- Select a language for the TTS:
- UK English Female
- UK English Male
- US English Female
- US English Male
- Australian Female
- Australian Male
- Language selected: (auto detect) - EN
Play all audios:
ABSTRACT BACKGROUND Breast fibroadenoma (FA) and phyllodes tumour (PT) often have variations of gene mediator complex subunit 12 (_MED12_) and mutations in the telomerase reverse
transcriptase promoter region (_TERTp_). _TERTp_ mutation is usually tested by Sanger sequencing. In this study, we compared Sanger sequencing and droplet-digital PCR (ddPCR) to measure
_TERTp_ mutations in FA and PT samples. METHODS FA and PT samples were collected from 82 patients who underwent surgery at our institution from 2005 to 2016. _MED12_ mutations for all cases
and _TERTp_ mutations for 17 tumours were detected by Sanger sequencing. ddPCR was performed to analyse _TERTp_ mutation in all cases. RESULTS A total of 75 samples were eligible for
analysis. Sanger sequencing detected _MED12_ mutations in 19/44 FA (42%) and 21/31 PT (68%). Among 17 Sanger sequencing-tested samples, 2/17 (12%) were _TERTp_ mutation-positive. In ddPCR
analyses, a significantly greater percentage of PT (19/31, 61%) was _TERTp_ mutation-positive than was FA (13/44, 30%; _P_ = 0.0046). The mutation positivity of _TERTp_ and _MED12_ did not
correlate, in either FA or PT. CONCLUSIONS ddPCR was more sensitive for detecting _TERTp_ mutation than Sanger sequencing, being able to elucidate tumorigenesis in FA and PT. SIMILAR CONTENT
BEING VIEWED BY OTHERS SYSTEMATIC REVIEW AND FEASIBILITY STUDY ON PRE-ANALYTICAL FACTORS AND GENOMIC ANALYSES ON ARCHIVAL FORMALIN-FIXED PARAFFIN-EMBEDDED BREAST CANCER TISSUE Article Open
access 06 August 2024 GENETIC DIFFERENCES BETWEEN BENIGN PHYLLODES TUMORS AND FIBROADENOMAS REVEALED THROUGH TARGETED NEXT GENERATION SEQUENCING Article 16 March 2021 COMPARATIVE ANALYSIS OF
EGFR MUTATIONS IN CIRCULATING TUMOR DNA AND PRIMARY TUMOR TISSUES FROM LUNG CANCER PATIENTS USING BEAM_ING_ PCR Article Open access 08 January 2025 BACKGROUND Breast fibroepithelial
tumours, which include fibroadenoma (FA) and phyllodes tumour (PT), are characterised by the biphasic proliferation of both epithelial and stromal components.1,2,3,4,5 FA is common benign
tumour that is often observed in young women. It expresses oestrogen receptor (ER)-α in epithelium and ERβ in stromal components, and is known to be hormone dependent.6,7 Small FA (<3 cm)
is usually followed up without resection, and 16–37% of FA cases reportedly regress or completely resolve spontaneously.8,9,10 PT is much less common than FA; it comprises only 2–3% of
fibroepithelial breast tumours and accounts for <1% of all breast tumours. PT is histologically classified as benign, borderline or malignant type. Resection is generally recommended for
PT because it often grows rapidly and can potentially become malignant. Occasionally, PT enlarges to huge sizes that require total mastectomy, and malignant PT has high risks of both local
recurrences and distant metastasis compared with the other types of PT.1,2,3,10,11,12,13,14,15 FA and PT, especially when they remain relatively small, are morphologically so similar that
differentiating them histologically is sometimes difficult. Some studies have suggested that FA could potentially progress to PT, especially FA with monoclonal stromal components; however,
the mechanisms underlying their initiation and progression are unclear.10,11,16,17 Although FA and PT have been genomically analysed,18,19,20,21 little had been known about their genetic
abnormalities. However, since next-generation sequencing (NGS) became widely used in research, several genetical alterations have been revealed in both tumours. The discovery of highly
recurrent Mediator complex subunit 12 (_MED12_) somatic mutations in breast FA was surprising; almost nothing had previously been known about these mutations.22 As for PT, in addition to
_MED12_,10,11,12,13,23,24,25,26,27 the telomerase reverse transcriptase (_TERT_) promoter has been shown to have repeated mutations in these tumours.28,29,30,31,32,33,34 In 2014, Lim et al.
first found highly frequent _MED12_ exon 2 mutations in FA (58/98, 59%) using exome analysis.22 The _MED12_ gene, located on the X chromosome, encodes MED12 protein, a member of the
multiprotein mediator complex that regulates transcription of all RNA polymerase II-dependent genes.35 Reportedly, up to 60% of FA, 80% of benign and borderline PT and 40% of malignant PT
harbours somatic mutations in exon 2 of the _MED12_ gene,10,11,12,13,22,23,24,25,26,27 which suggests that FA and PT have much more in common in their origin or development than previously
thought. However, the underlying mechanisms, and how this mutation generates or induces the progression of FA and PT, are unknown. Since the discovery of frequent _MED12_ mutations in FA and
PT, next-generation sequencing has been used to search for other gene mutations in these tumours. Many mutations found in PT, such as in _RARA_, _EGFR_, _RB1_ or _TP53_, are very uncommon
compared with _MED12_ mutations,12,21,27,28,29,30 whereas mutations in the _TERT_ promoter region (_TERTp_) are reportedly more frequent: 0–7% in FA, and 27–70.6% in PT.29,30,31,32,33,34
_TERTp_ is considered to be a critical regulatory element for telomerase expression.35,36 Hotspots for mutations in this region in PT and FA are reported to be c.−146 C > T (C250T) and
c.−124 C > T (C228T),29,31,32,33,34 which are concordant with those in other tumours, including central nervous system tumours, thyroid cancers, bladder cancers and skin melanoma.36,37
Previous studies found correlations between _TERTp_ mutations (_TERTp__Mut_) and _MED12_ mutations (_MED12__Mut_),28,29,30,31 which suggests that these mutations interact, especially in the
development of PT. Recently, McEvoy et al. showed that droplet-digital polymerase chain reaction (ddPCR) is very sensitive in detecting _TERTp__Mut_ in melanoma compared with pyrosequencing
or Sanger sequencing (SS).38,39 Digital PCR was developed to yield absolute measures for nucleic acid concentrations by a combination of limiting dilution, end-point PCR and Poisson
statistics.40 ddPCR is a newer, more precise and less subjective assay to quantify DNA amplification, based on water–oil emulsion droplet technology. In ddPCR, a sample is fractionated into
20,000 droplets, and PCR amplification of the template molecules occurs in each individual droplet. ddPCR has also been shown to obtain high levels of partitioning at a low cost.41,42
_TERTp__Mut_ was previously evaluated with ddPCR, but only in melanoma.38,39 We found no reports of studies that evaluated mutations in breast fibroepithelial lesions using ddPCR. In this
study, we used ddPCR to measure _TERTp__Mut_ in formalin-fixed, paraffin-embedded (FFPE) samples of resected FA or PT, and compared the results with those from conventional SS. We also
analysed the relationships among _TERTp__Mut_ and _MED12__Mut_ status and histopathological characteristics in FA and PT. METHODS TISSUE SAMPLES We collected FFPE samples of 54 FA and 31 PT
from 82 patients who underwent surgery at the University of Tokyo Hospital from 2005 to 2016. Three patients had two tumours excised at different times. All samples were diagnosed by two
expert pathologists. FA was classified as intracanalicular type, pericanalicular type, mastopathic type, organoid type,43,44 complex fibroadenoma45 or juvenile fibroadenoma;46 PT was
subclassified according to the WHO classification as benign, borderline or malignant lesions.2 All patients consented to the use of their stored tumour tissue. This study protocol was
approved by the ethics committee at the University of Tokyo Hospital, Tokyo, Japan. DNA EXTRACTION For each specimen, two or three 10-µm FFPE sections were cut from a single representative
block per case. Macrodissection was performed with a scalpel as needed to adjust the tumour content to be visually more than 20%. Microdissection was not performed. DNA was isolated using a
GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany), in accordance with the manufacturer’s instructions. Purified DNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific,
Wilmington, DE, USA), with 0.5–14.0 µg of DNA recovered per section. SANGER SEQUENCING TO DETECT MED12 MUTATION FA and PT were analysed for mutations in exon 2 of _MED12_ by SS for all
samples. Exon 2 was amplified with the following primers: (exon 2 forward) 5′-AACTAAACGCCGCTTTCCTG-3′, (exon 2 reverse) 5′-TTCCTTCAGCCTGGCAGAG-3′10,47 (Supplementary Table S1). The PCR
products were purified using agarose gel electrophoresis, labelled with Big Dye Terminator (Applied Biosystems, Foster City, CA, USA) with bidirectional primers and subjected to 3130 × l
Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) in accordance with standard protocols. The Catalogue of Somatic Mutations in Cancer (COSMIC) database48 was used to identify
already-known somatic mutations and mutation types. SANGER SEQUENCING FOR DETECTING TERTP MUTATIONS _TERTp_ mutations (C228T and C250T) were analysed in FA and PT with the same extracted DNA
as used for the _MED12__Mut_ analysis. We performed SS on 17 samples, which had been surgically resected in 2015 and 2016. _TERTp_ was amplified with the following primers: (promoter
forward) 5′-AGCGCTGCCTGAAACTCG-3′, (promoter reverse) 5′-CCTGCCCCTTCACCTTCCAG-3′31,34 (Supplementary Table S1). Q-Solution (Qiagen) was added to the reaction to amplify _TERTp_ by PCR, in
accordance with previous reports.38,39 The SS method was the same as that for the _MED12__Mut_ analysis described above. DDPCR FOR DETECTING TERTP MUTATIONS Analysis of _TERTp_ mutations
(C228T and C250T) with ddPCR was performed for all of the samples, using a previously described ddPCR method.38,39 Primers were the same as those used in SS (Supplementary Table S1). Two
probes were used to detect the mutations: one (the mutant probe) was designated as “/56-FAM/CCC + C + T + T + CCGG/3IABkFQ/” to detect both C228T and C250T mutations (as both mutations
result in the same sequencing string), and the other (the wild probe) was designated as “/5HEX/CCCC + C + T + CCGG/3IABkFQ/,” to recognise the C228 locus (Supplementary Fig. S1). The probes
were custom-synthesised by Integrated DNA Technologies (Coralville, IA, USA). PCR reactions were performed in 20-mL reactions that contained 10 µL of Bio-Rad 2× ddPCR Supermix for Probes (no
dUTP) (Bio-Rad, Hercules, CA, USA), 250 nmol/L probe, 900 nmol/L primers, 10–290 ng of DNA and water. Reaction mixtures were partitioned into emulsions of ~20,000 droplets in oil using a
QX200 Droplet Generator (Bio-Rad). The droplets were then transferred to a 96-well PCR plate, heat-sealed and placed in a thermal cycler (Bio-Rad PX1). Droplets were generated and analysed
using the QX200 Droplet Digital PCR System (Bio-Rad). The thermal cycling conditions were 1 cycle at 95 °C (2.5 °C/s ramp) for 10 min, 40 cycles at 94 °C (2.5 °C/s ramp) for 30 s and at 57
°C followed by 98 °C (2.5 °C/s ramp) for 10 min. Samples were held at 4 °C until further processing. After PCR, the PCR plates were loaded on a Bio-Rad QX200 droplet reader, and ddPCR
absolute quantifications of mutant and wild-type alleles were estimated by modelling them as a Poisson distribution, using Bio-Rad QuantaSoft version 1.6.6 software. Thresholds were defined
based on signals from empty droplets, wild-type DNA controls and mutant-positive controls, as described in the Droplet Digital Application Guide (Bio-Rad). STATISTICAL ANALYSIS All analyses
were performed using JMP Pro statistical software (ver. 14.1.0, SAS Institute, Tokyo, Japan). Fisher’s exact test or chi-squared test was used to analyse categorical data. Proportions were
compared by two-sample tests. The _t_ test, Mann–Whitney _U_ test or Kruskal–Wallis test was used to analyse continuous variables. _P_ < 0.05 was considered significant. RESULTS Among 85
FFPE tissue samples, 75 (44 FA and 31 PT) were eligible for analysis. Ten samples, including five FA samples with insufficient extracted DNA and five FA samples in which the _MED12_ amplicon
could not be amplified to perform SS, were excluded. We successfully performed ddPCR to detect _TERTp_ mutations in all PT samples and SS for _TERTp_ regions in all but one PT sample.
PATIENT CHARACTERISTICS Histological classifications of FA, histological grades of PT, ages and tumour sizes of each case are listed in Table 1. Among 44 FA, 18 samples were typed as
intracanalicular, 6 as pericanalicular, 8 as mastopathic, 7 as organoid, 4 as complex and 1 as juvenile fibroadenoma. Among 31 PT samples, 17 were classified as benign, 9 as borderline and 5
as malignant. The FA ranged in size from 9 to 125 mm, and the PT ranged from 23 to 130 mm. The Mann–Whitney _U_ test showed that patients with PT were significantly older (_P_ = 0.006) and
had larger tumours (_P_ < 0.001) than those with FA. MED12 MUTATIONS IN FA AND PT A significantly higher percentage of PT (21/31, 68%) was _MED12__Mut+_ than was FA (19/44, 42%, _P_ =
0.035, two-sample test of proportions). Among FA, the intracanalicular type showed _MED12_ mutations more frequently than the other types (_P_ = 0.046, chi-squared test). The frequency of
_MED12_ mutations did not significantly differ among histological grades of PT (_P_ = 0.81, chi-squared test; Supplementary Table S2). Table 2 shows the types and locations of _MED12_
mutations. All of the missense mutations have already been reported in COSMIC,47 but many deletion and deletion/insertion patterns were newly discovered in this study (shown in red in Table
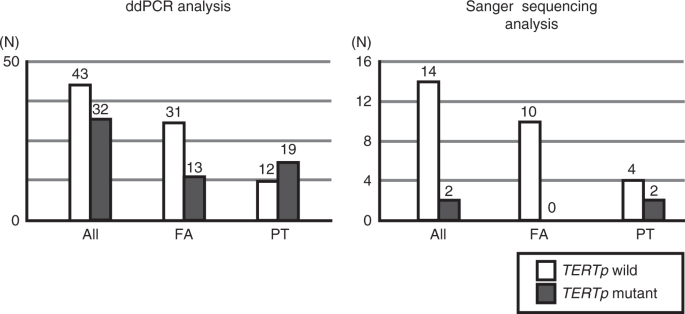
2). TERTP MUTATIONS IN FA AND PT In this study, the only mutation site for _TERTp_ was C228T; we saw no case of C250T. In 16 cases examined for _TERTp__Mut+_ by both SS and ddPCR, SS found
that 12.5% (2/16) were _TERTp__Mut+_, whereas ddPCR found 37.5% (6/16; _P_ = 0.037, two-sample test of proportions). Both cases determined as _TERTp__Mut+_ by SS were PT; no FA case carrying
_TERTp_ mutations was found (Fig. 1 and Supplementary Table S3). ddPCR analyses of all tumours found that 42.7% (32/75) were _TERTp__Mut+_, which was significantly more frequent in PT
(19/31; 61.3%) than in FA (13/44; 29.5%; _P_ = 0.009, Fisher’s exact test, Fig. 1 and Table 3). The two samples that were found to be _TERTp__Mut+_ by SS (16a and 16i) were strongly positive
in ddPCR analysis, whereas those with discrepant results between SS and ddPCR (e.g. 16b and 16k) were weakly positive by ddPCR (Fig. 2). Correlations between _TERTp-_mutation status and
clinicopathological variables in FA and PT are shown in Table 3. Among FA, _TERTp__Mut+_ tumours were significantly larger than _TERTp__Mut–_ ones (_P_ = 0.015, Mann–Whitney _U_ test), but
this was not the case in PT (_P_ = 1.0). None of the other variables, such as patients’ age, FA classification, PT grade and _MED12-_mutation status, differed significantly between
_TERTp__Mut+_ and _TERTp__Mut−_ tumours. FRACTIONAL ABUNDANCE OF FAS AND PTS WITH TERTP MUTATION A total of 70 FA and PT samples with _TERTp_ mutation were detected by ddPCR at a fractional
abundance from 0.7% to 92% (Supplementary Table S4). PT had significantly higher fractional abundance of _TERTp_ mutation than FA: the mean fractional abundance in FA was 20% (range:
1.0–89%), while that in PT was 40% (range: 0.7–92%) (_P_ = 0.049, Mann–Whitney _U_ test). Among the cases analysed by both SS and ddPCR, fractional abundance was relatively high in the two
concordant cases (28.8% and 52.3%; Supplementary Table S5). Those cases that were both _MED12-_mutation-positive with SS and _TERTp-_mutation-positive with ddPCR did not show high fractional
abundance compared with the others (_P_ = 0.067, _t_ test; Supplementary Table S4). CONCORDANCE OF MED12 AND TERTP MUTATIONS BETWEEN METACHRONOUS MULTIPLE TUMOURS FROM THE SAME INDIVIDUALS
In this study, three patients had metachronous multiple tumours. All three cases harboured the same _MED12_ mutation in the primary and secondary lesions (Table 4). However, only one case,
of malignant PT, was _TERTp__Mut+_ in both primary and secondary tumours, whereas the other two cases carried this mutation only in the second tumour. DISCUSSION In this study, we describe a
method to detect _TERT_ promoter mutations in FA and PT using ddPCR. _TERT_ promoter mutation has been reported to be one of the most prevalent mutations in PT other than _MED12_ mutation.
ddPCR not only succeeded in detecting _TERT_ promoter mutations, but also revealed this mutation to be much more frequent among breast fibroepithelial tumours, especially in FA, than was
previously known. The advantages of ddPCR are its high sensitivity, quantitative measurement and low running cost. As ddPCR can detect mutations that have few patterns of single-base
substitution, it is entirely appropriate for _TERTp_ mutations, because only two hotspots have been reported in this region.29,31,32,33,34 ddPCR can distinguish mutations with a very low
frequency of allele variants that SS would not have detected. Here, we show the potential high sensitivity of ddPCR in detecting _TERTp__Mut+_ FA and PT, as in a previous report on melanoma
by McEvoy et al.39 Around the mutation sites of C228T and C250T, _TERTp_ region contains guanine and cytosine in high frequency. Such GC-rich regions affect the generation of secondary
products during PCR, and as a consequence, inhibit DNA polymerases from copying these sites correctly. Besides, the two mutation hotspots, C228T and C250T, locate close to each other. These
inconvenient factors reduce the sensitivity in sequencing analyses, including NGS. Such disadvantageous potentials remain challenging in ddPCR research; however, Corless BC et al.49 showed
excellent sensitivity and specificity in detecting _TERTp_ mutations by ddPCR. According to them, in ddPCR analysis of mutations in GC-rich regions, shorter amplicon length provides better
detection sensitivity. They mentioned that DNA in FFPE samples is shortly fragmented due to the chemical reaction between formalin and nucleic acid, which helps primers bind to the target
regions and generate short amplicons, leading to the efficient PCR reaction.49 Although FFPE samples are often considered to be unfavourable in sequencing analysis due to the low quality of
nucleic acid conditions, they might be even advantageous in ddPCR analysis for _TERTp_-mutation detection. There have been a few reports comparing the detection of genetic mutations with SS
and digital PCR, including one from McEvoy et al.39 and another from Sho et al.50 McEvoy et al. examined mutations in _BRAF_, _NRAS_ and _TERTp_ in melanoma. According to them, the frequency
of mutation detection in SS was 20% (8/40) in _BRAF_, 0% (0/40) in _NRAS_ and 12.5% (5/40) in _TERTp_, whereas that with ddPCR was 55% (22/40) in _BRAF_, 10% (41/40) in _NRAS_ and 37.5%
(15/40) in _TERTp_.39 Sho et al. performed SS on resected specimens of the pancreas, and digital PCR on preoperative endoscopic ultrasound–fine-needle aspiration specimens. All 22 cases
positive for mutation by SS were also determined to be mutation-positive by digital PCR, and digital PCR detected five more mutations. This report demonstrates the utility of digital PCR in
cytology samples, which have a much lower tumour burden than surgical specimens.50 In these reports, digital PCR determined as mutation-positive for all cases in which SS detected the
mutation, and this result is consistent with ours. In this study, we used SS instead of NGS to detect _TERTp_ mutations in some cases. NGS’s deep sequencing is well known to be more
sensitive than Sanger sequencing, and it would be interesting to know what the results of NGS analysis would be in these samples. However, conducting NGS targeting only the _TERT_ promoter
region is too costly. In addition, because one of our future goals is to apply our method to clinical practice, we judged it impractical to perform additional NGS in this study from the
beginning of the research. With our _TERTp__Mut_ analysis by ddPCR, the frequency of this mutation was shown with histological grade, from FA (29%) and benign PT (59%) to borderline PT (67%,
Table 3). Nault et al. analysed _TERTp_ mutations in cirrhotic dysplastic nodules, which are premalignant lesions of hepatocellular carcinoma (HCC), and in early and progressed HCC. They
reported a strong relationship between _TERTp_ mutations and hepatocarcinogenesis: the mutations were identified in 6% of low-grade dysplastic nodules, 19% of high-grade nodules, 61% of
early HCC and 42% of small and progressed HCC.51 Our results in breast FA and PT were quite similar to their results in HCC, in that the frequency of _TERTp_ mutations increased as tumours
went from benign to malignant, which implies a sequential development by some FA into PT. Interestingly, our study showed a positive correlation between FA size and frequency of _TERTp_
mutations (_P_ = 0.015, Table 3). Some FA, especially large ones, may be genetically similar to benign PT, although morphologically distinguishable from them. The relationship between
_MED12_ and _TERTp_ mutations varies in previous reports.29,30,31,33,34 Piscuoglio et al.,29 Pareja et al.30 and Liu et al.34 reported that just 50% and slightly over of PT were
_MED12__Mut+_ (13/25, 5/9 and 3/6), whereas Yoshida et al.31 and Garcia-Dios et al.33 reported that almost all of them were simultaneously _MED12__Mut+_ (29/30 and 12/13, respectively). Only
one study found _TERTp_ mutations in FA:31 4 cases out of 58 (7%) and all of the _TERTp__Mut+_ FA also harboured _MED12_ mutations. Our study found no significant correlation between
_MED12_ and _TERTp_ mutations, in either FA or PT. Although _MED12_ mutations are most frequently observed in intracanalicular-type FA,10,11,21,23,24,28 _TERTp_ mutations detected in our
study had no statistical relationship with FA classification or PT grade (Table 3). These two mutations might independently affect the genesis or development of FA and PT. Among the 72
patients in our study, three experienced recurrences (Table 4). When we compared the first resected tumour with later ones, two of them, an FA and a benign PT, showed _TERTp__Mut_ status
change from negative to positive. Considering that all cases harboured the same pattern of _MED12_ mutations within each case, the secondary resected tumours seemed to be genetically
identical to the primary tumours, suggesting that they were truly recurrent tumours. Garcia-Dios et al. also reported evidence of recurrent PT that acquired _TERTp_ mutations,33 but our
result indicates that _TERTp_ mutations can be acquired during tumour growth or recurrence in both FA and PT. We detected _TERTp_ mutations at a much higher rate in FA than were seen in
previous studies,29,31,33 but some might doubt the accuracy of the FA diagnoses. In the current study, two expert pathologists diagnosed all of the tumours. The distinction between FA and PT
is often challenging, and in fact, we had eight cases, all eventually diagnosed as benign PT, on which the pathologists did not initially agree whether they were intracanalicular-type FA or
benign PT (data not shown). _TERTp_ mutations were found in five of these controversial tumours (63%), which is similar to the mutation rate of PT as a whole (61%), but much higher than
that of FA (30%, Table 3). We consider that these controversial cases were credibly distinguished from other FA, and would be similarly diagnosed by most pathologists. Although we found
ddPCR to be more sensitive than SS for _TERTp_ mutations, technical challenges remain. Sufficient quality and quantity of DNA is essential for detecting low-frequency mutations; therefore,
small tumours or hyalinised FA may be inadequate for mutation analysis with ddPCR. Another challenge to successful analysis is artefacts from formalin-fixed samples when using sensitive
molecular assays causing false-positive calls.52,53 We set thresholds based on signals from empty droplets, wild-type DNA and mutant-positive controls, and determined samples as negative
when they showed fewer than ten mutation calls, to decrease the possibility of error due to false positives. In other words, detecting very low-frequency mutations with fewer than ten calls
is highly challenging. Additional studies are required to resolve these problems and improve accuracy. To examine the influence of FFPE-derived artefacts of DNA in ddPCR analysis, it would
be more robust to prepare an FFPE-derived negative and a positive control for both C228T and C250T mutation-positive controls. We set distilled water as a negative control, and used DNA
collected from a cell line as a positive control, so the DNA of these controls did not derive from FFPE. However, in our positive control, only C228T positivity was confirmed; thus, it did
not show what type of dot plot would be obtained by ddPCR when C250T had a mutation in our study, although this was depicted in previous reports. These matters can be considered limitations
of this work. Another limitation is the primers that we used in this study, which were the same as those reported by Yoshida et al.31 and Liu et al.34 The nucleotide sequences of these
primers differ from those in the study by McEvoy et al.,39 albeit by only a few bases. As there is no previous report describing the performance of ddPCR analysis using our primers, it may
have been necessary to verify the findings of the study by using the primers that McEvoy et al. used for validation. Extracted tumour DNA in the present study contained mixtures of that from
epithelium, stroma or somewhat normal mammary cells and lymphocytes surrounding the tumour. Earlier studies found mutations for _TERTp_ and _MED12_ only in PT stromal components,29,31,32 so
strictly speaking, DNA extraction from only the stromal components through microdissection would be necessary to identify _TERTp_ mutations in FA and PT stroma, and to measure the allele
frequency by ddPCR. Extracting DNA and measuring _TERTp_ mutations from only the tumour’s stromal components may solve the problem of false positivity and -negativity more precisely, thus
enhancing studies of relationships of _TERTp__Mut_ allelic frequency with tumour growth rate and malignancy in FA and PT. Further study with targeted tumour cells is warranted. In
conclusion, we have presented the first assessment by ddPCR of _TERTp_ mutations in FFPE breast FA and PT, and detected these mutations at a higher rate than previously reported. Our new
findings reconfirm the genomic similarity of FA to PT and may help elucidate the biology of these tumours. We have shown ddPCR to be a robust method of detecting _TERTp_ mutations, which
suggest a wider clinical potential for this technology. A large-scale study is needed to determine whether _TERTp_ mutation detection by ddPCR can be predictive, and has prognostic value for
the surgical treatment of FA and PT. REFERENCES * Tavassoli, F. A. & Eusebi, V. Biphasic tumors. in _AFIP Atlas of Tumor Pathology_, Vol 10 (eds Tavassoli, F. A & Eusebi, V.)
315–340 (American Registry of Pathology, Washington, DC, 2009). * Tan, P. H., Tse, G., Lee, A., Simpson, J. F. & Hanby, A. M. Fibroepithelial tumours. in _WHO Classification of Tumours
of the Breast_, 4th edn. (eds Lakhani, S. R., Ellis, I. O., Schnitt, S. J., Tan, P. H. & van de Vijver, M. J.) 142–147 (IARC Press, Lyon, France, 2012). * Yang, X., Kandil, D., Cosar, E.
F. & Khan, A. Fibroepithelial tumors of the breast: pathologic and immunohistochemical features and molecular mechanisms. _Arch. Pathol. Lab. Med._ 138, 25–36 (2014). Article PubMed
Google Scholar * Tan, P. H. & Ellis, I. O. Myoepithelial and epithelial-myoepithelial, mesenchymal and fibroepithelial breast lesions: updates from the WHO Classification of Tumours of
the Breast. _J. Clin. Pathol._ 66, 465–470 (2012). Article CAS Google Scholar * Brogi, E. Fibroepithelial neoplasms. in _Rosen’s Breast Pathology_, 4th edn. (eds Hoda, S. A., Brogi, E.,
Koerner, F. C. & Rosen, P. R.) 213–270 (Lippincott Williams and Willkins, Philadelphia, PA, 2009). * Sapino, A., Bosco, M., Cassoni, P., Castellano, I., Arisio, R., Cserni, G. et al.
Estrogen receptor-beta is expressed in stromal cells of fibroadenoma and phyllodes tumors of the breast. _Mod. Pathol._ 19, 599–606 (2006). Article CAS PubMed Google Scholar * Tan, W.
J., Chan, J. Y., Thike, A. A., Lim, J. C., Md Nasir, N. D., Tan, J. S. et al. MED12 protein expression in breast fibroepithelial lesions: correlation with mutation status and oestrogen
receptor expression. _J. Clin. Pathol._ 69, 858–865 (2016). Article CAS PubMed Google Scholar * Carty, N. J., Carter, C., Rubin, C., Ravichandran, D., Royle, G. T. & Taylor, I.
Management of fibroadenoma of the breast. _Ann. R. Coll. Surg. Engl._ 77, 127–130 (1995). CAS PubMed PubMed Central Google Scholar * Dixon, J. M. Cystic disease and fibroadenoma of the
breast: natural history and relation to breast cancer risk. _Br. Med. Bull._ 47, 258–271 (1991). Article CAS PubMed Google Scholar * Mishima, C., Kagara, N., Tanei, T., Naoi, Y.,
Shimoda, A., Shimazu, K. et al. Mutational analysis of MED12 in fibroadenomas and phyllodes tumors of the breast by means of targeted next-generation sequencing. _Breast Cancer Res. Treat._
152, 305–312 (2015). Article CAS PubMed Google Scholar * Yoshida, M., Sekine, S., Ogawa, R., Yoshida, H., Maeshima, A., Kanai, Y. et al. Frequent MED12 mutations in phyllodes tumours of
the breast. _Br. J. Cancer_ 112, 1703–1708 (2015). Article CAS PubMed PubMed Central Google Scholar * Cani, A. K., Hovelson, D. H., McDaniel, A. S., Sadis, S., Haller, M. J., Yadati, V.
et al. Next-Gen sequencing exposes frequent MED12 mutations and actionable therapeutic targets in phyllodes tumors. _Mol. Cancer Res._ 13, 613–619 (2015). Article CAS PubMed PubMed
Central Google Scholar * Nagasawa, S., Maeda, I., Fukuda, T., Wu, W., Hayami, R., Kojima, Y. et al. MED12 exon 2 mutations in phyllodes tumors of the breast. _Cancer Med._ 4, 1117–1121
(2015). Article CAS PubMed PubMed Central Google Scholar * Ben Hassouna, J., Damak, T., Gamoudi, A., Chargui, R., Khomsi, F., Mahjoub, S. et al. Phyllodes tumors of the breast: a case
series of 106 patients. _Am. J. Surg._ 192, 141–147 (2016). Article Google Scholar * Guerrero, M. A., Ballard, B. R. & Grau, A. M. Malignant phyllodes tumor of the breast: review of
the literature and case report of stromal overgrowth. _Surg. Oncol._ 12, 27–37 (2003). Article PubMed Google Scholar * Noguchi, S., Yokouchi, H., Aihara, T., Motomura, K., Inaji, H.,
Imaoka, S. et al. Progression of fibroadenoma to phyllodes tumor demonstrated by clonal analysis. _Cancer_ 76, 1779–1785 (1995). Article CAS PubMed Google Scholar * Noguchi, S., Aihara,
T., Motomura, K., Inaji, H., Imaoka, S., Koyama, H. et al. Phyllodes tumor of the breast: pathology, genesis, diagnosis, and treatment. _Breast Cancer_ 3, 79–92 (1996). Article CAS PubMed
Google Scholar * Millikan, R., Hulka, B., Thor, A., Zhang, Y., Edgerton, S., Zhang, X. et al. p53 mutations in benign breast tissue. _J. Clin. Oncol._ 13, 2293–2300 (1995). Article CAS
PubMed Google Scholar * Franco, N., Picard, S. F., Mege, F., Arnould, L. & Lizard-Nacol, S. Absence of genetic abnormalities in fibroadenomas of the breast determined at p53 gene
mutations and microsatellite alterations. _Cancer Res._ 61, 7955–7958 (2001). CAS PubMed Google Scholar * Vorkas, P. A., Poumpouridou, N., Agelaki, S., Kroupis, C., Georgoulias, V. &
Lianidou, E. S. PIK3CA hotspot mutation scanning by a novel and highly sensitive high-resolution small amplicon melting analysis method. _J. Mol. Diagn._ 12, 697–704 (2010). Article CAS
PubMed PubMed Central Google Scholar * Loke, B. N., Md Nasir, N. D., Thike, A. A., Lee, J. Y. H., Lee, C. S., The, B. T. et al. Genetics and genomics of breast fibroadenomas. _J. Clin.
Pathol._ 71, 381–387 (2018). Article CAS PubMed Google Scholar * Lim, W. K., Ong, C. K., Tan, J., Thike, A. A., Ng, C. C., Rajasegaran, V. et al. Exome sequencing identifies highly
recurrent MED12 somatic mutations in breast fibroadenoma. _Nat. Genet._ 46, 877–880 (2014). Article CAS PubMed Google Scholar * Pfarr, N., Kriegsmann, M., Sinn, P., Klauchen, F., Endris,
V., Herpel, E. et al. Distribution of MED12 mutations in fibroadenomas and phyllodes tumors of the breast–implications for tumor biology and pathological diagnosis. _Genes Chromosomes
Cancer_ 54, 444–452 (2015). Article CAS PubMed Google Scholar * Piscuoglio, S., Murray, M., Fusco, N., Marchiò, C., Loo, F. L., Martelotto, L. G. et al. MED12 somatic mutations in
fibroadenomas and phyllodes tumours of the breast. _Histopathology_ 67, 719–729 (2015). Article PubMed PubMed Central Google Scholar * Lien, H. C., Huang, C. S., Yang, Y. W. & Jeng,
Y. M. Mutational analysis of _MED12_ exon 2 in a spectrum of fibroepithelial tumours of the breast: implications for pathogenesis and histogenesis. _Histopathology_ 68, 433–441 (2016).
Article PubMed Google Scholar * Ng, C. C., Tan, J., Ong, C. K., Lim, W. K., Rajasegaran, V., Nasir, N. D. et al. MED12 is frequently mutated in breast phyllodes tumours: a study of 112
cases. _J. Clin. Pathol._ 68, 685–691 (2015). Article CAS PubMed Google Scholar * Tan, J., Ong, C. K., Lim, W. K., Ng, C. C., Thike, A. A., Ng, L. M. et al. Genomic landscapes of breast
fibroepithelial tumors. _Nat. Genet._ 47, 1341–1345 (2015). Article CAS PubMed Google Scholar * Kim, J. Y., Yu, J. H., Nam, S. J., Kim, S. W., Lee, S. K., Park, W. Y. et al. Genetic and
clinical characteristics of phyllodes tumors of the breast. _Transl. Oncol._ 11, 18–23 (2018). Article PubMed Google Scholar * Piscuoglio, S., Ng, C. K., Murray, M., Burke, K. A.,
Edelweiss, M., Geyer, F. C. et al. Massively parallel sequencing of phyllodes tumours of the breast reveals actionable mutations, and TERT promoter hotspot mutations and TERT gene
amplification as likely drivers of progression. _J. Pathol._ 238, 508–518 (2016). Article CAS PubMed PubMed Central Google Scholar * Pareja, F., Geyer, F. C., Kumar, R., Selenica, P.,
Piscuoglio, S., Ng, C. K. Y. et al. Phyllodes tumors with and without fibroadenoma-like areas display distinct genomic features and may evolve through distinct pathways. _NPJ Breast Cancer_
3, 40 (2017). Article PubMed PubMed Central CAS Google Scholar * Yoshida, M., Ogawa, R., Yoshida, H., Maeshima, A., Kanai, Y., Kinoshita, T. et al. TERT promoter mutations are frequent
and show association with MED12 mutations in phyllodes tumors of the breast. _Br. J. Cancer_ 113, 1244–1248 (2015). Article CAS PubMed PubMed Central Google Scholar * Tsang, J. Y. S.,
Hui, Y. K., Lee, M. A., Lacambra, M., Ni, Y. B., Cheung, S. Y. et al. Association of clinicopathological features and prognosis of TERT alterations in phyllodes tumor of breast. _Sci. Rep._
8, 3881 (2018). Article PubMed PubMed Central CAS Google Scholar * Garcia-Dios, D. A., Levi, D., Shah, V., Gillett, C., Simpson, M. A., Hanby, A. et al. MED12, TERT promoter and RBM15
mutations in primary and recurrent phyllodes tumours. _Br. J. Cancer_ 118, 277–284 (2018). Article CAS PubMed PubMed Central Google Scholar * Liu, S. Y., Joseph, N. M., Ravindranathan,
A., Stohr, B. A., Greenland, N. Y., Vohra, P. et al. Genomic profiling of malignant phyllodes tumors reveals aberrations in FGFR1 and PI-3 kinase/RAS signaling pathways and provides insights
into intratumoral heterogeneity. _Mod. Pathol._ 29, 1012–1027 (2016). Article CAS PubMed Google Scholar * Clark, A. D., Oldenbroek, M. & Boyer, T. G. Mediator kinase module and
human tumorigenesis. _Crit. Rev. Biochem. Mol. Biol._ 50, 393–426 (2015). CAS PubMed PubMed Central Google Scholar * Yuan, P., Cao, J. L., Abuduwufuer, A., Wang, L. M., Yuan, X. S., Lv,
W. et al. Clinical characteristics and prognostic significance of TERT promoter mutations in cancer: a cohort study and a meta-analysis. _PLoS ONE_ 11, e0146803 (2016). Article PubMed
PubMed Central CAS Google Scholar * Vinagre, J., Almeida, A., Pópulo, H., Batista, R., Lyra, J., Pinto, V. et al. Frequency of TERT promoter mutations in human cancers. _Nat. Commun._ 4,
2185 (2013). Article PubMed CAS Google Scholar * McEvoy, A. C., Calapre, L., Pereira, M. R., Giardina, T., Robinson, C., Khattak, M. A. et al. Sensitive droplet digital PCR method for
detection of TERT promoter mutations in cell free DNA from patients with metastatic melanoma. _Oncotarget_ 8, 78890–78900 (2017). Article PubMed PubMed Central Google Scholar * McEvoy,
A. C., Wood, B. A., Ardakani, N. M., Pereira, M. R., Pearce, R., Cowell, L. et al. droplet digital PCR for mutation detection in formalin-fixed, paraffin-embedded melanoma tissues: a
comparison with Sanger sequencing and pyrosequencing. _J. Mol. Diagn._ 20, 240–252 (2018). Article CAS PubMed Google Scholar * Vogelstein, B. & Kinzler, K. W. Digital PCR. _Proc.
Natl Acad. Sci. USA_ 96, 9236–9241 (1999). Article CAS PubMed Google Scholar * Hindson, B. J., Ness, K. D., Masquelier, D. A., Belgrader, P., Heredia, N. J., Makarewicz, A. J. et al.
High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. _Anal. Chem._ 83, 8604–8610 (2011). Article CAS PubMed PubMed Central Google Scholar * Taylor,
S. C., Carbonneau, J., Shelton, D. N. & Boivin, G. Optimization of droplet digital PCR from RNA and DNA extracts with direct comparison to RT-qPCR: clinical implications for
quantification of oseltamivir-resistant subpopulations. _J. Virol. Methods_ 224, 58–66 (2015). Article CAS PubMed Google Scholar * Kuroda, H., Takeuchi, I., Ohnishi, K., Sakamoto, G.,
Akiyama, F., Toyozumi, Y. et al. Clinical and pathologic features of fibroadenoma of the mastopathic type. _Surg. Today_ 36, 590–595 (2006). Article PubMed Google Scholar * Mori, I., Han,
B., Wang, X., Taniguchi, E., Nakamura, M., Nakamura, Y. et al. Mastopathic fibroadenoma of the breast: a pitfall of aspiration cytology. _Cytopathology_ 17, 233–238 (2006). Article CAS
PubMed Google Scholar * Sklair-Levy, M., Sella, T., Alweiss, T., Craciun, I., Libson, E. & Mally, B. Incidence and management of complex fibroadenomas. _Am. J. Roentgenol._ 190,
214–218 (2008). Article Google Scholar * Wechselberger, G., Schoeller, T. & Piza-Katzer, H. Juvenile fibroadenoma of the breast. _Surgery_ 132, 106–107 (2002). Article PubMed Google
Scholar * Je, E. M., Kim, M. R., Min, K. O., Yoo, N. J. & Lee, S. H. Mutational analysis of MED12 exon 2 in uterine leiomyoma and other common tumors. _Int. J. Cancer_ 131, E1044–E1047
(2012). Article CAS PubMed Google Scholar * Forbes, S. A., Beare, D., Gunasekaran, P., Leung, K., Bindal, N., Boutselakis, H. et al. COSMIC: exploring the world’s knowledge of somatic
mutations in human cancer. _Nucleic Acids Res_. 43, D805–D811 (2015). Article CAS PubMed Google Scholar * Corless, B. C., Chang, G. A., Cooper, S., Syeda, M. M., Shao, Y., Iman Osman, I.
et al. Development of novel mutation-specific droplet digital PCR assays detecting TERT promoter mutations in tumor and plasma samples. _J. Mol. Diagn._ 21, 274–285 (2019). Article CAS
PubMed PubMed Central Google Scholar * Sho, S., Court, C. M., Kim, S., Braxton, D. R., Hou, S., Muthusamy, V. R. et al. Digital PCR improves mutation analysis in pancreas fine needle
aspiration biopsy specimens. _PLoS ONE_ 26, e0170897 (2017). Article CAS Google Scholar * Nault, J. C., Calderaro, J., Di Tommaso, L., Balabaud, C., Zafrani, E. S., Bioulac-Sage, P. et
al. Telomerase reverse transcriptase promoter mutation is an early somatic genetic alteration in the transformation of premalignant nodules in hepatocellular carcinoma on cirrhosis.
_Hepatology_ 60, 1983–1992 (2014). Article CAS PubMed Google Scholar * Ye, X., Zhu, Z. Z., Zhong, L., Lu, Y., Sun, Y., Yin, X. et al. High T790M detection rate in TKI-naive NSCLC with
EGFR sensitive mutation: truth or artifact? _J. Thorac. Oncol._ 8, 1118–1120 (2013). Article PubMed Google Scholar * Watanabe, M., Kawaguchi, T., Isa, S., Ando, M., Tamiya, A., Kubo, A.
et al. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. _Clin. Cancer
Res._ 21, 3552–3560 (2015). Article CAS PubMed Google Scholar Download references ACKNOWLEDGEMENTS We thank Marla Brunker, from Edanz Group (www.edanzediting.com/ac), for editing a draft
of this paper. AUTHOR INFORMATION AUTHORS AND AFFILIATIONS * Department of Breast and Endocrine Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan Kazutaka Otsuji,
Masahiko Tanabe & Yasuyuki Seto * Department of Next-Generation Pathology Information and Networking, Faculty of Medicine, The University of Tokyo, Tokyo, Japan Takeshi Sasaki *
Department of Gastrointestinal Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan Yasuyuki Seto Authors * Kazutaka Otsuji View author publications You can also
search for this author inPubMed Google Scholar * Takeshi Sasaki View author publications You can also search for this author inPubMed Google Scholar * Masahiko Tanabe View author
publications You can also search for this author inPubMed Google Scholar * Yasuyuki Seto View author publications You can also search for this author inPubMed Google Scholar CONTRIBUTIONS
K.O. and T.S. conceived the study. T.S., M.T. and Y.S. directed the study and supervised the research. K.O. collected tumour specimens. K.O. and T.S. confirmed the histopathological findings
and interpreted the clinical data. K.O. performed Sanger sequencing and droplet-digital PCR analyses. K.O. wrote the paper, with assistance and final approval of all authors. CORRESPONDING
AUTHOR Correspondence to Takeshi Sasaki. ETHICS DECLARATIONS ETHICS APPROVAL AND CONSENT TO PARTICIPATE This study protocol was approved by the ethics committee at the University of Tokyo
Hospital, Tokyo, Japan. Written informed consent was obtained from all patients. The study was performed in accordance with the Declaration of Helsinki. CONSENT TO PUBLISH Not applicable.
DATA AVAILABILITY All data supporting the study are available on request. No proprietary materials, except patient tissues, were used. COMPETING INTERESTS The authors declare no competing
interests. FUNDING INFORMATION This study was supported by “Project for the Advanced Genome-Based Medicine” from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
ADDITIONAL INFORMATION NOTE This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to
a Creative Commons Attribution 4.0 International (CC BY 4.0). PUBLISHER’S NOTE Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional
affiliations. SUPPLEMENTARY INFORMATION SUPPLEMENTARY INFORMATION RIGHTS AND PERMISSIONS This article is licensed under a Creative Commons Attribution 4.0 International License, which
permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to
the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless
indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory
regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit
http://creativecommons.org/licenses/by/4.0/. Reprints and permissions ABOUT THIS ARTICLE CITE THIS ARTICLE Otsuji, K., Sasaki, T., Tanabe, M. _et al._ Droplet-digital PCR reveals frequent
mutations in _TERT_ promoter region in breast fibroadenomas and phyllodes tumours, irrespective of the presence of _MED12_ mutations. _Br J Cancer_ 124, 466–473 (2021).
https://doi.org/10.1038/s41416-020-01109-8 Download citation * Received: 23 December 2019 * Revised: 10 September 2020 * Accepted: 16 September 2020 * Published: 13 October 2020 * Issue
Date: 19 January 2021 * DOI: https://doi.org/10.1038/s41416-020-01109-8 SHARE THIS ARTICLE Anyone you share the following link with will be able to read this content: Get shareable link
Sorry, a shareable link is not currently available for this article. Copy to clipboard Provided by the Springer Nature SharedIt content-sharing initiative

:max_bytes(150000):strip_icc():focal(216x0:218x2)/benedict-cumberbatch-1-435-4-20cc736017b24435a3498a49d7c22b0e.jpg)




