- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT Although the land snail _Cepaea nemoralis_ is one of the most thoroughly investigated colour polymorphic species, there have been few recent studies on the inheritance of the shell
traits. Previously, it has been shown that the shell polymorphism is controlled by a series of nine or more loci, of which five make a single ‘supergene’ containing tightly linked colour and
banding loci and more loosely linked pigmentation, spread band and punctate loci. However, one limitation of earlier work was that putative instances of recombination between loci within
the supergene were not easily verified. We therefore generated a new set of _C. nemoralis_ crosses that segregate for colour, banding and pigmentation, and several other unlinked shell
phenotype loci. The snails were genotyped using a set of RAD-seq-derived loci that flank the supergene, and instances of recombination tested by comparing inferred supergene genotype against
RAD-marker genotype. We found no evidence that suspected ‘recombinant’ individuals are recombinant between loci within the supergene. As point estimates of recombination between both
colour/banding, and colour/pigmentation loci are zero, incomplete penetrance and epistasis are a better explanation for the apparent ‘recombinant’ phenotype of some snail shells. Overall,
this work, therefore, shows that the architecture of the supergene may not be as previously supposed. It also provides a resource for fine mapping of the supergene and other major shell
phenotype loci. SIMILAR CONTENT BEING VIEWED BY OTHERS FINE MAPPING OF THE _CEPAEA NEMORALIS_ SHELL COLOUR AND MID-BANDED LOCI USING A HIGH-DENSITY LINKAGE MAP Article Open access 27
September 2023 THE FIRST HIGH-DENSITY GENETIC MAP OF COMMON COCKLE (_CERASTODERMA EDULE_) REVEALS A MAJOR QTL CONTROLLING SHELL COLOR VARIATION Article Open access 10 October 2022 A
SUPERGENE UNDERLIES LINKED VARIATION IN COLOR AND MORPHOLOGY IN A HOLARCTIC SONGBIRD Article Open access 25 November 2021 INTRODUCTION Historically, some of the most important animals in
studying colour polymorphism have been the land snails _Cepaea nemoralis_ and the sister taxon, _C. hortensis_, because it is straightforward to collect them and record the frequencies of
the different morphs in different locations and habitats (Cain and Sheppard 1950; Cain and Sheppard 1952; Cain and Sheppard 1954; Jones et al. 1977). There is also the benefit that the major
loci that determine the polymorphism show simple Mendelian inheritance (Cook 1967; Jones et al. 1977). However, while ongoing and long-term studies on these animals continue to provide
compelling evidence for the fundamental role of natural selection in promoting and maintaining variation in natural populations, as well as the impact of modern-day habitat change (Cameron
and Cook 2012; Cook 2017; Silvertown et al. 2011), the last research on the inheritance of the loci that determine the polymorphism dates to the late 1960s. This is a problem because now
that there is finally some progress towards identifying the genes involved (Kerkvliet et al. 2017; Mann and Jackson 2014; Richards et al. 2013), it is important that laboratory crosses are
available, to validate prior knowledge on the inheritance and for use in fine mapping recombination break-points. Previous work has shown that the shell polymorphism is controlled by a
series of nine or more loci, of which five or more make a single ‘supergene’, containing linked shell ground colour (_C_), banding (_B_), band/lip pigmentation (_P_/_L_), spread band (_S_)
and punctate (or ‘interrupted’; _I_) loci. In most studies, colour and banding have been found to be tightly linked, with recombination typically towards the lower end of 0–2% (Cain et al.
1960; Cook 1967; Cook and King 1966). The exceptions are a study by Fisher and Diver (1934), which reported recombination of ~20% between _C/B_, and two crosses in Cain et al. (1960) which
showed recombination of ~16%, also between _C/B_. Although there have been fewer studies, pigmentation, spread band and punctate are believed more loosely linked, showing rates of
recombination between 3 and 15% (Cain et al. 1960; Cain et al. 1968; Cook 1967). The main other loci that make up the shell phenotype are various forms of band-suppressing loci, all unlinked
to the supergene, including the mid-band locus, _U_ (unifasciata), and another that suppresses the first two bands, _T_ (trifasciata). One unavoidable limitation of prior works was that
putative instances of recombination between loci within the supergene could not be verified, except by breeding further generations of snails from the ‘recombinant’ offspring to confirm the
underlying genotype. This was rarely possible, perhaps due to logistics combined with the fact that many pairs do not produce offspring. Nonetheless, it was recognised that incomplete
penetrance might be an alternative explanation for the phenotype of recombinants. Chance arrangements of alleles at other loci might sometimes interact to prevent expression of a particular
phenotype, causing individuals to appear as if they are ‘recombinant’ (Cook and King 1966). To further understand the frequency of recombination within the supergene, and to generate further
material for fine mapping, we made a new set of _C. nemoralis_ crosses that segregate for several shell phenotype loci. The offspring were then genotyped using a set of linked RAD-seq loci
that flank either side of the supergene (Richards et al. 2013), and instances of recombination confirmed or refuted by comparing inferred supergene genotype against RAD-marker genotype. The
underlying idea is that individuals that show recombination within the supergene should also be recombinant by RAD-marker. Overall, we found that the phenotype of ‘recombinant’ individuals
is better explained by incomplete penetrance and epistasis. This work, therefore, provides a method to identify recombination events that either flank the supergene or are between loci
within the supergene. The results also show that recombination within the supergene may be considerably rarer than supposed. MATERIALS AND METHODS THE CULTURE OF _CEPAEA_ Snails were fed a
hydrated grass pellet, oat and chalk mix, supplemented with lettuce, as described previously (Davison 2000). Generally, large juvenile virgin snails were raised to adulthood in isolation and
then introduced to a partner. Pairs of snails were then kept in tanks with ~4 cm soil until egg laying began. As _C. nemoralis_ is a simultaneous hermaphrodite, offspring from both parents
were used. Egg batches were isolated, and the offspring reared to adulthood under the same feeding regime, with the time from egg to adult being ~6 months. Parents and adult offspring, or
large subadult offspring were preserved frozen. The majority of snails were raised to adulthood, and so the colour, banding, band pigmentation and lip colour phenotype was scored and then
the shell genotype inferred. Complications in scoring some characters necessitated minor deviations from the scheme put forward by Cain (1988), summarised in Table 1. To distinguish
phenotype from genes and genotypes, symbols for the latter are in italics. We have previously shown that colour variation is multimodal but continuously variable in natural populations,
necessitating the use of quantitative methods to measure it (Davison et al. 2018). However, in simple crosses it is straightforward to bin the individuals into one of two types; quantitative
measures are not necessary. Therefore, the shell ground colour phenotype was scored as either yellow (Y), pink (P) or brown (B), and where possible, the corresponding genotype inferred
(dominance is _C__B_ > _C__P_ > _C__Y_), including whether in coupling or repulsion phase with other loci (Table 1). Similarly, banding phenotype was generally scored as either
unbanded (O; 00000), mid-banded (M; 00300) or having several bands (B; most frequently 12345, but all combinations except 00300) and the genotype at the banding locus was inferred (unbanded
dominant; _B__O_ > _B__B_). Several crosses also segregated for the mid-band locus, so the genotype at that locus was also inferred (mid-band dominant; _U__3_ > _U__-_). One cross
potentially segregated for two other band-suppressing loci, _T_ (first two bands missing: 00345; _T_345 > _T_-) and another possible locus, which we called _X_ (first band missing or very
faint: 02345; X2345 > X-). It is not clear from previous studies as to whether the lip pigmentation is a separate locus, or is instead allelomorphic with the band pigmentation locus. As
a precaution, the two were therefore treated separately. Thus, band pigmentation phenotype was scored as normal (N) or hyalozonate (H) and the corresponding genotype inferred (_P__N_ >
_P__H_). Hyalozonate shells typically have unpigmented bands and lip (see Discussion); discrete bands can still be recognised because the background colour is paler than the shell ground
colour. In some crosses, the lip pigmentation phenotype was two distinct types, either normal (L), or white lip (A; albolabiate), and so the corresponding genotype was inferred (_L__L_ >
_L__A_). In other crosses, lip pigmentation showed quantitative variation and so was difficult to score. One cross also showed variation in spread-banding, another locus of the supergene,
for which the spread band allele, _S__S_, is dominant to normal banding, _S__-_. Some of the adults used were wild-collected from either the UK, Ireland or Spain; others were derived from
prior laboratory crosses (Table 2). The adults used in crosses 10, 11, 12 and 13 were derived from offspring of cross 9, so the shell genotype could be inferred with extra confidence. This
was aided by full-sib inbreeding in producing crosses 10, 11 and 12, and another round of inbreeding to produce cross 13. For each cross, Mendelian segregation ratios were tested using
chi‐square goodness‐of‐fit tests. DNA EXTRACTIONS AND RAD-MARKER GENOTYPING OF PARENTS AND OFFSPRING For future mapping of the supergene and other shell-character loci, we wished to identify
individuals that show evidence of recombination, ideally either close to a shell-character locus, or between loci within the supergene. As we have previously isolated RAD-seq markers that
flank the supergene (Richards et al. 2013), we could use those markers to confirm or refute individuals that show a phenotype that might have arisen due to recombination within the
supergene. To this end, genomic DNA was extracted from frozen snail tissue, as described previously (Richards et al. 2013), using foot because it is a good source of high molecular weight
DNA. For most samples, slices of snail tissue were incubated at 65 °C in extraction solution (3% CTAB, 100 mM Tris‐HCl, pH 7.5, 25 mM EDTA, pH 8, 2 M NaCl) with 0.2 mg/mL proteinase K and 80
μg/mL RNase. Upon lysis, a chloroform extraction was performed, then three volumes of CTAB dilution solution added (1% CTAB, 50 mm Tris‐HCl, pH 7.5, 10 mM EDTA, pH 8). Samples were mixed
until a precipitate appeared, then the supernatant removed. The pellet was washed twice in 0.4 M NaCl in TE (0.4 M NaCl, 10 mM Tris‐HCl, pH 7.5, 1 mm EDTA, pH 8), re-dissolved in 1.42 M NaCl
in TE (1.42 M NaCl, 10 mM Tris‐HCl, pH 7.5, 1 mM EDTA, pH 8), then precipitated in ethanol, centrifuged and dried. Latterly, a few samples were lysed in lysis buffer (10 mM Tris, 0.1 M
EDTA, 0.5% SDS), then extracted using the standard phenol-chloroform protocol. Then, a subset of the individuals in several of the crosses were genotyped using custom assays derived from
RAD-seq loci that flank either side of the supergene. For this, standard PCR was carried out either using Amplitaq Gold polymerase (Invitrogen), 1.5 mM MgCl2, using a cycle of 95 °C for 10
min, followed by 35 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min or Clontech Advantage 2 PCR with an initial denaturation of 95 °C for 1 min, followed by 35 cycles of 95 °C
for 15 s, 65 °C for 1 min, 68 °C for 1 min, and 72 °C for 1 min. Primers and custom genotyping assays were based on the previously characterised RAD-seq loci (Richards et al. 2013), and
varied according to the cross. The primers were RAD06F 5’-GCCTATCCGTCATTGTTGGT-3’ RAD06R 5’-GTCAAGGCTTGCTTCTTTGG-3’, RAD9F 5’-TTTCTCGGAACGACGGAGT-3’, RAD9R 5’-GGTCTCGTCAATGGCACTTT-3’, RAD11F
5′-AAGAAGCGTCCTTCTGGAAA-3’, RAD11R 5’-CACCTTCCCCATTCTTCAAA-3’. For each cross it was necessary to custom design an assay, so that segregation of markers could be observed in the offspring.
The enzymes used with each assay and each cross are detailed in Supplementary Table 1. RESULTS SEGREGATION OF MENDELIAN LOCI THAT DETERMINE SHELL PHENOTYPE Shell colour, locus _C_, showed
segregation in crosses 1–13 (Table 2; Supplementary Table 2), only deviating significantly from expected Mendelian segregation ratios in cross 12, with fewer yellow shells than expected.
Crosses 1–6 and 8 showed segregation for the band presence/absence locus, _B_, with no deviations from expected Mendelian ratios. Crosses 6 and 9–13 showed segregating variation for the
mid-band phenotype, coded by the unlinked _U_ locus. The observed phenotype frequencies did not differ from the expected frequencies. Crosses 10 to 13 showed segregation for the pigmentation
(hyalozonate; _P_) locus, with no deviations from expected Mendelian ratios. Cross 8 showed segregation for the putative lip colour locus, _L_. The offspring phenotype frequencies would be
consistent with single locus, assuming that _L_ is part of the supergene and treating lip colour phenotypes as either normal (N) or albolabiate lip (A); there was no deviation from expected
Mendelian ratios. Offspring in several of the other crosses, especially 10–13, showed considerable and apparently continuous variation in lip colour. We, therefore, tried to score the lip
phenotype in the conventional manner, having a phenotype as normal, pale, or albolabiate, and reconcile this with a knowledge of the parental phenotypes and genotypes (parents in crosses 8
and 9). No scheme that we devised fitted a simple Mendelian model. This fits with previous studies (Cain et al. 1968; Cook 2003). Unfortunately, it was not possible to quantitatively measure
the lip colour, as we have done for shell ground colour (Davison et al. 2018), because the coloured part of the lip was frequently too small and also on a curved surface. Cross 14 showed
segregating variation in the spread band phenotype. However, as one parent was homozygous for the dominant spread band allele _S__S_, the cross was non-informative for recombination with
other supergene loci. Finally, cross 5 showed segregating variation for the locus that suppresses the first two bands, converting a five-banded snail (12345) to three-banded (00345). The
offspring phenotype frequencies would be consistent with single locus, with _T__345_ dominant to _T__-_, with both parents being heterozygote, except that this would require both parents to
have a _T__345_ allele; apparently not possible because one of the parents is 12345. As five of the offspring with suppressed bands have 02345 phenotype and eleven a 0:345 (: = trace)
phenotype, then the results are consistent with their being two band-supressing loci, one that causes the 00345 phenotype and another that causes the 02345/0:345 phenotype (see Supplementary
Table 2 for inferred genotypes). PUTATIVE RECOMBINANTS BETWEEN COLOUR, BANDING AND LIP AND BAND PIGMENTATION LOCI Previously, the colour (P/Y) and banding (B/O) phenotype for six crosses
(1, 4, 5, 6, 7, 8) and 398 offspring was reported, and the genotype of flanking RAD-seq loci reported for cross 1 (Richards et al. 2013). In this new work, we raised a further 486 offspring
from eight more crosses, and genotyped six further crosses using flanking RAD-seq loci. The combined data set of parent and offspring phenotypes, alongside inferred genotypes, is presented
here together, summarised in Table 2, and presented in full in Supplementary Table 2. Offspring in several crosses produced snails with phenotypes that could be explained by a recombination
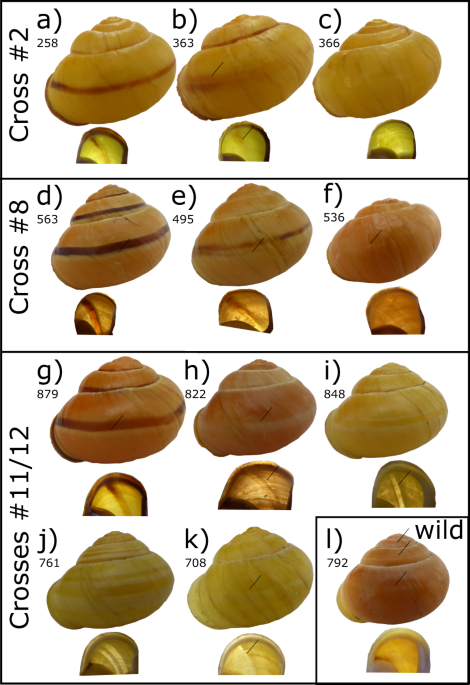
event within the supergene of the heterozygous parent (Fig. 1). Cross 2 produced three yellow unbanded snails (e.g., Fig. 1c), a phenotype that might be produced by recombination between the
colour (_C_) and banding (_B_) loci; one of these individuals, a sub-adult with a damaged shell, has a very faint trace of a band. A few other snails in the same cross have much-reduced
banding (e.g., Fig. 1b). Cross 8 produced a single pink unbanded snail, a phenotype that is also best explained by recombination between the colour (_C_) and banding (_B_) loci (Fig. 1f).
Very few of the snails in this cross have reduced banding. This cross also segregated for the lip pigmentation locus, _L_. As the recombinant snail has a pigmented lip (see Fig. 1f, lip
image), then this cross in theory informs upon the order of loci within the supergene (but see below). Cross 5 produced a single pink mid-banded snail. This phenotype is very difficult to
explain by recombination, based on the known genotypes (Supplementary Table 1). As the pink colour of this snail is qualitatively different from the other pink-banded snails in this cross,
the best explanation is that it is a likely a contaminant from another cross. The remaining crosses produced offspring that suggest possible recombination between the colour _C_ and band
pigmentation _P_ loci. Crosses 11, 12 and 13 produced several unbanded pink individuals, with pigmented lips (e.g., Fig. 1h). These were initially scored as hyalozonate, because the mid-band
was evident but not pigmented. However, closer inspection revealed that the banding phenotype of these shells is not the same as the yellow hyalozonate shells. Specifically, the unbanded
pink individuals retain the white highlighting pigment of a normal shell, but lack the dark pigment (compare Fig. 1h with 1g). This difference is especially evident when viewed from the
underside: hyalozonate shells have cleared bands which are entirely lacking pigment whereas the white pigment of ‘unbanded’ snails shows a silhouette (compare Fig. 1h with 1i, j). Not all of
the hyalozonate shells show such a clear pattern (e.g., Fig. 1k). For comparison, in a wild collected pink hyalozonate, the cleared bands are only evident on the upper whorls (Fig. 1l); in
another shell they are not evident at all. GENOTYPING OF OFFSPRING USING RAD-SEQ DERIVED LOCI Individual offspring from crosses 1, 2, 8, 9, 10, 11, 12, and 13 were genotyped using custom
assays derived from RAD-seq loci that flank either side of the supergene, using RAD06/RAD11 on one side and RAD09 on the other side (Table 3; Supplementary Table 2). Unfortunately, the
RAD-seq loci in cross 5 lacked polymorphism so no assay was possible. To confirm or refute individual recombination events, we inspected the genotype of the putative recombinant offspring.
In theory, individuals for which we have inferred recombination within the supergene should also show recombination by one of the RAD-markers. None of the three individuals from cross 2
showed evidence of recombination from the flanking loci RAD11 and RAD09. Similarly, the single individual in cross 8 did not show evidence of recombination for the same RAD-seq loci. All
eight putative recombinants in cross 12 and all three in cross 13 showed no evidence of recombination using RAD06 and RAD09; in cross 11, three individuals did not show evidence of
recombination, with one single individual (804) showing recombination between RAD06 and the supergene and apparent recombination between the colour and pigmentation loci (_C_/_P_). If this
were correct then it would inform the order of loci within the supergene. However, the phenotype of this snail is exactly the same as the other refuted recombinants (Fig. 1h). It is most
likely not a hyalozonate, and therefore a coincidence that it also shows recombination between RAD06 and the supergene. Thus, overall, while there is incomplete evidence in some cases, we
were not able to confidently confirm any recombination events within the supergene. This puts the point estimate on recombination between _C_ and _B_ at 0/376 (<0.27%), between _C_ and
_P_ at 0/170 (<0.60%) and between _C_ and _L_ at 0/25 (<4%). Therefore, the upper confidence limit for the mean rate of recombination, assuming that the probability of observing zero
recombination events is 5%, is 0.80% for _C/B_, 1.76% for _C/P_, and 12.0% for _C_/_L_. Of course, as there are no recombination events, then this work does not inform the order of loci
within the supergene; the higher upper limits for _C_/_P_ and _C_/_L_ are due to smaller sample sizes. DISCUSSION For future mapping and the precise identification of the supergene, it would
be useful to identify individuals that are known to have a recombination break-point close to, or within the supergene. In this study, we initially identified four putative recombination
events between the colour and banding loci (_C/B_) and fourteen putative recombination events between the colour and pigmentation loci (_C/P_). This is as expected because historic studies
have indicated that _C/B_ are more tightly linked than _C/P_. We also used genotyping of RAD-seq loci that flank the supergene in several of the crosses to reveal individuals that show
recombination between a linked RAD-seq marker and the supergene. These RAD-seq markers may, therefore, be used for future recombination break-point mapping. However, the same RAD-seq
genotyping falsified the putative inferences for recombination _within_ the supergene. This is also supported by a close analysis of the phenotype of the shells, including variable
penetrance of the mid-band phenotype (Fig. 1; crosses 2, 8, 11–13) and a comparison of true hyalozonate shells against shells that partially lack pigmentation (Fig. 1; crosses 8, 11–13).
Therefore, in contrast to previous studies that have reported rates of recombination between _C/B_ of 0–2% and between _C_/_B_ and the pigmentation locus, P, of 3–15%, we found zero
recombinants, putting upper limits on the rate of recombination at 0.8 and 1.8%, respectively. This does not mean that previous inferences of recombination within the supergene were
incorrect. However, as the ‘recombinants’ in this study are better explained by other means, then we would suggest that there is an absence of modern-day evidence for recombination within
the _C. nemoralis_ supergene. The structure of the supergene may not be as has previously been supposed. INCOMPLETE PENETRANCE AND EPISTASIS Four individuals in crosses 2 and 8 were
initially identified as putative recombinants, because they lacked the mid-band, or had only very faint traces of a band. If these snails had been true recombinant individuals then they
would most likely be homozygous for colour and heterozygous for banding (genotype _B__0__B__b_). Incomplete penetrance of the dominant _B__0_ allele could mean that some individuals show
evidence of banding (e.g., Fig. 1b, e). Instead, the genotyping shows that these individuals are not recombinant, and so must be genetically homozygous for colour and banding (_B__b__B__b_).
Therefore, the best explanation for their phenotype is that other loci are interacting epistatically to prevent full penetrance of homozygous banding alleles. Similarly, up to fourteen
individuals were identified that were putative recombinants between the colour and pigmentation loci. If they had been true recombinants, then they would most likely be heterozygous for
colour and homozygous for pigmentation (_C__P__C__Y__P__H__P__H_). However, close analysis of the phenotype and the genotyping together show that they are not recombinants and therefore,
more likely they were heterozygous for both colour and pigmentation (_C__P__C__Y__P__N__P__H_). These same snails are homozygous for the banding locus (_B__b__B__b_), but segregate for
mid-band phenotype (_U__3__U__-_, _U__-__U__-_); the putative recombinants were always mid-banded (_U__3__U__-_), rather than fully banded (_U__-__U__-_). The same explanation may apply, as
above. Other loci sometimes interact to prevent full penetrance of the mid-banded phenotype. The observation that the absence of a mid-band does not always have a simple genetic basis may
shed some light on previous findings. For example, both Fisher and Diver (1934) and Cain et al. (1960) reported individual crosses that showed elevated rates of recombination between the
_C_/_B_ loci. Cain et al. (1960) reported two crosses derived from the same mother that showed seven colour/banding recombinants in 43 snails, for which six were pink mid-banded snails. The
expectation is that putative recombinants would have been homozygote for the banding locus (_C__P__C__Y__B__b__B__b_) and non-recombinants heterozygous (_C__P__C__Y__B__0__B__b_). However,
if the six snails were not actually recombinants, then incomplete dominance of the band-suppressing allele (_B__0_), or else epistatic interactions with other loci, may be an alternative
explanation. Similarly, Fisher and Diver (1934) described unexpectedly high recombination (20%) between colour and banding in one cross. Unfortunately, they did not report whether the snails
used were mid-banded or not. However, in their specific case, doubt has been raised as to whether the individuals used were virgins before paired together (Cain et al. 1960; Ford 1971;
Lamotte 1954). In researching this work, we were fortunate to find copies of letters between Fisher and Diver in the archive of Bryan Clarke (Supplementary Material). In letters from
April/May 1934 that describe the preparation of the correspondence that was published in Nature in June 1934, there is clear admission that the snails used in the crosses were adult and not
virgin. The authors partly acknowledge that this may be a problem. Referring to possible previous matings (“experience”), Fisher writes that "on fairly strong ground, which is not
weakened by previous experience, but is not absolutely critical". Our interpretation of the text is that Fisher acknowledges that the snails may have previously mated, but discounts
this as being a problem, because the offspring ratios approximate to that expected with limited recombination. There are therefore perhaps two errors, which together invalidate the
conclusions of the published work. Epistasis could also explain other earlier data on recombination between the _C_/_B_ loci and the pigmentation locus. For example, in our study, we
initially scored some pink individuals as hyalozonate, even though they had a lightly pigmented lip, only later realising our error. Other authors may have made the same mistake.
Unfortunately, it is difficult to be certain from the previous literature whether pink hyalozonate recombinant individuals had an unpigmented lip, though if this was not the case then it
might have been explicitly noted (e.g. p404 in Cook 1967). However, just as it is hypothesised that epistasis makes brown shelled individuals less likely to be banded, then it is reasonable
to suppose that epistasis might mean that pink hyalozonates more rarely have a wholly unpigmented lip. Evidently, further crosses are required, especially with respect to the other major
loci, especially pigmentation, spread band and punctate loci in the supergene, and the various band-modifying genes. It is possible that some of these phenotypes, especially those for
band-modification (Wolda 1969), are under multi-factorial control and/or dependent upon genetic background. A further general consideration is that the genetics may differ depending upon the
location of origin of the snails. In our study, the snails are derived from the UK, Ireland or Spain, and hybrids between them. As a previous mitochondrial DNA study has shown that the
_Cepaea_ snails in the West of Ireland are at least partly derived from snails from the Pyrenees, and genetically divergent from those of the UK, then perhaps location should be more
properly considered (Grindon and Davison 2013). FUTURE PROGRESS Overall, this work provides a resource for fine mapping of the supergene, and the other major shell phenotype loci. On the one
hand, we have shown that phenocopies may be a problem in using the shell phenotype alone to detect recombination events within the supergene. On the other hand, the genotyping methods that
we have introduced enable a means to avoid this problem. Jones et al. (1977) (in)famously questioned whether understanding polymorphism in _Cepaea_ is “a problem with too many solutions?”
The intention of that work was to emphasise the perfect case study provided by _Cepaea_. We hope that these crosses may soon be used with new long-read DNA sequencing methods to assemble the
_C. nemoralis_ genome and to identify the supergene. Perhaps soon, polymorphism in _Cepaea_ may instead be considered “a solution to many problems.” DATA ARCHIVING All relevant data is in
the tables and Supplementary Material. REFERENCES * Cain AJ (1988) The scoring of polymorphic colour and pattern variation and its genetic basis in molluscan shells. Malacologia 28:1–15
Google Scholar * Cain AJ, King JMB, Sheppard PM (1960) New data on the genetics of polymorphism in the snail _Cepaea nemoralis_ L. Genetics 45:393–411 CAS PubMed PubMed Central Google
Scholar * Cain AJ, Sheppard PM (1950) Selection in the polymorphic land snail _Cepaea nemoralis_. Heredity 4:275–294 Article CAS Google Scholar * Cain AJ, Sheppard PM (1952) The effects
of natural selection on body colour in the land snail _Cepaea nemoralis_. Heredity 6:217–231 Article Google Scholar * Cain AJ, Sheppard PM (1954) Natural selection in _Cepaea_. Genetics
39:89–116 CAS PubMed PubMed Central Google Scholar * Cain AJ, Sheppard PM, King JMB (1968) Studies on _Cepaea_. I. Genetics of some morphs and varieties of _Cepaea nemoralis_ (L). Philos
Trans R Soc Lond B Biol Sci 253:383–396 Article Google Scholar * Cameron RAD, Cook LM (2012) Correlated phenotypic responses to habitat difference in _Cepaea nemoralis_ (L.). Folia
Malacol 20:255–263 Article Google Scholar * Cook LM (1967) The genetics of _Cepaea nemoralis_. Heredity 22:397–410 Article Google Scholar * Cook LM (2003) A colony of pale-lipped Cepaea
nemoralis. J Conchol 38:73–78 Google Scholar * Cook LM (2017) Reflections on molluscan shell polymorphisms. Biol J Linn Soc 121:717–730 Article Google Scholar * Cook LM, King JMB (1966)
Some data on the genetics of shell-character polymorphism in the snail _Arianta arbustorum_. Genetics 53:415–& CAS PubMed PubMed Central Google Scholar * Davison A (2000) The
inheritance of divergent mitochondria in the land snail, _Cepaea nemoralis_. J Mollusca Stud 66:143–147 Article Google Scholar * Davison A, Jackson H, Murphy EW, Reader T (2018) Discrete
or indiscrete? Redefining the colour polymorphism of the land snail _Cepaea nemoralis_. Heredity (in press) * Fisher RA, Diver C (1934) Crossing-over in the land snail _Cepaea nemoralis_, L.
Nature (Lond) 133:834–835 Article Google Scholar * Ford EB (1971) The genetic work of Cyril Diver. In: Merrett P (ed) Captain Cyril Diver (1892-1969) A memoir. The Nature Conservancy,
Dorset Google Scholar * Grindon AJ, Davison A (2013) Irish _Cepaea nemoralis_ land snails have a cryptic Franco-Iberian origin that is most easily explained by the movements of Mesolithic
humans PLoS ONE 8:e65792 Article CAS Google Scholar * Jones JS, Leith BH, Rawlings P (1977) Polymorphism in _Cepaea_: a problem with too many solutions? Annu Rev Ecol Syst 8:109–143
Article Google Scholar * Kerkvliet J, de Boer T, Schilthuizen M, Kraaijeveld K (2017) Candidate genes for shell colour polymorphism in _Cepaea nemoralis_. Peerj 5:e3715 Article Google
Scholar * Lamotte M (1954) Sur le déterminisme génétique du polymorphisme chez _Cepaea nemoralis_ L. Comptes Rendus De l'Acad Des Sci 239:365–367 CAS Google Scholar * Mann K, Jackson
DJ (2014) Characterization of the pigmented shell-forming proteome of the common grove snail _Cepaea nemoralis._ BMC Genomics 15:249.
https://bmcgenomics.biomedcentral.com/articles/10.1186/1471-2164-15-249 Article Google Scholar * Richards PM, Liu MM, Lowe N, Davey JW, Blaxter ML, Davison A (2013) RAD-Seq derived markers
flank the shell colour and banding loci of the _Cepaea nemoralis_ supergene. Mol Ecol 22:3077–3089 Article CAS Google Scholar * Silvertown J, Cook L, Cameron R, Dodd M, McConway K,
Worthington J et al (2011) Citizen science reveals unexpected continental scale evolutionary change in a model organism PLOS ONE 6:e18927 Article CAS Google Scholar * Wolda H (1969)
Genetics of polymorphism in the land snail. Cepaea nemoralis Genet 40:475–502 Google Scholar Download references ACKNOWLEDGEMENTS This work was supported by the University of Nottingham;
the Biotechnology and Biological Sciences Research Council [grant number BB/M008770/1], via a studentship to Daniel Ramos Gonzalez; and the Dept. of Education, Universities and Research of
the Basque Government [grant numbers PRE_2015_2_0191, EP_2015_1_33], via a visiting fellowship awarded to Amaia Caro Aramendia. Thanks to both Sheila Keeble and Julie Rodgers for help with
the care of snails, and to Laurence Cook as well as three anonymous referees for comments on the manuscript, and to Anne Clarke and the University of Nottingham for access to the archive of
Professor Bryan Clarke. AUTHOR INFORMATION Author notes * These authors contributed equally: Daniel Ramos Gonzalez, Amaia Caro Aramendia AUTHORS AND AFFILIATIONS * School of Life Sciences,
University of Nottingham, Nottingham, NG7 2RD, UK Daniel Ramos Gonzalez, Amaia Caro Aramendia & Angus Davison * Departamento de Zoología y Biología Celular Animal, Universidad del País
Vasco, Paseo Univ 7, Vitoria, 01006, Spain Amaia Caro Aramendia Authors * Daniel Ramos Gonzalez View author publications You can also search for this author inPubMed Google Scholar * Amaia
Caro Aramendia View author publications You can also search for this author inPubMed Google Scholar * Angus Davison View author publications You can also search for this author inPubMed
Google Scholar CORRESPONDING AUTHOR Correspondence to Angus Davison. ETHICS DECLARATIONS CONFLICT OF INTEREST The authors declare that they have no conflict of interest. ADDITIONAL
INFORMATION PUBLISHER’S NOTE: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY
TABLE 1 SUPPLEMENTARY TABLE 2 SUPPLEMENTARY MATERIAL RIGHTS AND PERMISSIONS OPEN ACCESS This article is licensed under a Creative Commons Attribution 4.0 International License, which permits
use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the
Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated
otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds
the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/. Reprints and
permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Gonzalez, D.R., Aramendia, A.C. & Davison, A. Recombination within the _Cepaea nemoralis_ supergene is confounded by incomplete
penetrance and epistasis. _Heredity_ 123, 153–161 (2019). https://doi.org/10.1038/s41437-019-0190-6 Download citation * Received: 19 November 2018 * Revised: 09 January 2019 * Accepted: 13
January 2019 * Published: 14 February 2019 * Issue Date: August 2019 * DOI: https://doi.org/10.1038/s41437-019-0190-6 SHARE THIS ARTICLE Anyone you share the following link with will be able
to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing
initiative