
of pancreatic cancer in Japanese Population. _PLoS ONE_ 5, e11824 (2010). Article ADS PubMed PubMed Central CAS Google Scholar * Campa, D. et al. Lack of replication of seven
pancreatic cancer susceptibility loci identified in two Asian populations. _Cancer Epidemiol. Prev. Biomark._ 22, 320–323 (2013). Article CAS Google Scholar * Li, D. et al. Pathway
analysis of genome-wide association study data highlights pancreatic development genes as susceptibility factors for pancreatic cancer. _Carcinogenesis_ 33, 1384–1390 (2012). Article CAS
PubMed PubMed Central Google Scholar * Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. _Proc. Natl Acad.
Sci. USA_ 102, 15545–15550 (2005). Article ADS CAS PubMed PubMed Central Google Scholar * The Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic
ductal adenocarcinoma. _Cancer Cell_ 32, 185–203 (2017). Article PubMed Central CAS Google Scholar * Li, D. Diabetes and pancreatic cancer. _Mol. Carcinog._ 51, 64–74 (2012). Article
CAS PubMed PubMed Central Google Scholar * Suzuki, K. et al. Identification of 28 novel susceptibility loci for type 2 diabetes in the Japanese population. _Nat. Genet._ 51, 379–386
(2019). Article CAS PubMed Google Scholar * Kanai, M. et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. _Nat. Genet._
50, 390–400 (2018). Article CAS PubMed Google Scholar * de Leeuw, C. A. et al. MAGMA: generalized gene-set analysis of GWAS data. _PLoS Comput. Biol._ 11, e1004219 (2015). Article
PubMed PubMed Central CAS Google Scholar * Klein, A. P. et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. _Nat. Commun._ 9, 556 (2018).
Article ADS PubMed PubMed Central CAS Google Scholar * Wen, W. et al. Meta-analysis identifies common variants associated with body mass index in east Asians. _Nat. Genet._ 44, 307–311
(2012). Article CAS PubMed PubMed Central Google Scholar * Ohno, H. & Hase, K. Glycoprotein 2 (GP2). _Gut Microbes_ 1, 407–410 (2010). Article PubMed PubMed Central Google
Scholar * Lonsdale, J. et al. The Genotype-Tissue Expression (GTEx) project. _Nat. Genet._ 45, 580–585 (2013). Article CAS Google Scholar * Mao, Y. et al. RNA sequencing analyses reveal
novel differentially expressed genes and pathways in pancreatic cancer. _Oncotarget_ 8, 42537–42547 (2017). Article PubMed PubMed Central Google Scholar * Grote, V. A. et al. Diabetes
mellitus, glycated haemoglobin and C-peptide levels in relation to pancreatic cancer risk: a study within the European Prospective Investigation into Cancer and Nutrition (EPIC) cohort.
_Diabetologia_ 54, 3037–3046 (2011). Article CAS PubMed Google Scholar * Lu, Y. et al. New-onset type 2 diabetes, elevated HbA1c, anti-diabetic medications, and risk of pancreatic
cancer. _Br. J. Cancer_ 113, 1607–1614 (2015). Article CAS PubMed PubMed Central Google Scholar * Carreras-Torres, R. et al. The role of obesity, type 2 diabetes, and metabolic factors
in pancreatic cancer: a Mendelian randomization study. _J. Natl Cancer Inst_. 109, djx012 (2017). * Risch, H. A. Diabetes and pancreatic cancer: both cause and effect. _J. Natl Cancer Inst._
111, 1–2 (2019). Article PubMed Google Scholar * Karantza, V. Keratins in health and cancer: more than epithelial cell markers. _Oncogene_ 30, 127–138 (2011). Article CAS PubMed
Google Scholar * Casanova, M. L. et al. Exocrine pancreatic disorders in transgenic mice expressing human keratin 8. _J. Clin. Invest._ 103, 1587–1595 (1999). Article CAS PubMed PubMed
Central Google Scholar * Ku, N. O. et al. Keratin 8 mutations in patients with cryptogenic liver disease. _N. Engl. J. Med._ 344, 1580–1587 (2001). Article CAS PubMed Google Scholar *
Nakatochi, M. et al. Prediction model for pancreatic cancer risk in the general Japanese population. _PLoS ONE_ 13, e0203386 (2018). Article PubMed PubMed Central CAS Google Scholar *
Nagai, A. et al. Overview of the BioBank Japan Project: study design and profile. _J. Epidemiol._ 27, S2–S8 (2017). Article PubMed PubMed Central Google Scholar * Delaneau, O. et al.
Improved whole-chromosome phasing for disease and population genetic studies. _Nat. Methods_ 10, 5–6 (2013). Article CAS PubMed Google Scholar * Loh, P. R. et al. Reference-based phasing
using the Haplotype Reference Consortium panel. _Nat. Genet._ 48, 1443–1448 (2016). Article CAS PubMed PubMed Central Google Scholar * Das, S. et al. Next-generation genotype
imputation service and methods. _Nat. Genet._ 48, 1284–1287 (2016). Article CAS PubMed PubMed Central Google Scholar * Howie, B. N. et al. A flexible and accurate genotype imputation
method for the next generation of genome-wide association studies. _PLoS Genet._ 5, e1000529 (2009). Article PubMed PubMed Central CAS Google Scholar * Wang, K., Li, M. &
Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. _Nucleic Acids Res._ 38, e164 (2010). Article PubMed PubMed Central CAS Google
Scholar * Boyle, A. P. et al. Annotation of functional variation in personal genomes using RegulomeDB. _Genome Res._ 22, 1790–1797 (2012). Article CAS PubMed PubMed Central Google
Scholar * Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. _Nat. Protoc._ 8, 2281–2308 (2013). Article CAS PubMed PubMed Central Google Scholar * Labun, K. et al.
CHOP v3: expanding the CRISPR web toolbox beyond genome editing. _Nucleic Acids Res._ 47, W171–W174 (2019). Article CAS PubMed PubMed Central Google Scholar * Feng, J. et al. GFOLD: a
generalized fold change for ranking differentially expressed genes from RNA-seq data. _Bioinformatics_ 28, 2782–2788 (2012). Article CAS PubMed Google Scholar * Akiyama, M. et al.
Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. _Nat. Genet._ 49, 1458–1467 (2012). Article CAS Google Scholar * Burgess, S., Davies,
N. M. & Thompson, S. G. Bias due to participant overlap in two-sample Mendelian randomization. _Genet. Epidemiol._ 40, 597–608 (2016). Article PubMed PubMed Central Google Scholar *
Yavorska, O. O. & Burgess, S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. _Int. J. Epidemiol._ 46, 1734–1739 (2017).
Article PubMed PubMed Central Google Scholar * Verbanck, M. et al. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between
complex traits anddiseases. _Nat. Genet._ 50, 693–698 (2018). Article CAS PubMed PubMed Central Google Scholar Download references ACKNOWLEDGEMENTS We thank Mayuko Masuda, Kikuko Kaji,
Kazue Ando, Etsuko Ohara, Sumiyo Asakura, Keiko Hanai, and Yoko Mitsuda for assistance with data collection. We would like to express our gratefulness to the staff of BioBank Japan for their
outstanding assistance. This work was supported by the Ministry of Health, Labor, and Welfare of Japan (H21-11-1) and the Ministry of Education, Culture, Sports, Science, and Technology of
Japan (Nos. 16H06277, 17K09095, 17H04127, 26293145, 26253041, and 17015018). HERPACC, a part of the JaPAN consortium, was supported by Grants-in-Aid for Scientific Research for Priority
Areas of Cancer (No. 17015018) and Innovative Areas (No. 221S0001); JSPS KAKENHI grants (Nos. 26253041, 15H02524, 16H06277, and 18H03045) from the Japanese Ministry of Education, Science,
Sports, Culture and Technology; and a Grant-in-Aid for the Third Term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health, Labor and Welfare of Japan and the Cancer
Biobank Aichi. This study was partially supported by the BioBank Japan project and the Tohoku Medical Megabank project, which is supported by the Ministry of Education, Culture, Sports,
Sciences and Technology of Japan and the Japan Agency for Medical Research and Development. The JPHC Study has been supported by the National Cancer Center Research and Development Fund
(23-A31[toku], 26-A-2, and 29-A-4) since 2011 and was supported by a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor and Welfare of Japan from 1989 to 2010. The
case–cohort study within the JPHC Study was supported by the National Cancer Center Research and Development Fund (28-A-19 and 31-A-18) and by Practical Research for Innovative Cancer
Control (JP16ck0106095h0003 and JP19ck0106266h0003) from the Japan Agency for Medical Research and Development, AMED. The J-MICC study was supported by Grants-in-Aid for Scientific Research
for Priority Areas of Cancer (No. 17015018) and Innovative Areas (No. 221S0001) and a JSPS KAKENHI grant (No. 16H06277) from the Japanese Ministry of Education, Science, Sports, Culture and
Technology. The JEPA study was supported by Grants-in-Aid for Scientific Research (S) and a JSPS KAKENHI grant (No. 15H05791) from the Japanese Ministry of Education, Science, Sports,
Culture and Technology. The Yale-Shanghai study was supported by NIH/NCI 5R01 CA114421. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
The MEC replication study was supported by NIH/NCI CA209798, and CA164973. AUTHOR INFORMATION Author notes * These authors contributed equally: Yingsong Lin, Masahiro Nakatochi, Yasuyuki
Hosono, Hidemi Ito. AUTHORS AND AFFILIATIONS * Department of Public Health, Aichi Medical University School of Medicine, Nagakute, Aichi, 480-1195, Japan Yingsong Lin, Chaochen Wang &
Shogo Kikuchi * Division of Public Health Informatics, Department of Integrated Health Sciences, Nagoya University Graduate School of Medicine, Nagoya, 461-8673, Japan Masahiro Nakatochi *
Department of Nursing, Nagoya University Graduate School of Medicine, Nagoya, 461-8673, Japan Masahiro Nakatochi * Division of Molecular Therapeutics, Aichi Cancer Center Research Institute,
Nagoya, 464-8681, Japan Yasuyuki Hosono, Yuko Hayashi & Hiromichi Ebi * Division of Cancer Information and Control, Aichi Cancer Center Research Institute, Nagoya, 464-8681, Japan
Hidemi Ito * Department of Descriptive Epidemiology, Nagoya University Graduate School of Medicine, Nagoya, 466-8550, Japan Hidemi Ito * Laboratory for Statistical Analysis, RIKEN Center for
Integrative Medical Sciences, Yokohama, 230-0045, Japan Yoichiro Kamatani, Masato Akiyama, Kazuyoshi Ishigaki, Ken Suzuki & Yukinori Okada * Laboratory of Complex Trait Genomics,
Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, 108-8639, Japan Yoichiro Kamatani * Division of Cancer
Epidemiology and Prevention, Aichi Cancer Center Research Institute, Nagoya, 464-8681, Japan Akihito Inoko, Yumiko Kasugai & Keitaro Matsuo * Department of Pathology, Aichi Medical
University School of Medicine, Nagakute, 480-1195, Japan Akihito Inoko * Genetics Division, National Cancer Center Research Institute, Tokyo, 104-0045, Japan Hiromi Sakamoto & Teruhiko
Yoshida * Data Science Division, Data Coordinating Center, Department of Advanced Medicine, Nagoya University Hospital, Nagoya, 461-8673, Japan Fumie Kinoshita & Yumiko Kobayashi * Chiba
Cancer Center, Chiba, 260-8717, Japan Hiroshi Ishii * Department of Hepato-biliary-pancreatic Medicine, The Cancer Institute Hospital of Japanese Foundation for Cancer Research, Tokyo,
135-8550, Japan Masato Ozaka, Takashi Sasaki, Masato Matsuyama & Naoki Sasahira * Department of Gastroenterology, Hepatobiliary and Pancreatic Medical Oncology Division, Kanagawa Cancer
Center, Yokohama, 241-8515, Japan Manabu Morimoto, Satoshi Kobayashi, Taito Fukushima, Makoto Ueno & Shinichi Ohkawa * Department of Gastroenterology, Tokyo Metropolitan Hiroo Hospital,
Tokyo, 150-0013, Japan Naoto Egawa * Department of Gastroenterology, Tokyo Metropolitan Komagome Hospital, Tokyo, 113-8677, Japan Sawako Kuruma * Hokkaido Chitose College of Rehabilitation,
Hokkaido, 066-0055, Japan Mitsuru Mori * Division of Hepatology and Pancreatology, Aichi Medical University School of Medicine, Nagakute, 480-1195, Japan Haruhisa Nakao * Sapporo
Shirakabadai Hospital, Sapporo, 062-0052, Japan Yasushi Adachi * Department of Pediatrics, Hyogo College of Medicine, Nishinomiya, Hyogo, 663-8501, Japan Masumi Okuda * Department of
Infectious Diseases, Kyorin University School of Medicine, Tokyo, 181-8611, Japan Takako Osaki & Shigeru Kamiya * Department of Gastroenterology, Aichi Cancer Center Hospital, Nagoya,
464-8681, Japan Kazuo Hara * Department of Gastroenterological Surgery, Aichi Cancer Center Hospital, Nagoya, 464-8681, Japan Yasuhiro Shimizu * Department of Genetics and Cell Biology,
Research Institute for Radiation Biology and Medicine, Hiroshima University, Hiroshima, 734-8553, Japan Tatsuo Miyamoto * Department of Human Genetics, Tokushima University Graduate School
of Medicine, Tokushima, 770-8503, Japan Tomohiro Kohmoto * Division of Molecular Genetics, Aichi Cancer Center Research Institute, Nagoya, 464-8681, Japan Tomohiro Kohmoto & Issei Imoto
* Department of Cancer Epidemiology, Nagoya University Graduate School of Medicine, Nagoya, 466-8550, Japan Yumiko Kasugai & Keitaro Matsuo * Division of Molecular Pathology, Institute
of Medical Science, The University of Tokyo, Tokyo, 108-8639, Japan Yoshinori Murakami & Makoto Hirata * Department of Ophthalmology, Graduate School of Medical Sciences, Kyushu
University, Fukuoka, 812-8582, Japan Masato Akiyama * Graduate School of Frontier Sciences, The University of Tokyo, Tokyo, 108-8639, Japan Koichi Matsuda * Department of Hepatobiliary and
Pancreatic Surgery, National Cancer Center Hospital, Tokyo, 104-0045, Japan Kazuaki Shimada * Department of Hepatobiliary and Pancreatic Oncology, National Cancer Center Hospital, Tokyo,
104-0045, Japan Takuji Okusaka * Center for Genomic Medicine, Graduate School of Medicine, Kyoto University, Kyoto, 606-8501, Japan Takahisa Kawaguchi, Meiko Takahashi & Fumihiko Matsuda
* Department of Epidemiology for Community Health and Medicine, Kyoto Prefectural University of Medicine, Kyoto, 602-8566, Japan Yoshiyuki Watanabe * Laboratory of Public Health, School of
Food and Nutritional Sciences, University of Shizuoka, Shizuoka, 422-8526, Japan Kiyonori Kuriki * Department of Public Health, Shiga University of Medical Science, Otsu, 520-2192, Japan Aya
Kadota * Department of Preventive Medicine, Nagoya University Graduate School of Medicine, Nagoya, 466-8550, Japan Rieko Okada & Kenji Wakai * Cancer Prevention Center, Chiba Cancer
Center Research Institute, Chiba, 260-8717, Japan Haruo Mikami * Department of International Island and Community Medicine, Kagoshima University Graduate School of Medical and Dental
Sciences, Kagoshima, 890-8544, Japan Toshiro Takezaki * Department of Public Health, Nagoya City University Graduate School of Medical Sciences, Nagoya, 467-8601, Japan Sadao Suzuki *
Division of Epidemiology, Center for Public Health Sciences, National Cancer Center, Tokyo, 104-0045, Japan Taiki Yamaji, Motoki Iwasaki, Norie Sawada & Atsushi Goto * Tohoku Medical
Megabank Organization, Tohoku University, Sendai, 980-8573, Japan Kengo Kinoshita, Nobuo Fuse & Fumiki Katsuoka * Iwate Tohoku Medical Megabank Organization, Iwate Medical University,
Iwate, 028-3694, Japan Atsushi Shimizu, Satoshi S. Nishizuka & Kozo Tanno * Department of Hygiene and Preventive Medicine, School of Medicine, Iwate Medicalm University, Iwate, 028-3694,
Japan Kozo Tanno * Department of Diabetes and Metabolic Diseases, Graduate School of Medicine, The University of Tokyo, Tokyo, 113-0033, Japan Ken Suzuki, Toshimasa Yamauchi & Takashi
Kadowaki * Laboratory for Endocrinology, Metabolism and Kidney Diseases, RIKEN Centre for Integrative Medical Sciences, Yokohama, 230-0045, Japan Ken Suzuki & Momoko Horikoshi *
Department of Statistical Genetics, Osaka University Graduate School of Medicine, Osaka, 565-0871, Japan Ken Suzuki & Yukinori Okada * Laboratory of Statistical Immunology, Immunology
Frontier Research Center (WPI-IFReC), Osaka University, Osaka, 565-0871, Japan Yukinori Okada * University of Hawaii Cancer Center, Honolulu, HI, 96813, USA Herbert Yu & Loic Le Marchand
* Laboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD, 20892, USA Jun Zhong &
Laufey T. Amundadottir * Department of Gastroenterological Surgery, Graduate School of Medicine, Osaka University, Osaka, 565-0871, Japan Yuichiro Doki & Hidetoshi Eguchi * Department of
Medical Data Science, Graduate School of Medicine, Osaka University, Osaka, 565-0871, Japan Hideshi Ishii * Department of Preventive Medicine, Keck School of Medicine, University of
Southern California, Los Angeless, CA, 90033, USA David Bogumil, Christopher A. Haiman & Veronica W. Setiawan * Norris Comprehensive Cancer Center, University of Southern California, Los
Angeles, CA, 90033, USA Christopher A. Haiman & Veronica W. Setiawan * Department of Surgery and Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka, 812-8582,
Japan Masaki Mori * Department of Chronic Disease Epidemiology, Yale School of Public Health, New Haven, CT, 06520, USA Harvey Risch * Center for Public Health Sciences, National Cancer
Center, Tokyo, 104-0045, Japan Shoichiro Tsugane * RIKEN Center for Integrative Medical Sciences, Yokohama, 230-0045, Japan Michiaki Kubo Authors * Yingsong Lin View author publications You
can also search for this author inPubMed Google Scholar * Masahiro Nakatochi View author publications You can also search for this author inPubMed Google Scholar * Yasuyuki Hosono View
author publications You can also search for this author inPubMed Google Scholar * Hidemi Ito View author publications You can also search for this author inPubMed Google Scholar * Yoichiro
Kamatani View author publications You can also search for this author inPubMed Google Scholar * Akihito Inoko View author publications You can also search for this author inPubMed Google
Scholar * Hiromi Sakamoto View author publications You can also search for this author inPubMed Google Scholar * Fumie Kinoshita View author publications You can also search for this author
inPubMed Google Scholar * Yumiko Kobayashi View author publications You can also search for this author inPubMed Google Scholar * Hiroshi Ishii View author publications You can also search
for this author inPubMed Google Scholar * Masato Ozaka View author publications You can also search for this author inPubMed Google Scholar * Takashi Sasaki View author publications You can
also search for this author inPubMed Google Scholar * Masato Matsuyama View author publications You can also search for this author inPubMed Google Scholar * Naoki Sasahira View author
publications You can also search for this author inPubMed Google Scholar * Manabu Morimoto View author publications You can also search for this author inPubMed Google Scholar * Satoshi
Kobayashi View author publications You can also search for this author inPubMed Google Scholar * Taito Fukushima View author publications You can also search for this author inPubMed Google
Scholar * Makoto Ueno View author publications You can also search for this author inPubMed Google Scholar * Shinichi Ohkawa View author publications You can also search for this author
inPubMed Google Scholar * Naoto Egawa View author publications You can also search for this author inPubMed Google Scholar * Sawako Kuruma View author publications You can also search for
this author inPubMed Google Scholar * Mitsuru Mori View author publications You can also search for this author inPubMed Google Scholar * Haruhisa Nakao View author publications You can also
search for this author inPubMed Google Scholar * Yasushi Adachi View author publications You can also search for this author inPubMed Google Scholar * Masumi Okuda View author publications
You can also search for this author inPubMed Google Scholar * Takako Osaki View author publications You can also search for this author inPubMed Google Scholar * Shigeru Kamiya View author
publications You can also search for this author inPubMed Google Scholar * Chaochen Wang View author publications You can also search for this author inPubMed Google Scholar * Kazuo Hara
View author publications You can also search for this author inPubMed Google Scholar * Yasuhiro Shimizu View author publications You can also search for this author inPubMed Google Scholar *
Tatsuo Miyamoto View author publications You can also search for this author inPubMed Google Scholar * Yuko Hayashi View author publications You can also search for this author inPubMed
Google Scholar * Hiromichi Ebi View author publications You can also search for this author inPubMed Google Scholar * Tomohiro Kohmoto View author publications You can also search for this
author inPubMed Google Scholar * Issei Imoto View author publications You can also search for this author inPubMed Google Scholar * Yumiko Kasugai View author publications You can also
search for this author inPubMed Google Scholar * Yoshinori Murakami View author publications You can also search for this author inPubMed Google Scholar * Masato Akiyama View author
publications You can also search for this author inPubMed Google Scholar * Kazuyoshi Ishigaki View author publications You can also search for this author inPubMed Google Scholar * Koichi
Matsuda View author publications You can also search for this author inPubMed Google Scholar * Makoto Hirata View author publications You can also search for this author inPubMed Google
Scholar * Kazuaki Shimada View author publications You can also search for this author inPubMed Google Scholar * Takuji Okusaka View author publications You can also search for this author
inPubMed Google Scholar * Takahisa Kawaguchi View author publications You can also search for this author inPubMed Google Scholar * Meiko Takahashi View author publications You can also
search for this author inPubMed Google Scholar * Yoshiyuki Watanabe View author publications You can also search for this author inPubMed Google Scholar * Kiyonori Kuriki View author
publications You can also search for this author inPubMed Google Scholar * Aya Kadota View author publications You can also search for this author inPubMed Google Scholar * Rieko Okada View
author publications You can also search for this author inPubMed Google Scholar * Haruo Mikami View author publications You can also search for this author inPubMed Google Scholar * Toshiro
Takezaki View author publications You can also search for this author inPubMed Google Scholar * Sadao Suzuki View author publications You can also search for this author inPubMed Google
Scholar * Taiki Yamaji View author publications You can also search for this author inPubMed Google Scholar * Motoki Iwasaki View author publications You can also search for this author
inPubMed Google Scholar * Norie Sawada View author publications You can also search for this author inPubMed Google Scholar * Atsushi Goto View author publications You can also search for
this author inPubMed Google Scholar * Kengo Kinoshita View author publications You can also search for this author inPubMed Google Scholar * Nobuo Fuse View author publications You can also
search for this author inPubMed Google Scholar * Fumiki Katsuoka View author publications You can also search for this author inPubMed Google Scholar * Atsushi Shimizu View author
publications You can also search for this author inPubMed Google Scholar * Satoshi S. Nishizuka View author publications You can also search for this author inPubMed Google Scholar * Kozo
Tanno View author publications You can also search for this author inPubMed Google Scholar * Ken Suzuki View author publications You can also search for this author inPubMed Google Scholar *
Yukinori Okada View author publications You can also search for this author inPubMed Google Scholar * Momoko Horikoshi View author publications You can also search for this author inPubMed
Google Scholar * Toshimasa Yamauchi View author publications You can also search for this author inPubMed Google Scholar * Takashi Kadowaki View author publications You can also search for
this author inPubMed Google Scholar * Herbert Yu View author publications You can also search for this author inPubMed Google Scholar * Jun Zhong View author publications You can also search
for this author inPubMed Google Scholar * Laufey T. Amundadottir View author publications You can also search for this author inPubMed Google Scholar * Yuichiro Doki View author
publications You can also search for this author inPubMed Google Scholar * Hideshi Ishii View author publications You can also search for this author inPubMed Google Scholar * Hidetoshi
Eguchi View author publications You can also search for this author inPubMed Google Scholar * David Bogumil View author publications You can also search for this author inPubMed Google
Scholar * Christopher A. Haiman View author publications You can also search for this author inPubMed Google Scholar * Loic Le Marchand View author publications You can also search for this
author inPubMed Google Scholar * Masaki Mori View author publications You can also search for this author inPubMed Google Scholar * Harvey Risch View author publications You can also search
for this author inPubMed Google Scholar * Veronica W. Setiawan View author publications You can also search for this author inPubMed Google Scholar * Shoichiro Tsugane View author
publications You can also search for this author inPubMed Google Scholar * Kenji Wakai View author publications You can also search for this author inPubMed Google Scholar * Teruhiko Yoshida
View author publications You can also search for this author inPubMed Google Scholar * Fumihiko Matsuda View author publications You can also search for this author inPubMed Google Scholar
* Michiaki Kubo View author publications You can also search for this author inPubMed Google Scholar * Shogo Kikuchi View author publications You can also search for this author inPubMed
Google Scholar * Keitaro Matsuo View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS Y.L., M.N., Y. Hosono, H. Ito, A.I., M.O., T.O, S. Kamiya,
and C.W. designed the study. Y.L., M.N., Y. Hosono, and Y. Kamatani wrote the manuscript. M.N., F.K., Y. Kobayashi., Y. Kamatani., M.A., K.I., and J.Z. performed the statistical analysis.
H.S., H.I., M.O., T.S., M. Matsuyama., N.S., M.M., S. Kobayashi, T.F., M.U., S.O., N.E., S. Kuruma, Mitsura Mori., H.N., Y.A., K.H., Y.S., Y.M., K. Matsuda, M. Hirata, K. Shimada, T.O.,
Y.W., K. Kuriki, A.K., R.O., H.M., T.T., S.S., K.W., T. Yamaji, M.I., N.S., A.G., S.T., K. Kinoshita, N.F., F.K., A.S., S.S.N., K.T., K. Suzuki, Y.O., M. Horikoshi, T. Yamauchi, T. Kadowaki,
H.Y., L.T.A., Y.D., H. Ishii, H.E., D.B., C.A.H., L.L.M., Masaki Mori, H.R., V.W.S., T. Yoshida, and M.K. contributed to data collection. Y. Hosono, A.I., T.M., Y. Hayashi, H.E., T.
Kohmoto, I.I., Y. Kasugai, M.K., T. Kawaguchi, M.T., and F.M. contributed to SNP genotyping and functional characterization. S. Kikuchi and K. Matsuo supervised the study. All authors
approved the final version of the manuscript. CORRESPONDING AUTHORS Correspondence to Yingsong Lin, Masahiro Nakatochi or Keitaro Matsuo. ETHICS DECLARATIONS COMPETING INTERESTS The authors
declare no competing interests. ADDITIONAL INFORMATION PEER REVIEW INFORMATION _Nature Communications_ thanks Paul Brennan and the other anonymous reviewer(s) for their contribution to the
peer review of this work. Peer review reports are available. PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION DESCRIPTION OF ADDITIONAL SUPPLEMENTARY FILES SUPPLEMENTARY DATA 1 SUPPLEMENTARY DATA 2 SUPPLEMENTARY DATA 3 SUPPLEMENTARY
DATA 4 SUPPLEMENTARY DATA 5 SUPPLEMENTARY DATA 6 SUPPLEMENTARY DATA 7 SUPPLEMENTARY DATA 8 SUPPLEMENTARY DATA 9 SUPPLEMENTARY DATA 10 PEER REVIEW FILE REPORTING SUMMARY SOURCE DATA SOURCE
DATA RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and
reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes
were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If
material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain
permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS
ARTICLE Lin, Y., Nakatochi, M., Hosono, Y. _et al._ Genome-wide association meta-analysis identifies _GP2_ gene risk variants for pancreatic cancer. _Nat Commun_ 11, 3175 (2020).
https://doi.org/10.1038/s41467-020-16711-w Download citation * Received: 10 September 2019 * Accepted: 15 May 2020 * Published: 24 June 2020 * DOI: https://doi.org/10.1038/s41467-020-16711-w
SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy
to clipboard Provided by the Springer Nature SharedIt content-sharing initiative




