- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
Access through your institution Buy or subscribe It is now well-established that antigen engagement and microenvironmental cytokine signals direct the functional plasticity of effector CD4+
T cells to shape the outcome of immune responses.5 In contrast, we have recently begun to appreciate that certain pathogens can reprogram the lineage commitment of effector CD4+ T cells to
other T-cell lineages in the periphery. One example is that human immunodeficiency virus (HIV) can induce the generation of various lineage-intermediate populations, including CD4−CD8− and
CD4+CD8+ T-cell subsets. Interestingly, one recent study shows that a unique population of MHC-II-restricted CD4−CD8+ T cells, which is derived from effector CD4+ T cells, displays strong
proliferative capacity and antiviral function in HIV-infected patients.6 It is of great interest to understand the regulatory mechanisms governing the conversion of effector CD4+ T cells to
this CD4−CD8+ T-cell subset, which may benefit the development of a novel strategy to reinvigorate antiviral responses in patients with HIV infection. To explore how MHC-II-restricted
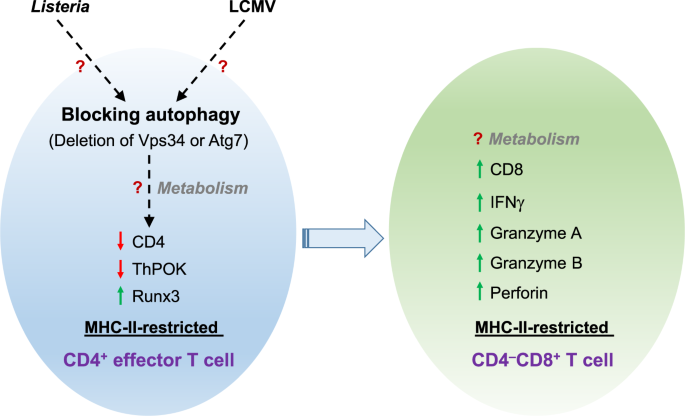
CD4−CD8+ T cells are induced during pathogen infection, Robins et al. used the MHC-II-restricted antigen LLO190–205, a major epitope of _Listeria monocytogenes_ that is recognized by CD4+ T
cells, and transgenic TCR118 mice that contain CD4+ T cells specifically recognizing LLO190–205–MHC-II molecules. Upon in vitro stimulation, they found that TCR118 transgenic CD4+ T cells
downregulate CD4 expression and concomitantly upregulate CD8 expression without altering the MHC-II restriction of LLO190-205. After in vivo _L. monocytogenes_ infection, the authors show
that a portion of antigen-experienced TCR118 transgenic CD4+ T cells give rise to MHC-II-restricted CD4−CD8+ T cells, reminiscent of the unique CD8+ T-cell subset observed in HIV-infected
patients.6 The authors also observed a similar conversion of antigen-specific CD4+ T cells to MHC-II-restricted CD4−CD8+ T cells upon acute infection with lymphocytic choriomeningitis virus
(LCMV). Moreover, these MHC-II-restricted CD4−CD8+ T cells express high levels of cytotoxic molecules granzymes A and B and perforin, as well as the cytokine IFNγ, suggesting that they are
functionally similar to effector CD8+ T cells. These results indicate that the generation of MHC-II-restricted CD4−CD8+ T cells likely represents a general response to infectious
microorganisms. This is a preview of subscription content, access via your institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 12 digital issues and
online access to articles $119.00 per year only $9.92 per issue Learn more Buy this article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to
local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in * Learn about institutional subscriptions * Read our FAQs * Contact customer support REFERENCES * Singer,
A., Adoro, S. & Park, J. H. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. _Nat. Rev. Immunol._ 8, 788–801 (2008). Article CAS Google
Scholar * He, X. et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. _Nature_ 433, 826–833 (2005). Article CAS Google Scholar *
Setoguchi, R. et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. _Science_ 319, 822–825 (2008). Article CAS Google Scholar * Robins,
E. et al. Conversion of effctor CD4+ T cells to a CD8+ MHCII-recognizing lineage. _Cell Mol. Immunol_. (2019). * DuPage, M. & Bluestone, J. A. Harnessing the plasticity of CD4(+) T cells
to treat immune-mediated disease. _Nat. Rev. Immunol._ 16, 149–163 (2016). Article CAS Google Scholar * Ranasinghe, S. et al. Antiviral CD8(+) T cells restricted by human leukocyte
antigen class II exist during natural HIV infection and exhibit clonal expansion. _Immunity_ 45, 917–930 (2016). Article CAS Google Scholar * Shan, Q. et al. The transcription factor
Runx3 guards cytotoxic CD8(+) effector T cells against deviation towards follicular helper T cell lineage. _Nat. Immunol._ 18, 931–939 (2017). Article CAS Google Scholar * Xu, X. et al.
Autophagy is essential for effector CD8(+) T cell survival and memory formation. _Nat. Immunol._ 15, 1152–1161 (2014). Article CAS Google Scholar * Chapman, N. M., Boothby, M. R. &
Chi, H. Metabolic coordination of T cell quiescence and activation. _Nat. Rev. Immunol._ 20, 55–70 (2020). Article CAS Google Scholar * Araki, K. et al. mTOR regulates memory CD8 T-cell
differentiation. _Nature_ 460, 108–112 (2009). Article CAS Google Scholar Download references ACKNOWLEDGEMENTS The authors thank N. Chapman for the careful reading and editing of the
paper. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Pediatrics and the Herman B Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN,
USA Kai Yang * Department of Microbiology and Immunology, Indiana University School of Medicine, Indianapolis, IN, USA Kai Yang * Department of Immunology, St. Jude Children’s Research
Hospital, Memphis, TN, USA Hongbo Chi Authors * Kai Yang View author publications You can also search for this author inPubMed Google Scholar * Hongbo Chi View author publications You can
also search for this author inPubMed Google Scholar CORRESPONDING AUTHORS Correspondence to Kai Yang or Hongbo Chi. ETHICS DECLARATIONS COMPETING INTERESTS The authors declare no competing
interests. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Yang, K., Chi, H. Autophagy modulates CD4+ T-cell lineage recommitment upon pathogen
infection. _Cell Mol Immunol_ 17, 682–683 (2020). https://doi.org/10.1038/s41423-020-0368-0 Download citation * Received: 03 January 2020 * Accepted: 03 January 2020 * Published: 11 February
2020 * Issue Date: July 2020 * DOI: https://doi.org/10.1038/s41423-020-0368-0 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable
link Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative