- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
New strains of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with several dominant mutations in the spike protein have been identified recently, and crucial issues associated
with the possible reinfection of recovered patients and the efficiencies of vaccines designed based on epidemic strains in early 2020. Here, we provide evidence that the sera collected from
convalescent COVID-19 patients in early 2020 and rhesus macaques vaccinated with receptor-binding domain (RBD)-based nanoparticles efficiently neutralize viral variants of D614G and B.1.1.7
but weakly neutralize those of 501Y.V2, displaying a warning to recovered patients and developed vaccines. During the last 2 months, two new epidemic SARS-CoV-2 variants named 501Y.V2 and
B.1.1.7 have rapidly spread across South Africa and the United Kingdom, respectively.1,2 In the 501Y.V2 variant, the spike protein harbors three pivotal mutations (K417N, E484K and N501Y)
within the RBD; this variant emerged in early August and became the dominant lineage in early November in South Africa.1 The spike protein of the B.1.1.7 lineage harbors 8 mutations: one
mutation (N501Y) within the RBD, three mutations (ΔH69/V70, ΔY144 and A570D) within S1, and four mutations (P681H, T716I, S982A, and D1118H) within S2.2 The B.1.1.7 variant was isolated in
late September and accounted for over 60% of cases in the United Kingdom in early December.3 Both 501Y.V2 and B.1.1.7 variants harbor the early epidemic D614G mutation within the spike
protein.4 Several reports demonstrate that the D614G mutation enhances SARS-CoV-2 infectivity and transmission by increasing the functional spike density on the virion.5,6,7,8 However, the
D614G mutation does not alter the binding affinity to hACE2 or the susceptibility to neutralizing antibodies.5,7,9 The mutant virus with glycine at residue 614 (Spike-G614 virus) also
becomes more sensitive to sera from Spike-D614-vaccinated mice, nonhuman primates (NHPs) and humans, and convalescent sera as well as from RBD-specific monoclonal antibodies.6 Another shared
mutation of 501Y.V2 and B.1.1.7 is N501Y within the RBD. Mutational scanning of the RBD and correlated experiments in mice indicate that N501F and N501Y enhance ACE2 binding affinity and
increase virulence, respectively.10,11 Although both 501Y.V2 and B.1.1.7 exhibit high transmissibility, there are no reports that indicate whether mutations within both lineages enhance
infectivity and impair antibody neutralization. Here, we performed neutralizing assays with convalescent sera from COVID-19-infected patients collected before April 15, 2020 and sera from
RBD nanoparticle vaccine-immunized rhesus macaques against various mutated spike-packaged pseudotyped viruses. Our results demonstrated that most of the current vaccines based on the
original D614 spike sequence should have potent protection against the new round of pandemic-mutated SARS-CoV-2. The evolution of the 501Y.V2 and B.1.1.7 lineages have been speculated to be
driven by convalescent plasma therapy for chronically infected individuals.12 Immunodeficient or immunosuppressed patients who are chronically infected with SARS-CoV-2 are treated with
convalescent plasma. The deficiency of natural immune responses in these patients and the treatment of high antibody concentrations in convalescent plasma give high selective pressures on
the intrapatient virus population, which could drive the genetic divergences of 501Y.V2 and B.1.1.7. In addition, many mutations within the spike protein of 501Y.V2 and B.1.1.7 are located
in the RBD region, which harbors many pivotal epitopes that elicit neutralizing antibodies.13 To evaluate whether the current dominant SARS-CoV-2 variants could escape neutralization from
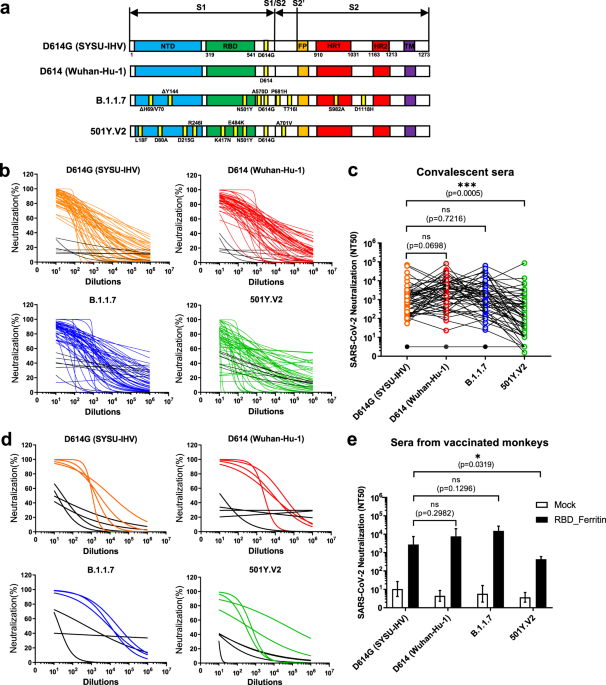
wild-type strain-derived antibodies, we first constructed various spike protein-expressing plasmids derived from G614 SARS-CoV-2 (SYSU-IHV), D614 virus (Wuhan-Hu-1), B.1.1.7, and 501Y.V2 to
package the pseudotyped SARS-CoV-2 S/HIV-1 viruses (Fig. 1a). The luciferase gene is incorporated into the HIV-1 vector and can be expressed after pseudotyped virus infection.14 We incubated
various pseudotyped SARS-CoV-2 S/HIV-1 viruses with convalescent sera from 49 COVID-19 patients,15 followed by infection and luciferase reporter assays (Table S1). All 49 sera were
collected in the cities Guangzhou and Zhuhai in South China before April 15, 2020, which could exclude the possible epidemic of mutated SARS-CoV-2 strains. The antibodies in convalescent
sera efficiently neutralized pseudotyped SARS-CoV-2 D614G and B.1.1.7. Although these sera were also capable of neutralizing 501Y.V2, the neutralizing titers were significantly low against
this strain (Fig. 1b and c). Another major concern is whether these variants would turn to resistance to the protection of existing vaccines, as most of the current vaccines were based on
the spike protein of viruses that were prevalent earlier in the epidemic.16 Recently, we reported an efficient ferritin-based nanoparticle vaccine that simultaneously presented 24 copies of
the RBD and elicited robust protective immune responses against the wild-type SARS-CoV-2 strain.17 Various pseudotyped viruses were incubated with sera from rhesus macaques vaccinated with
RBD nanoparticles to evaluate their neutralizing potential against the new variants. Consistent with the sera from convalescent COVID-19 patients, the sera from rhesus macaques efficiently
neutralized variants D614G and B.1.1.7; however, their neutralization efficiency against 501Y.V2 was significantly low (Fig. 1d and e). Based on the above results, we believe that the risk
of recovered individuals being infected once again by the mutants D614G and B.1.1.7 is much lower than that by the mutant 501Y.V2. Although the spike protein of both B.1.1.7 and 501Y.V2
acquired multiple mutations, part of which were even within the RBD, some mutations impaired the binding of neutralizing antibodies in the sera from early convalescent individuals, while
some mutations did not. With the decrease in neutralizing antibodies in the individuals who recovered from COVID-19 early in 2020, they might become especially susceptible to infection by
the 501Y.V2 stain. Supportively, we found that the sera from RBD nanoparticle-vaccinated rhesus macaques efficiently neutralized the B.1.1.7 pseudotyped virus but weakly neutralized the
501Y.V2 pseudotyped virus. The RBD nanoparticle vaccine we tested was established based upon the RBD of a strain that was prevalent early in the epidemic (Wuhan-Hu-1). While the single
mutation N501Y in the RBD region of B.1.1.7 does not affect the efficiency of neutralizing antibodies, the combination of the 3 mutations (K417N, E484K, and N501Y) in the RBD region of
501Y.V2 might result in the decreased interaction between the neutralizing antibodies and spike protein. Taken together, these results show that as mutations accumulate in the RBD, spike
proteins may acquire an antigenic shift that enable SARS-CoV-2 variants to eventually resist the current vaccines. Therefore, intensive monitoring of virus mutations and timely adjustment of
the designed vaccines are required to control the viral pandemic. REFERENCES * Tegally, H. et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2
(SARS-CoV-2) lineage with multiple spike mutations in South Africa. _medRxiv_, 2020.2012.2021.20248640, https://doi.org/10.1101/2020.12.21.20248640 (2020). * Kemp, S. et al. Recurrent
emergence and transmission of a SARS-CoV-2 Spike deletion ΔH69/ΔV70. _bioRxiv_, 2020.2012.2014.422555, https://doi.org/10.1101/2020.12.14.422555 (2020). * Page, A. J. et al. Large scale
sequencing of SARS-CoV-2 genomes from one region allows detailed epidemiology and enables local outbreak management. _medRxiv_, 2020.2009.2028.20201475,
https://doi.org/10.1101/2020.09.28.20201475 (2020). * Volz, E. et al. Evaluating the Effects of SARS-CoV-2 Spike Mutation D614G on Transmissibility and Pathogenicity. _Cell_,
https://doi.org/10.1016/j.cell.2020.11.020 (2020). * Zhang, L. et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. _Nat. Commun._ 11, 6013,
https://doi.org/10.1038/s41467-020-19808-4 (2020). Article CAS PubMed PubMed Central Google Scholar * Weissman, D. et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to
Neutralization. _Cell Host & Microbe_, https://doi.org/10.1016/j.chom.2020.11.012 (2020). * Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and
transmission in vivo. _Science_ 370, 1464–1468 (2020). CAS PubMed PubMed Central Google Scholar * Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. _Nature_,
https://doi.org/10.1038/s41586-020-2895-3 (2020). * Garcia-Beltran, W. F. et al. COVID-19 neutralizing antibodies predict disease severity and survival. _Cell_,
https://doi.org/10.1016/j.cell.2020.12.015 (2020). * Gu, H. et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. _Science_ 369, 1603–1607 (2020). Article CAS Google
Scholar * Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. _Cell_ 182, 1295–1310.e1220 (2020). Article
CAS Google Scholar * Kemp, S. et al. Neutralising antibodies drive Spike mediated SARS-CoV-2 evasion. _medRxiv_, 2020.2012.2005.20241927, https://doi.org/10.1101/2020.12.05.20241927
(2020). * Yuan, M. et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. _Science_ 368, 630–633 (2020). Article CAS Google Scholar *
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. _Cell_ 181, 271–280.e278 (2020). Article CAS Google Scholar
* Liu, B. et al. Recovered COVID-19 patients with recurrent viral RNA exhibit lower levels of anti-RBD antibodies. _Cell. Mol. Immunol._ 17, 1098–1100 (2020). Article CAS Google Scholar
* Krammer, F. SARS-CoV-2 vaccines in development. _Nature_ 586, 516–527 (2020). Article CAS Google Scholar * Ma, X. et al. Nanoparticle vaccines based on the receptor binding domain
(RBD) and heptad repeat (HR) of SARS-CoV-2 elicit robust protective immune responses. _Immunity_ 53, 1315–1330.e1319 (2020). Article CAS Google Scholar Download references
ACKNOWLEDGEMENTS This work was supported by the National Special Research Program of China for Important Infectious Diseases (2018ZX10302103 and 2017ZX10202102), the Special 2019-nCoV
Program of Natural Science Foundation of China (NSFC) (82041002), the Special 2019-nCoV Project of National Key Research and Development Program of China (2020YFC0841400), the Special
2019-nCoV Project of Research and Development Program of Guangdong (2020B111123001), the Important Key Program of NSFC (81730060), and the Joint-innovation Program in Healthcare for Special
Scientific Research Projects of Guangzhou (201803040002) to H.Z. This work was also supported by the National Postdoctoral Program for Innovative Talents and the General Program of China
Postdoctoral Science Foundation (BX20190398 and 2019M663215) to X.M. AUTHOR INFORMATION Author notes * These authors contributed equally: Rong Li, Xiancai Ma AUTHORS AND AFFILIATIONS *
Institute of Human Virology, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, Guangdong, 510080, China Rong Li, Xiancai Ma, Jieyi Deng, Qier Chen, Weiwei Liu, Zhilin Peng,
Yidan Qiao, Yingtong Lin, Xin He & Hui Zhang * Key Laboratory of Tropical Disease Control of Ministry of Education, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou,
Guangdong, 510080, China Rong Li, Xiancai Ma, Jieyi Deng, Qier Chen, Weiwei Liu, Zhilin Peng, Yidan Qiao, Yingtong Lin, Xin He & Hui Zhang * Guangdong Engineering Research Center for
Antimicrobial Agent and Immunotechnology, Zhongshan School of Medicine, Sun Yat-sen University, Guangzhou, Guangdong, 510080, China Rong Li, Xiancai Ma, Jieyi Deng, Qier Chen, Weiwei Liu,
Zhilin Peng, Yidan Qiao, Yingtong Lin, Xin He & Hui Zhang * Engineering Research Center of Gene Vaccine of Ministry of Education, Zhongshan School of Medicine, Sun Yat-sen University,
Guangzhou, Guangdong, 510080, China Rong Li, Xiancai Ma, Jieyi Deng, Qier Chen, Weiwei Liu, Zhilin Peng, Yidan Qiao, Yingtong Lin, Xin He & Hui Zhang Authors * Rong Li View author
publications You can also search for this author inPubMed Google Scholar * Xiancai Ma View author publications You can also search for this author inPubMed Google Scholar * Jieyi Deng View
author publications You can also search for this author inPubMed Google Scholar * Qier Chen View author publications You can also search for this author inPubMed Google Scholar * Weiwei Liu
View author publications You can also search for this author inPubMed Google Scholar * Zhilin Peng View author publications You can also search for this author inPubMed Google Scholar *
Yidan Qiao View author publications You can also search for this author inPubMed Google Scholar * Yingtong Lin View author publications You can also search for this author inPubMed Google
Scholar * Xin He View author publications You can also search for this author inPubMed Google Scholar * Hui Zhang View author publications You can also search for this author inPubMed Google
Scholar CONTRIBUTIONS Conceptualization, R.L., X.M., X.H., and H.Z.; methodology, R.L., X.M., and H.Z.; validation, X.H. and H.Z.; formal analysis, R.L., X.M., X.H., and H.Z.;
investigation, R.L., X.M., J.D., Q.C., W.L., Z.P., Y.Q., Y.L., X.H., and H.Z.; resources, R.L., X.M., and H.Z.; data curation, X.M., X.H., and H.Z.; writing–original draft, X.M. and H.Z.;
writing–reviewing and editing, X.M., X.H., and H.Z.; supervision, X.H. and H.Z.; project administration, X.M., X.H., and H.Z.; and funding acquisition, X.M. and H.Z. CORRESPONDING AUTHORS
Correspondence to Xin He or Hui Zhang. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing interests. SUPPLEMENTARY INFORMATION ADDITIONAL INFORMATION RIGHTS AND
PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Li, R., Ma, X., Deng, J. _et al._ Differential efficiencies to neutralize the novel mutants B.1.1.7 and 501Y.V2 by
collected sera from convalescent COVID-19 patients and RBD nanoparticle-vaccinated rhesus macaques. _Cell Mol Immunol_ 18, 1058–1060 (2021). https://doi.org/10.1038/s41423-021-00641-8
Download citation * Received: 09 January 2021 * Accepted: 19 January 2021 * Published: 12 February 2021 * Issue Date: April 2021 * DOI: https://doi.org/10.1038/s41423-021-00641-8 SHARE THIS
ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not currently available for this article. Copy to clipboard
Provided by the Springer Nature SharedIt content-sharing initiative









