- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
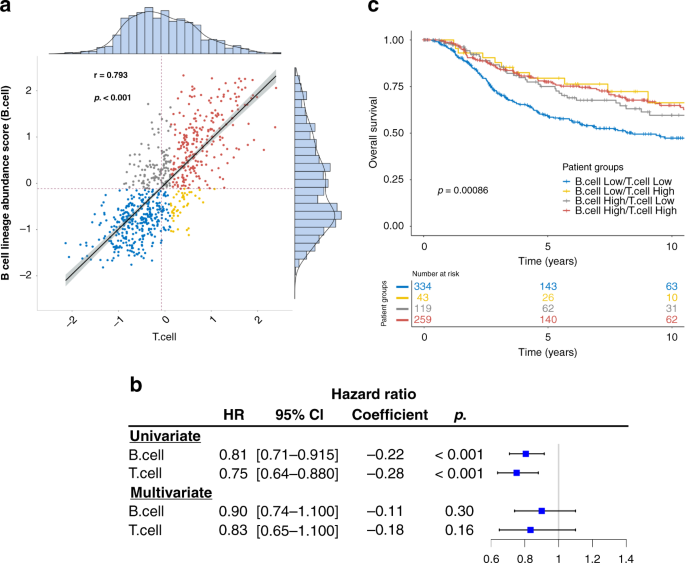
ABSTRACT BACKGROUND B lymphocytes have multifaceted functions in the tumour microenvironment, and their prognostic role in human cancers is controversial. Here we aimed to identify tumour
microenvironmental factors that influence the prognostic effects of B cells. METHODS We conducted a gene expression analysis of 3585 patients for whom the clinical outcome information was
available. We further investigated the clinical relevance for predicting immunotherapy response. RESULTS We identified a novel B cell-related gene (BCR) signature consisting of nine cytokine
signalling genes whose high expression could diminish the beneficial impact of B cells on patient prognosis. In triple-negative breast cancer, higher B cell abundance was associated with
favourable survival only when the BCR signature was low (HR = 0.68, _p_ = 0.0046). By contrast, B cell abundance had no impact on prognosis when the BCR signature was high (HR = 0.93, _p_ =
0.80). This pattern was consistently observed across multiple cancer types including lung, colorectal, and melanoma. Further, the BCR signature predicted response to immune checkpoint
blockade in metastatic melanoma and compared favourably with the established markers. CONCLUSIONS The prognostic impact of tumour-infiltrating B cells depends on the status of cytokine
signalling genes, which together could predict response to cancer immunotherapy. Access through your institution Buy or subscribe This is a preview of subscription content, access via your
institution ACCESS OPTIONS Access through your institution Subscribe to this journal Receive 24 print issues and online access $259.00 per year only $10.79 per issue Learn more Buy this
article * Purchase on SpringerLink * Instant access to full article PDF Buy now Prices may be subject to local taxes which are calculated during checkout ADDITIONAL ACCESS OPTIONS: * Log in
* Learn about institutional subscriptions * Read our FAQs * Contact customer support SIMILAR CONTENT BEING VIEWED BY OTHERS B CELL HETEROGENEITY, PLASTICITY, AND FUNCTIONAL DIVERSITY IN
CANCER MICROENVIRONMENTS Article 29 June 2021 ESTABLISHING A PROGNOSTIC MODEL WITH IMMUNE-RELATED GENES AND INVESTIGATING _EPHB6_ EXPRESSION PATTERN IN BREAST CANCER Article Open access 24
February 2025 A BALANCE SCORE BETWEEN IMMUNE STIMULATORY AND SUPPRESSIVE MICROENVIRONMENTS IDENTIFIES MEDIATORS OF TUMOUR IMMUNITY AND PREDICTS PAN-CANCER SURVIVAL Article Open access 05
November 2020 DATA AVAILABILITY All original data sets used in this article are publicly available with accession codes provided in Supplemental Table 5. REFERENCES * Tsou P, Katayama H,
Ostrin EJ, Hanash SM. The emerging role of B cells in tumor immunity. Cancer Res. 2016;76:5597–601. Article CAS Google Scholar * Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV,
Chudakov DM. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol. 2020;27:1–14. * Wouters MCA, Nelson BH. Prognostic significance of
tumor-infiltrating B cells and plasma cells in human cancer. Clin Cancer Res. 2018;24:6125–35. Article CAS Google Scholar * Cillo AR, Kürten CHL, Tabib T, Qi Z, Onkar S, Wang T, et al.
Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity. 2020;52:183.e9–99.e9. Article Google Scholar * Shalapour S, Font-Burgada J, Di Caro G, Zhong Z,
Sanchez-Lopez E, Dhar D, et al. Immunosuppressive plasma cells impede T-cell-dependent immunogenic chemotherapy. Nature. 2015;521:94–8. Article CAS Google Scholar * Lu Y, Zhao Q, Liao
J-Y, Song E, Xia Q, Pan J, et al. Complement signals determine opposite effects of B cells in chemotherapy-induced immunity. Cell. 2020;180:1081.e24–97.e24. Article Google Scholar * Shen
M, Sun Q, Wang J, Pan W, Ren X. Positive and negative functions of B lymphocytes in tumors. Oncotarget. 2016;7:55828–39. Article Google Scholar * Sautès-Fridman C, Petitprez F, Calderaro
J, Fridman WH. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer. 2019;19:307–25. Article Google Scholar * Helmink BA, Reddy SM, Gao J, Zhang S, Basar R,
Thakur R, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–55. Article CAS Google Scholar * Gentles AJ, Newman AM, Liu CL, Bratman SV,
Feng W, Kim D, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–45. Article CAS Google Scholar * Milne K, Köbel M,
Kalloger SE, Barnes RO, Gao D, Gilks CB, et al. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors.
PLoS ONE. 2009;4:e6412. Article Google Scholar * Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, et al. The Cancer Genome Atlas
Pan-Cancer analysis project. Nat Genet. 2013;45:1113–20. Article Google Scholar * Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data
repository. Nucleic Acids Res. 2002;30:207–10. Article CAS Google Scholar * Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic
architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. Article CAS Google Scholar * Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, et
al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. Article Google Scholar * Newman AM, Liu
CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. Article CAS Google Scholar * R Development
Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2008. * Altman DG. Practical statistics for medical research. Boca Raton,
FL: CRC Press; 1990. 624 p. * Stouffer SA, Suchman EA, Devinney LC, Star SA, Williams Jr. RM. The American soldier: adjustment during army life (studies in social psychology in World War
II). Vol. 1. Oxford: Princeton Univ. Press; 1949. xii, 599 p. * Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat
Soc Ser B (Methodol). 1995;57:289–300. Google Scholar * Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36:1–48. Article Google Scholar * Liu D,
Schilling B, Liu D, Sucker A, Livingstone E, Jerby-Amon L, et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat
Med. 2019;25:1916–27. Article CAS Google Scholar * Allen EMV, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic
melanoma. Science. 2015;350:207–11. Article Google Scholar * Schwartz LH, Litière S, de Vries E, Ford R, Gwyther S, Mandrekar S, et al. RECIST 1.1 – update and clarification: from the
RECIST Committee. Eur J Cancer. 2016;62:132–7. Article Google Scholar * Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer
immunotherapy response. Nat Med. 2018;24:1550–8. Article CAS Google Scholar * Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ–related mRNA profile predicts
clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–40. * Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, et al. Pan-cancer immunogenomic analyses reveal
genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–62. Article CAS Google Scholar * Nishino M, Ramaiya NH, Hatabu H, Hodi FS.
Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14:655–68. Article CAS Google Scholar * Shukla SA, Bachireddy P, Schilling
B, Galonska C, Zhan Q, Bango C, et al. Cancer-germline antigen expression discriminates clinical outcome to CTLA-4 blockade. Cell. 2018;173:624.e8–33.e8. Article Google Scholar * Maaten
LJP, van der, Hinton GE. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9:2579–605. Google Scholar * Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, et al.
Batf controls the global regulators of class switch recombination in both B and T cells. Nat Immunol. 2011;12:536–43. Article CAS Google Scholar * Xu Z, Pone EJ, Al-Qahtani A, Park S-R,
Zan H, Casali P. Regulation of aicda expression and AID activity: relevance to somatic hypermutation and class switch DNA recombination. Crit Rev Immunol. 2007;27:367–97. Article CAS
Google Scholar * Mintz MA, Felce JH, Chou MY, Mayya V, Xu Y, Shui J-W, et al. The HVEM-BTLA axis restrains T cell help to germinal center B cells and functions as a cell-extrinsic
suppressor in lymphomagenesis. Immunity. 2019;51:310.e7–23.e7. Article Google Scholar * Derré L, Rivals J-P, Jandus C, Pastor S, Rimoldi D, Romero P, et al. BTLA mediates inhibition of
human tumor-specific CD8+ T cells that can be partially reversed by vaccination. J Clin Invest. 2010;120:157–67. Article Google Scholar * Harrop JA, McDonnell PC, Brigham-Burke M, Lyn SD,
Minton J, Tan KB, et al. Herpesvirus entry mediator ligand (HVEM-L), a novel ligand for HVEM/TR2, stimulates proliferation of T cells and inhibits HT29 cell growth. J Biol Chem.
1998;273:27548–56. Article CAS Google Scholar * Pasero C, Barbarat B, Just-Landi S, Bernard A, Aurran-Schleinitz T, Rey J, et al. A role for HVEM, but not lymphotoxin-beta receptor, in
LIGHT-induced tumor cell death and chemokine production. Eur J Immunol. 2009;39:2502–14. Article CAS Google Scholar * Li Q, Lao X, Pan Q, Ning N, Yet J, Xu Y, et al. Adoptive transfer of
tumor reactive B cells confers host T-cell immunity and tumor regression. Clin Cancer Res. 2011;17:4987–95. Article CAS Google Scholar * Al-Shibli KI, Donnem T, Al-Saad S, Persson M,
Bremnes RM, Busund L-T. Prognostic effect of epithelial and stromal lymphocyte infiltration in non–small cell lung cancer. Clin Cancer Res. 2008;14:5220–7. Article CAS Google Scholar *
Goswami S, Chen Y, Anandhan S, Szabo PM, Basu S, Blando JM, et al. ARID1A mutation plus CXCL13 expression act as combinatorial biomarkers to predict responses to immune checkpoint therapy in
mUCC. Sci Transl Med. 2020. https://doi.org/10.1126/scitranslmed.abc4220. * Torres A, Casanova JF, Nistal M, Regadera J. Quantification of immunocompetent cells in testicular germ cell
tumours. Histopathology. 1997;30:23–30. Article CAS Google Scholar * Neesse A, Michl P, Frese KK, Feig C, Cook N, Jacobetz MA, et al. Stromal biology and therapy in pancreatic cancer.
Gut. 2011;60:861–8. Article Google Scholar Download references FUNDING This research was partially supported by the National Institutes of Health grant R01 CA222512. AUTHOR INFORMATION
AUTHORS AND AFFILIATIONS * Department of Radiation Oncology, Stanford University School of Medicine, Stanford, CA, USA Arian Lundberg, Bailiang Li & Ruijiang Li * Department of Radiation
Oncology, University of California San Francisco, San Francisco, CA, USA Arian Lundberg Authors * Arian Lundberg View author publications You can also search for this author inPubMed Google
Scholar * Bailiang Li View author publications You can also search for this author inPubMed Google Scholar * Ruijiang Li View author publications You can also search for this author
inPubMed Google Scholar CONTRIBUTIONS AL and RL contributed to the study concept and design. AL contributed to the acquisition and analyses of data. All authors interpreted the data and did
the manuscript drafting and critical revision. All authors read and approved the final manuscript. CORRESPONDING AUTHOR Correspondence to Ruijiang Li. ETHICS DECLARATIONS COMPETING INTERESTS
The authors declare no competing interests. ETHICS APPROVAL AND CONSENT TO PARTICIPATE Publicly available data—not applicable. CONSENT FOR PUBLICATION Publicly available data—not
applicable. ADDITIONAL INFORMATION PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations. SUPPLEMENTARY
INFORMATION SUPPLEMENTAL FIGURE 1. SUPPLEMENTAL FIGURE 2. SUPPLEMENTAL FIGURE 3. SUPPLEMENTAL FIGURE 4. SUPPLEMENTAL FIGURE 5. SUPPLEMENTAL FIGURE 6. SUPPLEMENTAL FIGURE 7. SUPPLEMENTAL
TABLE 1. SUPPLEMENTAL TABLE 2. SUPPLEMENTAL TABLE 3. SUPPLEMENTAL TABLE 4. SUPPLEMENTAL TABLE 5. RIGHTS AND PERMISSIONS Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE
Lundberg, A., Li, B. & Li, R. B cell-related gene signature and cancer immunotherapy response. _Br J Cancer_ 126, 899–906 (2022). https://doi.org/10.1038/s41416-021-01674-6 Download
citation * Received: 24 July 2021 * Revised: 24 November 2021 * Accepted: 09 December 2021 * Published: 17 December 2021 * Issue Date: 01 April 2022 * DOI:
https://doi.org/10.1038/s41416-021-01674-6 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link Sorry, a shareable link is not
currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative